Introduction
Lung cancer is a leading cause of cancer-related
deaths worldwide with over one million cases diagnosed yearly
(1). Lung cancer is morphologically
divided into non-small cell lung carcinoma (NSCLC) and small cell
lung carcinoma, in which NSCLC accounts for ~80% of all lung cancer
cases (2,3). Despite progress in the multimodality
treatment of lung cancer, prognosis is still poor with a 10–15%
5-year survival rate. Therefore, the identification of a reliable
biomarker for predicting recurrence and for identifying tumors is
important not only for understanding the molecular and cellular
processes involved, but also for searching for possible new
therapeutic molecular targets.
The proto-oncogene protein DEK was originally
identified as a fusion with the CAN/NUP214 nucleoporin in a subset
of acute myeloid leukemia patients (4,5). DEK
is abundantly expressed in proliferating cells, and the majority of
the protein is bound to chromatin, whereas a small fraction is
bound to RNA. The 43-kDa nuclear phosphoprotein is the only member
of its family, and contains a conserved central SAP DNA binding
domain with homology to SAF-A/B, acinus and PIAS, and a second DNA
binding motif within the C-terminus. Studies have suggested that
DEK may promote tumorigenesis, at least in part, by its ability to
interfere with cell division, DNA repair, inhibit cell
differentiation, senescence and apoptosis, and cooperate with
transforming oncogenes (6–9). Wise-Draper et al recently
reported that DEK expression promoted transformation in
vitro and in vivo and the DEK proto-oncogene is
upregulated in many human cancers including colon, breast, ovarian
and cervical cancer (5). The degree
of DEK upregulation often correlates to the severity of prognosis
as indicated by histopathological determination of a later stage
and grade, or poor differentiation characteristics (5,10). Our
previous studies showed that DEK protein was closely related to the
proliferation of serous ovarian tumor cells and an increased
proliferating index of Ki-67 in cervical cancer (11,12).
Furthermore, based on tumor tissue analyses, we found that DEK
expression was correlated with the prognosis of a variety of human
tumors, such as breast and colorectal cancer (13,14).
Thus, DEK is expected to be the new molecular target for cancer
therapy. However, the role of DEK in the prognostic evaluation and
its relationship to survival in NSCLC are unknown. The critical
role of DEK in numerous cancers impelled us to study the function
of DEK in NSCLC. Therefore, we performed immunofluorescence (IF)
staining in NSCLC A549 cells, and quantitative real-time RT-PCR
(qRT-PCR), western blotting and immunohistochemical (IHC) staining
of DEK in NSCLC and normal lung tissues, and found that DEK protein
was usually upregulated in NSCLC compared with the normal
counterparts. Multivariate analysis revealed that DEK may be an
independent biomarker for predicting NSCLC prognosis.
Materials and methods
Ethics statement
The present study complied with the Helsinki
Declaration and was approved by the Human Ethics and Research
Ethics Committees of the Medical College of Eastern Liaoning
University in China. Through the surgery consent form, patients
were informed that the resected specimens would be stored by the
hospital and potentially used for scientific research, and that
their privacy would be maintained. Follow-up survival data were
retrospectively collected through medical-record analyses.
IF staining for DEK protein in non-small
cell lung cancer cells (A549)
The lung cancer cell line, A549, was grown on
coverslips to 70% confluency and then fixed in 4% paraformaldehyde
for 10 min and permeabilized with 0.5% Triton X-100 for 10 min
after 24 h. Blocking was performed with 3% bovine serum albumin
fraction V (Solarbio, Beijing, China) for 1 h at room temperature.
After washing with phosphate-buffered saline (PBS), the cells were
incubated with mouse anti-human DEK (1:50; BD Biosciences
Pharmingen, San Diego, CA, USA) at 4°C overnight, followed by
incubation with Alexa Fluor 568 goat anti-mouse IgG (H+L) (A11004,
1:1,000; Life Technologies, Carlsbad, CA, USA) for 1 h at room
temperature. After washing with PBS, the cells were counterstained
with 4′,6-diamidino-2-phenylindole (DAPI), and the coverslips were
mounted with Antifade Mounting Medium (both from Beyotime,
Shanghai, China). Finally, immunofluorescence signals were
visualized and recorded using Leica SP5 II confocal microscope
(21).
Clinical samples
Fresh samples from 6 cases of NSCLC were paired with
adjacent non-cancerous tissues, and 196 cases of routinely
processed and paraffin-embedded NSCLC meeting strict follow-up
criteria were randomly selected from patients undergoing surgery
between 2004 and 2008 at the Department of Pathology and Tumor
Tissue Bank, the Medical College of Eastern Liaoning University.
Pathological parameters, including age, gender, smoking status,
tumor size, pathological stage, differentiation, subtype, CEA
level, metastasis status, disease-free and overall survival data,
were carefully reviewed. The patient ages ranged between 34 and 76
years, with a mean age of 64.6 years. The male to female ratio was
109:87. Tumors were staged according to the 6th edition of the
American Joint Committee on Cancer (15). Of the 196 NSCLC samples, 101 were
determined as early-stage (I–II) and 95 as late-stage (III–IV).
Forty-four samples were well differentiated, 94 were moderately
differentiated and 58 were poorly differentiated cancers. No
patients had received chemotherapy and radiotherapy before surgery.
By March 2013, 78 patients had died and 118 patients remained
alive. The median survival time was 71 months.
Western blotting
Fresh tissue samples of NSCLC were ground to powder
in liquid nitrogen and lysed with SDS-PAGE sample buffer. Equal
protein samples (20 µg) were separated on 12% SDS
polyacrylamide gels and transferred to PVDF membranes. The
membranes were blocked with 5% fat-free milk in tris-buffered
saline containing 0.1% Tween-20 for 1 h at room temperature. The
membranes were incubated with the DEK antibody (1:1,000; BD
Biosciences Pharmingen) overnight at 4°C, and then with horseradish
peroxidase-conjugated rabbit anti-mouse IgG. DEK expression was
detected using ECL Prime western blotting detection reagent
(Amersham) according to the manufacturer's instructions. β-actin
(Sigma, St. Louis, MO, USA) was used as a loading control. Protein
bands were quantified using a LANE 1D system (Sage, China).
RNA extraction and qRT-PCR
Total RNA from fresh tissues was extracted using
TRIzol reagent (Invitrogen, Carlsbad, CA, USA). First-strand cDNA
was synthesized by PrimeScript reverse transcriptase (Takara
Biotechnology, Dalian, China) and oligo(dT) following the
manufacturer's instructions. To examine expression, real-time PCR
was performed with a Bio-Rad sequence detection system according to
the manufacturer's instructions using a double-stranded
DNA-specific SYBR Premix Ex Taq™ II kit (Takara Biotechnology).
