Introduction
Renal cell carcinoma (RCC) is the most lethal
urologic tumor, with estimated 63,920 new cases and 13,860 deaths
in 2014 (1). Clear cell renal cell
carcinoma (ccRCC), originating from the renal proximal tubule, is
the most common subtype of RCC and accounts for ~75–80% of these
tumors with the highest rates of local invasion, metastasis,
mortality and refractory to current treatments (2,3).
Despite increased early detection of RCC and more frequent surgery,
30% of patients develop metastases after surgery (3–5).
Therefore, it is urgent to understand the underlying molecular
mechanisms involved in the pathogenesis of RCC for developing
cancer prevention strategies and possible guiding disease
management in the clinic.
MicroRNAs (miRNAs) are a class of small, single
stranded, non-coding RNA molecules of 19–24 nucleotides that bind
to complementary sequences in the 3′ untranslated regions (UTR) of
their target mRNAs and induce mRNA degradation or translational
repression (6,7). miRNAs have been implicated in the
regulation of various biological processes, such as cell
proliferation, migration, invasion, apoptosis, metabolism, and
cellular differentiation (8,9).
Furthermore, miRNAs have recently been reported to function as both
oncogenes and suppressors of tumor progression, and to play crucial
roles in human tumorigenesis and/or metastasis by directly
targeting oncogenes or tumor suppressor genes (10). Therefore, miRNA expression can
contribute to the diagnosis and classification of cancers, and
contribute to develop novel avenues for targeted therapy (11).
miR-22, a 22-nt non-coding RNA, has been found to be
frequently downregulated in several cancers, such as breast cancer
(12), osteosarcoma (13), esophageal squamous cell carcinoma
(14), gastric cancer (15) and colorectal cancer (16). Accumulating evidence shows that
miR-22 plays key roles in the regulation of cell growth, migration,
chemosensitivity and epithelial-mesenchymal transition in cancers
(17). For RCC, recently it was
reported that miR-22 was significantly downregulated in RCC tumor
tissues as compared with adjacent normal tissues (18). However, our knowledge of the
clinicopathological impact and the exact roles of the miR-22 in EOC
and the underlying molecular mechanisms remains unclear.
The aim of this study was to investigate the
involvement of miR-22 in renal cancer. We found that the expression
level of miR-22 decreased in ccRCC tissues and was inversely
associated with histologic grade, tumor stage, lymph node
metastasis, and distant metastasis. Function studies demonstrated
that miR-22 acts as a tumor suppressor by affecting renal cancer
cell proliferation, apoptosis, migration and invasion. Furthermore,
SIRT1 was identified as a direct functional target of miR-22.
Overexpression of miR-22 activated p53 and its downstream target
p21 and PUMA, as well as the apoptosis markers cleaved CASP3 and
PARP, and inhibited epithelial-mesenchymal transition (EMT). These
findings indicated that miR-22 has a potential role in the therapy
of RCC.
Materials and methods
Human RCC clinical specimens
A total of 50 primary RCC tissues and matched
adjacent normal tissues were obtained from patients who underwent
radical nephrectomy in the TCM Nephropathy, the Affiliated
Hospital, Changchun University of Chinese Medicine between July
2012 to December 2014. None of the patients had received
chemotherapy or radiotherapy prior to surgery. After surgical
resection, tumor tissues and adjacent normal tissues were collected
and stored at −80°C until use. Use of clinical sample cohorts in
this study was approved by the Institution Research Ethics
Committee of Changchun University of Chinese Medicine. All patients
gave written consent for their information to be stored in the
hospital database and used for this study. The clinical and
pathological information from patient records was gathered, and is
listed in Table I.
 | Table ICorrelation between
clinicopathological features and miR-22 expression in 50 RCC
tissues. |
Table I
Correlation between
clinicopathological features and miR-22 expression in 50 RCC
tissues.
| Variables | No. of cases | miR-22 expression
| P-value |
|---|
Low
(n %) | High
(n %) |
|---|
| Age (years) | | | | P=0.918 |
| <55 | 23 | 11 (47.8) | 12 (52.2) | |
| ≥55 | 27 | 13 (48.1) | 14 (51.9) | |
| Gender | | | | P=0.632 |
| Male | 28 | 14 (50.0) | 14 (50.0) | |
| Female | 22 | 10 (45.4) | 12 (54.6) | |
| TNM stage | | | | P<0.01 |
| T1-T2 | 30 | 7 (23.3) | 23 (76.7) | |
| T3-T4 | 20 | 17 (85.0) | 3 (15.0) | |
| Tumor size
(cm) | | | | P=0.369 |
| <5 | 19 | 8 (42.1) | 11 (57.9) | |
| ≥5 | 31 | 16 (51.6) | 15 (48.9) | |
| Lymph node
metastasis | | | | P<0.01 |
| No | 32 | 7 (21.8) | 25 (78.2) | |
| Yes | 18 | 17 (94.3) | 1 (5.6) | |
Cell culture
Immortalized normal human proximal tubule epithelial
cell line HK-2 was purchased from the American Type Culture
Collection (ATCC; USA). Four human RCC cell lines 786-O, ACHN,
Caki-1, and Caki-2 were obtained from the Cell Bank of Type Culture
Collection of Chinese Academy of Sciences (Shanghai, China). All
cells were cultured in RPMI-1640 medium with 10% fetal bovine serum
(FBS) (both from Gibco), 50 U/ml penicillin or 50 µg/ml
streptomycin at 37°C in a 5% CO2 humidified
incubator.