Double-stranded DNA-specific expression was tested using the
comparative Ct method using 2−ΔΔCt. The DEK primers
were: 5′-AAACCTAGCCAGCTTCACGA-3′ and 5′-AGCCCC AACTCCAGAGAAAC-3′;
GAPDH, 5′-GGTCTCCTCTGA CTTCAACA-3′ and 5′-ATACCAGGAAATGAGCTTGA-3′.
All assays were performed in triplicate and repeated at least three
times.
Immunohistochemical analysis
Immunohistochemical analysis was performed using a
Dako LSAB kit (Dako A/S, Glostrup, Denmark). Briefly, to eliminate
endogenous peroxidase activity, 4-µm thick tissue sections
were deparaffinized, rehydrated and incubated with 3%
H2O2 in methanol for 15 min at room
temperature. The antigen was retrieved at 95°C for 20 min by
placing the slides in 0.01 M sodium citrate buffer (pH 6.0). The
slides were then incubated with the DEK antibody (1:50) at 4°C
overnight. After incubation with the biotinylated secondary
antibody at room temperature for 30 min, the slides were incubated
with streptavidin-peroxidase complex at room temperature for 30
min. Immunostaining was developed using 3,3′-diaminobenzidine, and
Mayer's hematoxylin was used for counterstaining. Tonsil sections
were used as positive controls and mouse IgG as isotope controls.
For negative controls, positive tissue sections were processed in
the same manner but the primary antibody (mouse anti-DEK) was
omitted.
All specimens were examined by two pathologists (Z.
Lin and S. Liu) who did not possess knowledge of the clinical data.
In case of discrepancies, a final score was established by
reassessment on a double-headed microscope. Briefly, immunostaining
for DEK was semi-quantitatively scored as '−' (none or <5%
positive cells), '+' (5–50% positive cells) and '++' (>50%
positive cells). Only a nuclear expression pattern was considered
as positive staining. For survival analysis, the DEK expression
level was denoted as either positive expression ('+' and '++') or
negative expression ('−').
Statistical analyses
Statistical analyses were performed using SPSS 17.0.
Correlations between DEK expression and clinicopathological
characteristics were evaluated using χ2 and Fisher's
exact tests. The disease-free and overall survival rates after
tumor removal were calculated using the Kaplan-Meier method, and
differences in survival curves were analyzed using log-rank tests.
Multivariate survival analysis was performed on all significant
characteristics measured by univariate survival analysis with the
Cox proportional hazard regression model. A P-value of <0.05 was
considered to indicate a statistically significant result.
Results
DEK expression in A549 cells and fresh
tissues of NSCLC
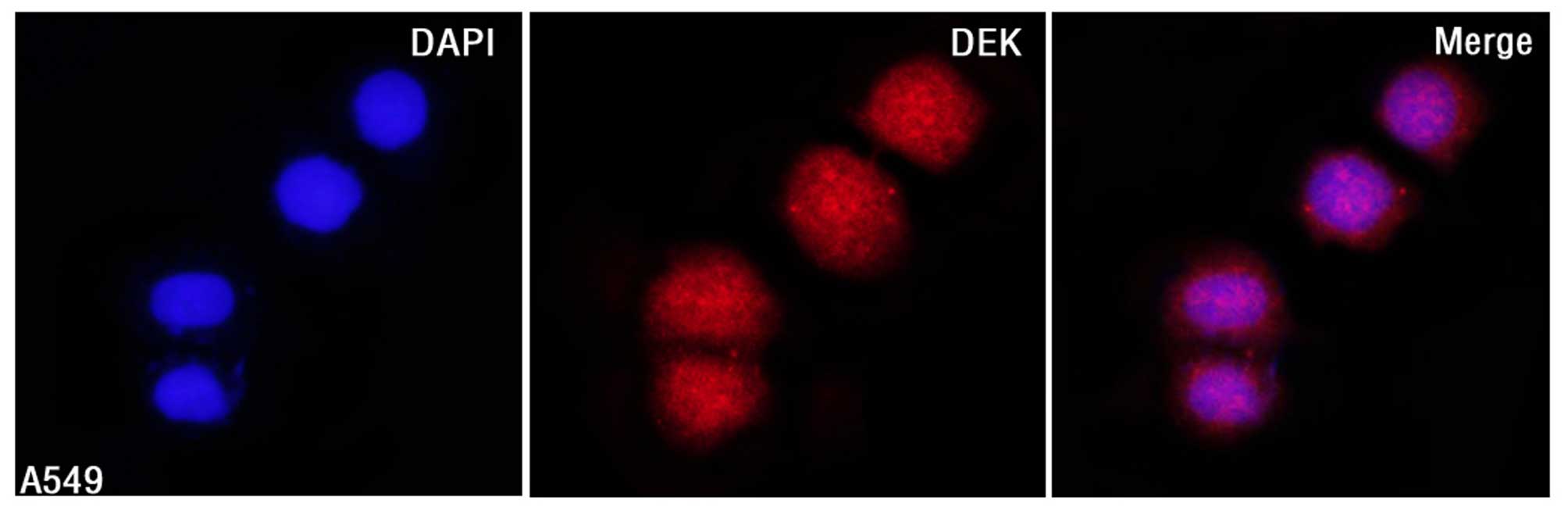
DEK protein mainly localized to the nuclei of A549
cells by IF staining (Fig. 1).
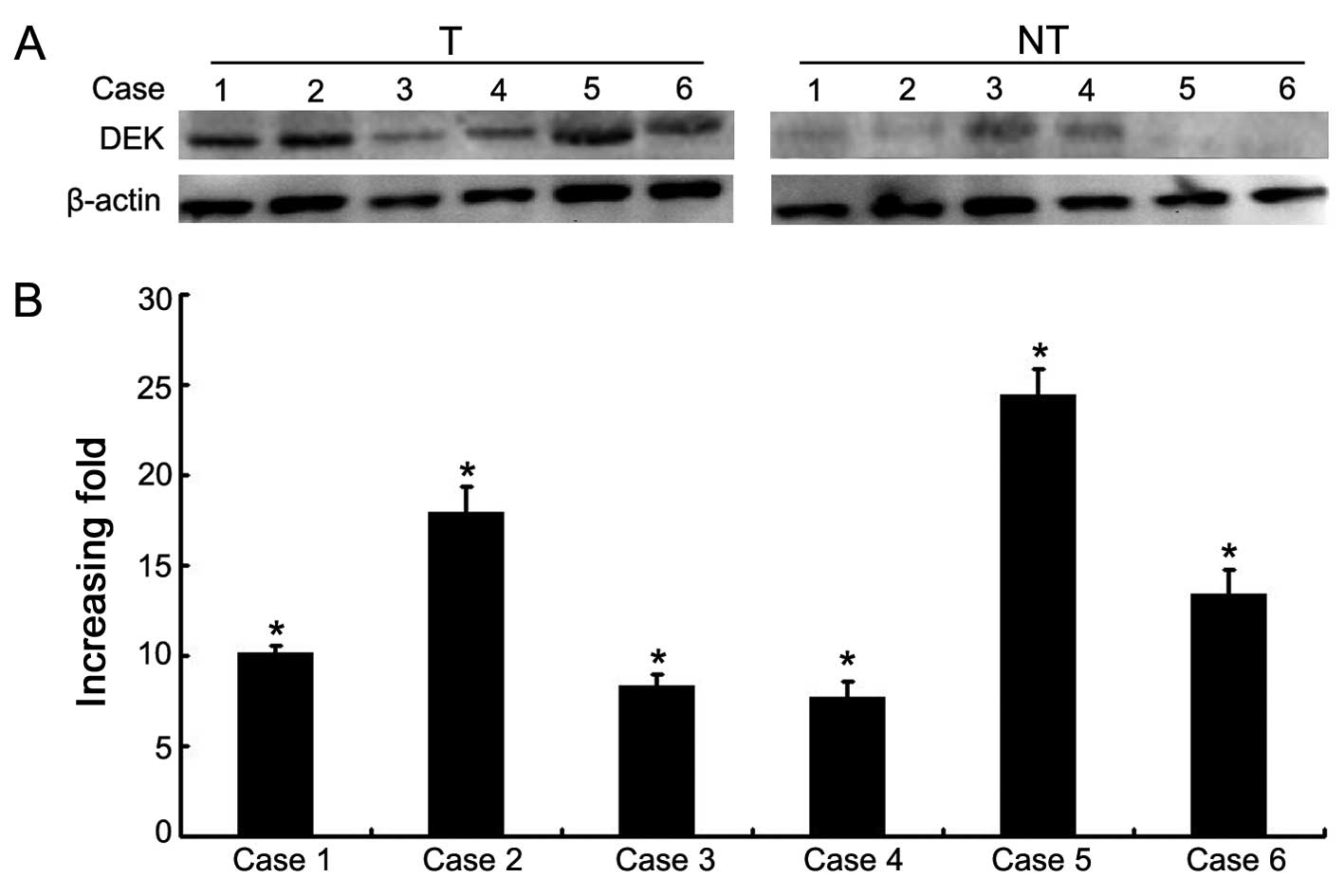
Moreover, the protein and mRNA expression levels of DEK were
determined for 6 NSCLC samples with matched adjacent non-tumor
fresh tissues. Western blot data showed that DEK protein was highly
expressed in the NSCLC tissues compared with this level in the
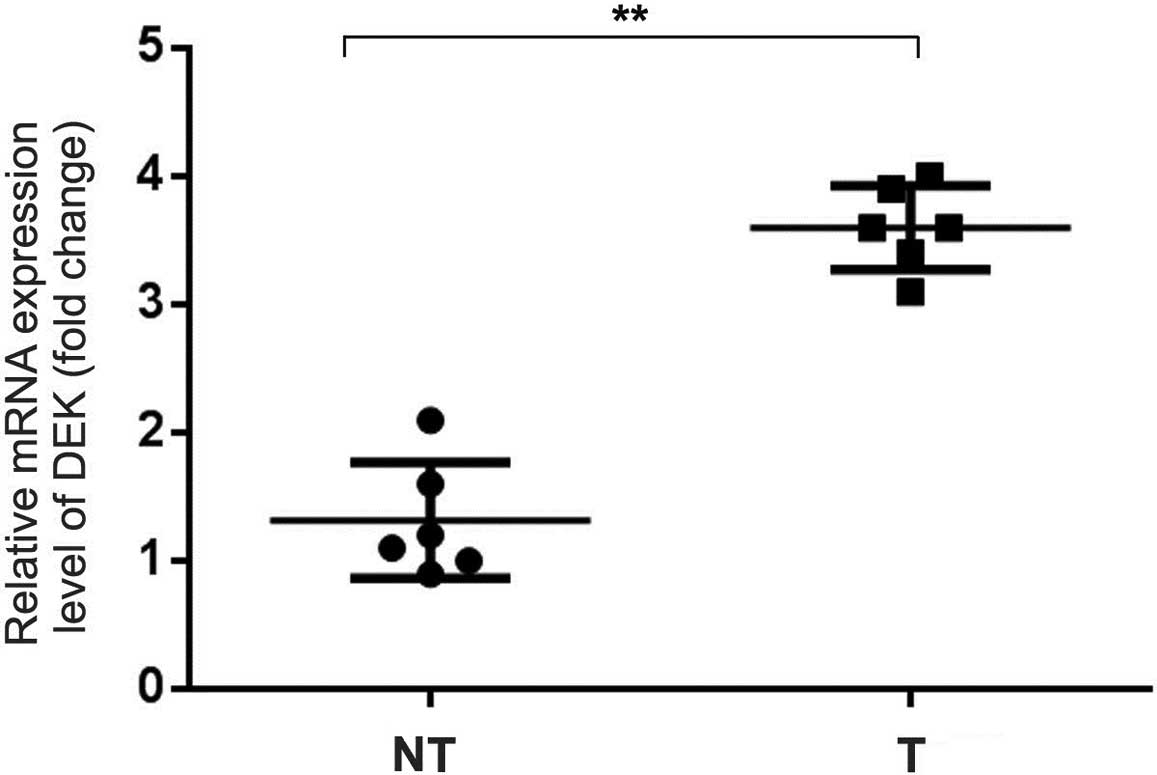
matched adjacent non-tumor tissues (Fig. 2). qRT-PCR data confirmed an
increased level of DEK mRNA expression in the NSCLC samples
compared with that in the adjacent non-tumor tissues (Fig. 3).