Quantitative reverse
transcription-polymerase chain reaction (qRT-PCR)
Total RNA was extracted from tissue and cultured
cells using TRIzol (Invitrogen, USA) according to the
manufacturer's instructions. For mRNA reverse transcription, cDNA
was synthesized using the PrimeScript RT reagent kit (Takara Bio,
Japan). Quantitative real-time polymerase chain reaction (RT-qPCR)
analysis of the SIRT1 expression in mRNA level was performed using
Fast SYBR Green Master Mix (Applied Biosystems, Foster City, CA,
USA) under ABI 7900 Sequence Detection system (Life Technologies,
NY, USA). GAPDH were used as endogenous controls for miRNA GAPDH.
The specific primers for GAPDH and SIRT1 are as follows: GAPDH
(sense), 5′-TCAACGACCACTTTGTCAAGCTCA-3′ and antisense,
5′-GCTGGTGGTCCAGGGGTCTTACT-3′; and SIRT1 (sense),
5′-GCCAGAGTCCAAGTTTAGAAGA-3′ and antisense,
5′-CCATCAGTCCCAAATCCAG-3′.
Total miRNA extraction was performed from tissue and
cells using the mirVana miRNA Isolation kit (Ambion, Austin, TX,
USA). For miRNA reverse transcription, cDNA was synthesized using
One Step PrimeScript miRNA cDNA Synthesis kit (Qiagen, Valencia,
CA, USA) according to the manufacturer's instructions.
Quantification of miR-22 expression was performed using the mirVana
qRT-PCR miRNA Detection kit (Ambion). U6 were used as endogenous
controls for miRNA. The comparative 2−∆∆Ct method was
used for relative quantification and statistical analysis. All
experiments were performed in triplicate.
Transient transfection of miRNA
mimics
The miRNA-22 mimic and corresponding negative
control mimics (miR-NC) were purchased from GenePharma (Shanghai,
China). Transfection of miR-22 mimic or miR-NC at the final
concentration of 100 nM into RCC cells was performed using
Lipofectamine 2000 (Invitrogen) according to the manufacturer's
procedure. Transfection efficiency was evaluated in each experiment
by qRT-PCR 48 h after transfection.
Cell proliferation and colony formation
assay
To assess cell proliferation, 5×103 786-O
cells were plated in 96-well plates overnight and transfected with
the miR-22 mimic or miR-NC. Proliferation was determined by
3-(4,5-dimethyl-thiazol-2-yl)-2-5 diphenyltetrazolium bromide (MTT;
Sigma, USA) assay at 24, 48, 72, and 96 h after transfection, and
the absorbance of samples was measured with a microplate reader
(Bio-Rad, Gaithersburg, MD, USA) at 490 nm. All experiments were
performed in three replicates and were repeated three times
independently.
For colony formation assay, 786-O cells were
transfected with miR-22 mimic or miR-NC in 60-mm culture dishes.
After 24 h, 1×103 cells were transferred to 6-well
plates and incubated for 2 weeks. Then, cells were rinsed with PBS
and fixed with 1% formaldehyde for 30 min at room temperature.
Fixed cells were stained with 1% crystal violet. Finally the clones
were photographed and counted. The percentage of the colony
formation was calculated by adjusting control (miR-NC) to 100%.
Cell cycle assay
To assess cell cycle distribution and cell
apoptosis, 2×104 786-O cells were plated in 6-well
plates and transfected with miR-22 or miR-NC. After transfection,
the cells were collected by trypsinization, fixed in 70% ice-cold
ethanol overnight, then washed with PBS and stained with propidium
iodide (PI, 50 mg/ml; Sigma) in PBS supplemented with RNase (50
mg/ml) for 30 min in the dark at room temperature (RT). Analyses of
cell cycle distribution were performed by flow cytometer following
the manufacturer's guidelines (BD Biosciences).
Apoptosis assay
Cell apoptosis analysis was performed with Annexin
V-FITC Apoptosis Detection kit I (BD Biosciences). In briefly,
2×104 786-O cells transfected with miR-22 mimic or
miR-NC were suspended in RPMI-1640 medium. The cells were
re-suspended in 500 µl cold binding buffer with 2 µl
Annexin V-FITC, and incubated for 15 min in the dark at room
temperature. Cells were re-suspended in 500 µl cold binding
buffer with 10 µl PI, incubated for 4 h and analyzed by flow
cytometry (BD Biosciences).