DEK protein expression in
paraffin-embedded NSCLC samples
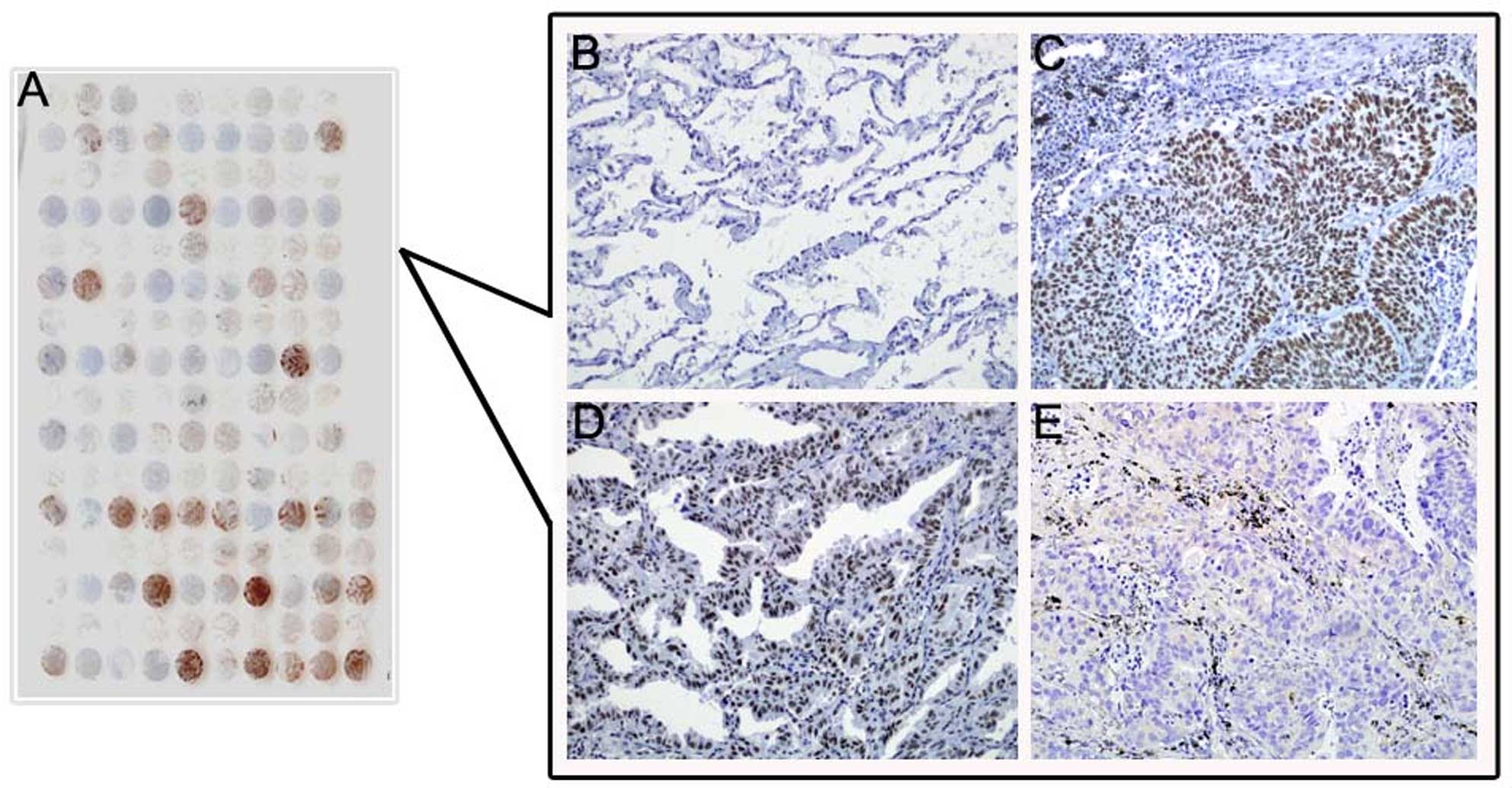
DEK protein expression showed a strict nuclear
staining pattern in NSCLC with immunohistochemistry, except that 3
cases of adenocarcinoma showed mainly a cytoplasmic staining
pattern. DEK protein was negative in normal lung tissues, but
usually upregulated in NSCLC. The positive rate of DEK protein was
66.33% (130/196) in NSCLC tissues, and was significantly higher
than that in either adjacent non-tumor tissues (26.67%, 8/30) or
normal lung tissues (0%, 0/20) (P<0.001) (Fig. 4 and Table I).
 | Table IDEK protein expression in the NSCLC
cases. |
Table I
DEK protein expression in the NSCLC
cases.
| | DEK protein
expression
| | |
|---|
| Tissues | No. of cases | − | + | ++ | Positive rate
(%) | P-value |
|---|
| NSCLC | 196 | 66 | 51 | 79 | 66.33 | 0.000b |
| Adjacent
non-tumor | 30 | 22 | 2 | 6 | 26.67 | 0.015a |
| Normal lung | 20 | 20 | 0 | 0 | 0 | |
Correlation between DEK expression and
clinicopathological features of NSCLC
To evaluate the role of DEK protein in NSCLC
progression, we analyzed correlations between DEK protein
expression and major clinicopathological features of NSCLC. The
results showed that DEK expression was significantly correlated to
differentiation and clinical stages of the NSCLC cases (P=0.001 and
P=0.016, respectively). However, DEK expression levels were not
correlated to age, gender, tumor size, CEA level, smoking status,
pathological subtype and metastasis of NSCLC (P>0.05) (Table II).
 | Table IICorrelation between DEK expression
and clinicopathological features of the NSCLC cases. |
Table II
Correlation between DEK expression
and clinicopathological features of the NSCLC cases.
| DEK protein
expression (%)
| | |
|---|
| Variables | +/++ | − | χ2 | P-value |
|---|
| Age (years) | | | 0.081 | 0.776 |
| <65 | 86 (65.65) | 45 (34.35) | | |
| ≥65 | 44 (67.69) | 21 (32.31) | | |
| Gender | | | 0.677 | 0.412 |
| Male | 75 (68.81) | 34 (31.19) | | |
| Female | 55 (63.22) | 32 (36.78) | | |
| Tumor size
(cm) | | | 0.535 | 0.466 |
| ≤3 | 52 (63.41) | 30 (39.59) | | |
| >3 | 78 (68.42) | 36 (31.58) | | |
|
Differentiation | | | 12.210 | 0.001b |
| Well | 20 (45.45) | 24 (54.55) | | |
| Moderately | 65 (69.15) | 29 (30.85) | | |
| Poorly | 45 (77.59) | 13 (22.41) | | |
| Pathological
subtype | | | 1.982 | 0.160 |
| SCC | 67 (62.04) | 41 (37.96) | | |
| AC | 63 (71.59) | 25 (28.41) | | |
| Clinical stage | | | 5.839 | 0.016a |
| I–II | 59 (58.42) | 42 (41.58) | | |
| III–IV | 71 (74.74) | 24 (25.26) | | |
| LN metastasis | | | 1.420 | 0.235 |
| Positive | 88 (69.29) | 39 (30.71) | | |
| Negative | 42 (60.87) | 27 (39.13) | | |
| CEA level | | | 2.589 | 1.108 |
| Normal | 63 (61.17) | 40 (38.83) | | |
| Increased | 67 (72.04) | 26 (27.96) | | |
| Smoking status | | | 0.177 | 0.675 |
| Yes | 101 (65.58) | 53 (34.42) | | |
| No | 29 (69.05) | 13 (30.95) | | |
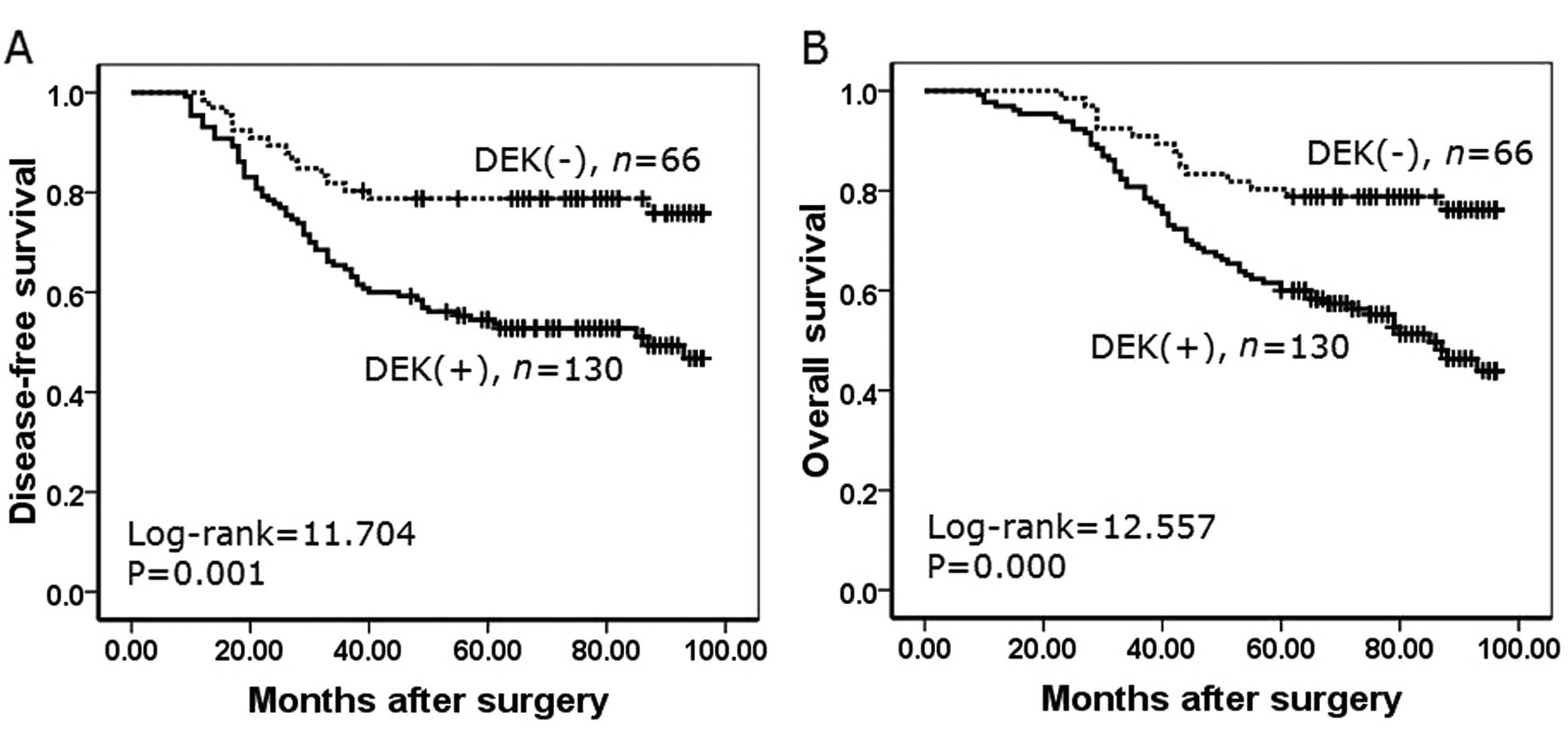
Correlation between the survival rates
and DEK expression using the Kaplan-Meier method
To further confirm the role of DEK expression in
NSCLC progression, we analyzed disease-free survival and overall
survival rates of 196 NSCLC cases using the Kaplan-Meier method. We
found that NSCLC patients with DEK expression had a lower
disease-free survival rate (log-rank=11.704, P=0.001) and a lower
overall survival rate (log-rank=12.557, P<0.001) than those
patients without DEK expression (Fig.
5).
To substantiate the importance of DEK expression in
NSCLC progression, we analyzed correlations between DEK expression
and clinical stages of NSCLC. In early-stage NSCLC, patients with
DEK expression had lower disease-free and overall survival rates
compared with patients without DEK expression (P=0.004 and P=0.003,
respectively) (Fig. 6A and B).
However, disease-free and overall survival rates were not
correlated to DEK expression status (P=0.136, and P=0.125,
respectively) in late-stage NSCLC (Fig.
6C and D).
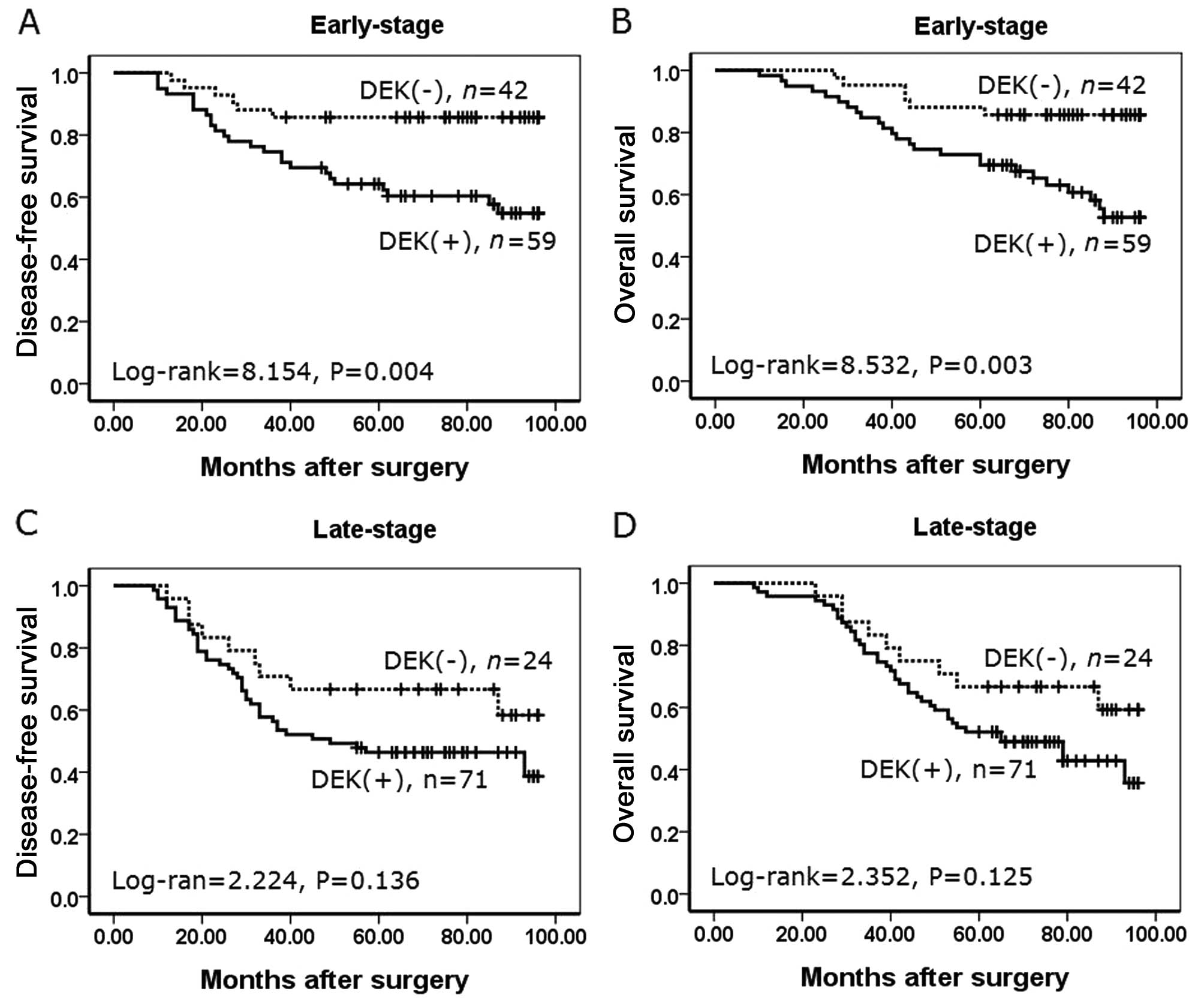 | Figure 6Kaplan-Meier analysis of disease-free
survival and overall survival rates in 196 NSCLC patients with or
without DEK expression in relation to clinical stage. (A) In the
early-stage, disease-free survival rate of patients with
DEK-positive expression was lower than the rate in patients with
DEK-negative expression (log-rank=8.154, P=0.004). (B) In the
early-stage, overall survival rate of patients with DEK-positive
expression was lower than the rate in patients with DEK-negative
expression (log-rank=8.532, P=0.003). (C) In the late-stage,
disease-free survival rate of patients was not correlated with DEK
expression (log-rank=2.224, P=0.136). (D) In the late-stage,
overall survival rate of patients was not correlated with DEK
expression (log-rank=2.352, P=0.125) (+, positive; −,
negative). |
DEK is an independent prognostic factor
in NSCLC using the Cox proportional hazard regression model
Univariate analysis showed that patients with NSCLC
tumors that expressed DEK had significantly lower overall survival
(P=0.005) than patients with NSCLC tumors that did not express DEK.
Additionally, patient age (P=0.043), pathological stage (P=0.000)
and lymph node metastasis (P=0.001) were associated with the
overall survival rate. Therefore, multivariate survival analysis
was performed using the Cox proportional hazards model for all the
significant variables found with univariate survival analysis. The
results suggested that clinical stage (HR, 1.689; 95% CI,
1.261–2.261; P<0.001), and lymph node metastasis (HR, 1.516; 95%
CI, 1.132–2.029; P=0.005) were independent prognostic factors for
overall survival rates in NSCLC. Importantly, DEK expression also
emerged as a significant independent prognostic factor in the
prognosis of NSCLC (HR, 1.388; 95% CI, 1.024–1.882; P=0.035)
(Table III).
 | Table IIIUnivariate and multivariate survival
analyses of the clinicopathological factors for the overall
survival rate of 196 patients with NSCLC. |
Table III
Univariate and multivariate survival
analyses of the clinicopathological factors for the overall
survival rate of 196 patients with NSCLC.
| | | | | 95% CI
| |
|---|
|
Characteristics | β | SE | Wald | HR | Lower | Upper | P-value |
|---|
| Univariate | | | | | | | |
| Gender | 0.106 | 0.144 | 0.547 | 1.112 | 0.839 | 1.475 | 0.460 |
| Age (years) | 0.309 | 0.152 | 4.102 | 1.361 | 1.010 | 1.835 | 0.043a |
| Smoking status | 0.132 | 0.174 | 0.571 | 1.141 | 0.811 | 1.606 | 0.450 |
| Tumor size | 0.279 | 0.146 | 3.663 | 1.322 | 0.993 | 1.761 | 0.056 |
| Clinical stage | 0.589 | 0.147 | 16.090 | 1.801 | 1.351 | 2.401 | 0.000b |
|
Differentiation | 0.194 | 0.101 | 3.680 | 1.214 | 0.996 | 1.479 | 0.055 |
| CEA | 0.030 | 0.143 | 0.044 | 1.030 | 0.778 | 1.364 | 0.834 |
| Pathological
subtype | 0.064 | 0.144 | 0.197 | 1.066 | 0.804 | 1.413 | 0.657 |
| LN metastasis | 0.472 | 0.148 | 10.226 | 1.603 | 1.201 | 2.142 | 0.001b |
| DEK | 0.423 | 0.152 | 7.739 | 1.527 | 1.133 | 2.057 | 0.005b |
| Multivariate | | | | | | | |
| Age (years) | 0.227 | 0.155 | 2.166 | 1.255 | 0.927 | 1.700 | 0.141 |
| Clinical stage | 0.524 | 0.149 | 12.384 | 1.689 | 1.261 | 2.261 | 0.000a |
| LN metastasis | 0.416 | 0.149 | 7.797 | 1.516 | 1.132 | 2.029 | 0.005a |
| DEK | 0.328 | 0.155 | 4.458 | 1.388 | 1.024 | 1.882 | 0.035a |
Discussion
DEK is located on chromosome 6p22.3, and was
initially described as the target of a recurrent t(6;9)
translocation in a subset of acute myeloid leukemia patients. Human
DEK protein consists of 375 amino acids with four distinct trenches
of acidic amino acids. As a highly conserved nuclear factor and the
only member of its protein class, it is preferentially expressed in
actively proliferating and malignant cells, where it can reach up
to 4 to 6 million copies/nucleus (16,17).
The ability of DEK to bind nucleic acid has led to
functional associations with several cellular processes, including
chromatin remodeling, transcriptional regulation, replication, mRNA
splicing and DNA repair (18–21).
Carro et al found that when DEK is upregulated, as observed
in numerous types of cancer, perturbations to the normal genome
architecture and integrity are likely contributors to oncogenesis
(22). Privette Vinnedge et
al reported that DEK depletion can result in cell death and
impaired DNA double-strand break repair (9). Therefore, cellular DEK expression is
tightly controlled to maintain proper cell function and viability.
Wise-Draper et al found that there was a significant delay
in the formation of papillomas in DEK-knockout mice compared with
wild-type and heterozygous littermate mice (5). Papillomas ultimately formed in the
DEK-knockout mice suggesting a role for DEK in tumor initiation in
this model.
The increasing list of tumor types, including acute
myeloid leukemia (23),
glioblastoma (24), cervical cancer
(25,26), melanoma (22) and ovarian cancer (11) among others (27–29),
showing high DEK protein expression raises the exciting possibility
of using DEK as a tumor marker (29). Datta et al reported that DEK
protein was present in voided urine of patients with both low- and
high-grade bladder cancers, suggesting that DEK could be used as a
biomarker for detection of this cancer using patient urine samples
(30). Privette Vinnedge et
al suggested that DEK promotes the pathogenesis of
ER+ breast cancer and that the targeted inhibition of
DEK may enhance the efficacy of conventional hormone therapies
(6). Our previous study showed that
DEK protein expression was closely related to the disease-free and
overall survival rates of patients with colorectal cancer, and its
overexpression was an independent risk factor for mortality in
colorectal cancer (13).
Few studies to date have reported an association
between DEK expression and clinicopathological parameters, as well
as the DEK prognostic role in lung cancer. Wang et al
analyzed DEK immunohistochemistry in 112 NSCLC cases and reported
that DEK-positive tumors were correlated with poor differentiation,
advanced p-TNM stage and nodal metastasis, and DEK expression in
lung adenocarcinoma was significantly higher compared with DEK
expression in squamous cell carcinoma (31). In the present study, qRT-PCR and
western blot data demonstrated that the levels of DEK mRNA and
protein were significantly higher in NSCLC samples compared with
adjacent non-tumor tissues. Furthermore, we performed
immunohistochemical staining and analysis in 196 cases of NSCLC,
and found that the positive rate of DEK protein expression was
66.33% in NSCLC, and was significantly higher than that noted in
either adjacent non-tumor or normal lung tissues, indicating that
DEK potentially plays an important role in the progression of
NSCLC. These findings are consistent with the current theory that
suggests that DEK upregulation is high in proliferating cells and
low in resting and terminally differentiated cells (4,32).
We examined DEK expression and the
clinicopathological features of NSCLC and found that DEK expression
was significantly correlated with poor differentiation (P<0.01)
and advanced clinical stage (P<0.05), but was not related to
age, gender, tumor size, nodal status, pathological subtype, CEA
level and the smoking status of patients with NSCLS (P>0.05).
However, Wang et al reported that DEK expression was
significantly related to nodal metastasis and pathological subtype
(31). The causes for the
differences in results may be due to case selection, the source of
the antibody used or staining method. Further study is needed to
explore the mechanisms of DEK upregulation in NSCLC
progression.
With regard to survival, we found that NSCLC
patients with DEK expression had a lower disease-free survival rate
(P=0.001) and overall survival rate (P<0.001) than patients
without DEK expression. In early-stage NSCLC, patients with DEK
expression had lower disease-free and overall survival rates
compared with those without DEK expression (P=0.004 and P=0.003,
respectively). However, DEK expression status was not related to
the survival of NSCLC patients with an advanced clinical stage.
Multivariate survival analysis demonstrated that DEK expression
emerged as a significantly independent hazard factor for overall
survival in NSCLC, along with clinical stage and metastasis.
In conclusion, DEK plays an important role in NSCLC
progression, and it may be an independent biomarker for evaluating
prognosis in patients with NSCLC.
Acknowledgments
The present study was supported by grants from the
National Natural Science Fund of China (no. 81272927), the Projects
for Research and Innovation of the Jilin Youth Leader and Team (no.
20130521017JH), and the Projects of Science Research of the
Education Department in Liaoning Province (no. L2015189).
References
|
1
|
Jemal A, Siegel R, Ward E, Murray T, Xu J
and Thun MJ: Cancer statistics, 2007. CA Cancer J Clin. 57:43–66.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Brambilla E, Travis WD, Colby TV, Corrin B
and Shimosato Y: The new World Health Organization classification
of lung tumours. Eur Respir J. 18:1059–1068. 2001. View Article : Google Scholar
|
|
3
|
Lee HW, Kim EH and Oh MH:
Clinicopathologic implication of ezrin expression in non-small cell
lung cancer. Korean J Pathol. 46:470–477. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
von Lindern M, Fornerod M, van Baal S,
Jaegle M, de Wit T, Buijs A and Grosveld G: The translocation
(6;9), associated with a specific subtype of acute myeloid
leukemia, results in the fusion of two genes, dek and can, and the
expression of a chimeric, leukemia-specific dek-can mRNA. Mol Cell
Biol. 12:1687–1697. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wise-Draper TM, Mintz-Cole RA, Morris TA,
Simpson DS, Wikenheiser-Brokamp KA, Currier MA, Cripe TP, Grosveld
GC and Wells SI: Overexpression of the cellular DEK protein
promotes epithelial transformation in vitro and in vivo. Cancer
Res. 69:1792–1799. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Privette Vinnedge LM, Ho SM,
Wikenheiser-Brokamp KA and Wells SI: The DEK oncogene is a target
of steroid hormone receptor signaling in breast cancer. PLoS One.
7:e469852012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cleary J, Sitwala KV, Khodadoust MS, Kwok
RP, Mor-Vaknin N, Cebrat M, Cole PA and Markovitz DM:
p300/CBP-associated factor drives DEK into interchromatin granule
clusters. J Biol Chem. 280:31760–31767. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Khodadoust MS, Verhaegen M, Kappes F,
Riveiro-Falkenbach E, Cigudosa JC, Kim DS, Chinnaiyan AM, Markovitz
DM and Soengas MS: Melanoma proliferation and chemoresistance
controlled by the DEK oncogene. Cancer Res. 69:6405–6413. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Privette Vinnedge LM, McClaine R, Wagh PK,
Wikenheiser-Brokamp KA, Waltz SE and Wells SI: The human DEK
oncogene stimulates β-catenin signaling, invasion and mammosphere
formation in breast cancer. Oncogene. 30:2741–2752. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hasiów-Jaroszewska B, Borodynko N and
Pospieszny H: Infectious RNA transcripts derived from cloned cDNA
of a pepino mosaic virus isolate. Arch Virol. 154:853–856. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Han S, Xuan Y, Liu S, Zhang M, Jin D, Jin
R and Lin Z: Clinicopathological significance of DEK overexpression
in serous ovarian tumors. Pathol Int. 59:443–447. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wu Q, Li Z, Lin H, Han L, Liu S and Lin Z:
DEK overexpression in uterine cervical cancers. Pathol Int.
58:378–382. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lin L, Piao J, Gao W, Piao Y, Jin G, Ma Y,
Li J and Lin Z: DEK over expression as an independent biomarker for
poor prognosis in colorectal cancer. BMC Cancer. 13:3662013.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu S, Wang X, Sun F, Kong J, Li Z and Lin
Z: DEK overexpression is correlated with the clinical features of
breast cancer. Pathol Int. 62:176–181. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wrona A and Jassem J: The new TNM
classification in lung cancer. Pneumonol Alergol Pol. 78:407–417.
2010.In Polish.
|
|
16
|
Kappes F, Burger K, Baack M, Fackelmayer
FO and Gruss C: Subcellular localization of the human
proto-oncogene protein DEK. J Biol Chem. 276:26317–26323. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kappes F, Scholten I, Richter N, Gruss C
and Waldmann T: Functional domains of the ubiquitous chromatin
protein DEK. Mol Cell Biol. 24:6000–6010. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Alexiadis V, Waldmann T, Andersen J, Mann
M, Knippers R and Gruss C: The protein encoded by the
proto-oncogene DEK changes the topology of chromatin and reduces
the efficiency of DNA replication in a chromatin-specific manner.
Genes Dev. 14:1308–1312. 2000.PubMed/NCBI
|
|
19
|
Campillos M, García MA, Valdivieso F and
Vázquez J: Transcriptional activation by AP-2alpha is modulated by
the oncogene DEK. Nucleic Acids Res. 31:1571–1575. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Soares LM, Zanier K, Mackereth C, Sattler
M and Valcárcel J: Intron removal requires proofreading of U2AF/3′
splice site recognition by DEK. Science. 312:1961–1965. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kavanaugh GM, Wise-Draper TM, Morreale RJ,
Morrison MA, Gole B, Schwemberger S, Tichy ED, Lu L, Babcock GF,
Wells JM, et al: The human DEK oncogene regulates DNA damage
response signaling and repair. Nucleic Acids Res. 39:7465–7476.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Carro MS, Spiga FM, Quarto M, Di Ninni V,
Volorio S, Alcalay M and Müller H: DEK Expression is controlled by
E2F and deregulated in diverse tumor types. Cell Cycle.
5:1202–1207. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Casas S, Nagy B, Elonen E, Aventín A,
Larramendy ML, Sierra J, Ruutu T and Knuutila S: Aberrant
expression of HOXA9, DEK, CBL and CSF1R in acute myeloid leukemia.
Leuk Lymphoma. 44:1935–1941. 2003. View Article : Google Scholar
|
|
24
|
Kroes RA, Jastrow A, McLone MG, Yamamoto
H, Colley P, Kersey DS, Yong VW, Mkrdichian E, Cerullo L, Leestma
J, et al: The identification of novel therapeutic targets for the
treatment of malignant brain tumors. Cancer Lett. 156:191–198.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kappes F, Khodadoust MS, Yu L, Kim DS,
Fullen DR, Markovitz DM and Ma L: DEK expression in melanocytic
lesions. Hum Pathol. 42:932–938. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu K, Feng T, Liu J, Zhong M and Zhang S:
Silencing of the DEK gene induces apoptosis and senescence in CaSki
cervical carcinoma cells via the up-regulation of NF-κB p65. Biosci
Rep. 32:323–332. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Paderova J, Orlic-Milacic M, Yoshimoto M,
da Cunha Santos G, Gallie B and Squire JA: Novel 6p rearrangements
and recurrent translocation breakpoints in retinoblastoma cell
lines identified by spectral karyotyping and mBAND analyses. Cancer
Genet Cytogenet. 179:102–111. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Privette Vinnedge LM, Kappes F, Nassar N
and Wells SI: Stacking the DEK: From chromatin topology to cancer
stem cells. Cell Cycle. 12:51–66. 2013. View Article : Google Scholar :
|
|
29
|
Lin L, Piao J, Ma Y, Jin T, Quan C, Kong
J, Li Y and Lin Z: Mechanisms underlying cancer growth and
apoptosis by DEK overexpression in colorectal cancer. PLoS One.
9:e1112602014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Datta A, Adelson ME, Mogilevkin Y,
Mordechai E, Sidi AA and Trama JP: Oncoprotein DEK as a tissue and
urinary biomarker for bladder cancer. BMC Cancer. 11:2342011.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang J, Sun L, Yang M, Luo W, Gao Y, Liu
Z, Qiu X and Wang E: DEK depletion negatively regulates
Rho/ROCK/MLC pathway in non-small cell lung cancer. J Histochem
Cytochem. 61:510–521. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kappes F, Damoc C, Knippers R, Przybylski
M, Pinna LA and Gruss C: Phosphorylation by protein kinase CK2
changes the DNA binding properties of the human chromatin protein
DEK. Mol Cell Biol. 24:6011–6020. 2004. View Article : Google Scholar : PubMed/NCBI
|