Scratch migration assay
To assess the effect of miR-22 on cell migration,
786-O cells transfected with miR-22 mimic or miR-NC were seeded
into 6-well plates and cultured overnight. Before scratching, cells
were starved in medium with 1% FBS for 24 h. Thereafter, similar
sized wounds were scratched into the monolayer using a sterile
plastic micropipette tip. Wounded monolayer cells were washed three
times by PBS to remove cell debris and then cultured. Migration of
cells into the wound was observed at 0 and 24 h using an IX51
inverted microscope (Olympus, Tokyo, Japan). Individual cells were
quantified as an average of at least five fields for each
experiment. Each experiment was performed three times
independently.
Invasion assay
For the Transwell invasion assay, 3×104
transfected cells suspended in serum-free medium were added to the
upper chamber. In the bottom chamber, medium containing 10% FBS was
added as a chemoattractant. After 24-h culture at 37°C, the cells
remaining in the upper chamber or on the upper membrane was removed
with a sterile swab. Invaded cells of the bottom chamber were fixed
with 70% ethanol for 30 min and stained with 0.2% crystal violet
(Sigma) for 10 min. Invading cells were counted by taking
photomicrographs in five random fields.
Vector construction and luciferase
reporter assay
A human SIRT1 3′UTR was amplified from renal cancer
cell line cDNAs and cloned into the XhoI/NotI site of
a psiCHECK-2 vector (Promega, Madison, WI, USA). For mutagenesis of
the miR-22-binding site, a QuikChange Site-Directed Mutagenesis kit
(Agilent Technologies, Palo Alto, CA, USA) was used following the
manufacturer's instructions.
For luciferase activity assay, 786-O cells were
seeded into 12-well plates overnight before transfection, and then
co-transfected with 200 ng of psiCHECK-2 vectors, which harbored
SIRT1 3′UTR wild-type or mutant constructs, and 200 nM of miR-22 or
miR-NC. Forty-eight hours after transfection, luciferase activity
was measured with a Dual-Luciferase assay kit (Promega) according
to the manufacturer's instructions. Renilla luciferase
activity was normalized to firefly luciferase activity.
Western blot analysis
Tissue sample and cells were collected and
homogenized with RIPA lysis buffer (150 mM NaCl, 0.5% sodium
deoxycholate, 0.1% SDS, 1% Igepal, 50 mM Tris-HCl pH 8.0, 2 mM
EDTA) supplemented with 1 mM DTT, 1 mM Pefabloc, 1 mM
NaV3, 10 mM NaF and 1X complete mini protease inhibitor
cocktail tablets. Total protein concentration was detected using a
bicinchoninic acid (BCA) protein assay kit (Boster, China). Thirty
micrograms of protein/lane were resolved on 8–15% sodium
dodecylsulfate-polyacrylamide gels (SDS-PAGE), and then transferred
onto the nitrocellulose membrane (Bio-Rad, Munich, Germany). The
membrane was incubated with 5% non-fat skim milk for 2 h at room
temperature. The membrane was incubated with the following
antibodies overnight at 4°C: SIRT1, E-cadherin, N-cadherin,
vimentin, p21, p53, Ac-p53(382), PUMA, PARP, GAPDH and cleaved
CASP3. GAPDH was used as the internal control. All antibodies were
from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Then the
membrane was incubated with the corresponding horseradish
peroxidase (HRP)-conjugated secondary antibody (Santa Cruz
Biotechnology) for 2 h at room temperature. Proteins were
visualized with ECL chemiluminescent kit (ECL Plus; Thermo
Scientific).
In vivo nude mouse tumorigenesis
assay
Male BALB/c nude mice (five weeks old, 18–20 g) were
obtained from Experiments Animal Center of Changchun Biological
Institute (Changchun, China), and maintained under specific
pathogen-free conditions. This study and all experimental protocols
were approved by the Animal Ethics Committee of Changchun
University of Chinese Medicine (Changchun, China).
About 2×106 logarithmically growing 786-O
cells stably expressing miR-22 or miR-NC, were suspended in 100
µl of medium (containing 10% Matrigel) then injected into
the flanks of mice (n=10), respectively. Tumor volume was measured
every five days by digital Vernier calipers, and was calculated
according to the following formula: [π/6 × length × width ×
height]. Thirty days after inoculation, mice were sacrificed, and
tumors were removed and weighed. Part of the tumor tissues was
harvested for analysis of the expression of SIRT1 by western blot
analysis.
Statistical analysis
All the data are shown as mean ± SD (standard
deviation) from at least three independent experiments. The
difference was determined by two-tailed Student's t-test.
Statistical analysis was performed with GraphPad Prism 5.0
(GraphPad Software, San Diego, CA, USA). In all cases, P<0.05
was considered statistically significant.
Results
miR-22 expression is downregulated in
renal cancer tissue and cell lines
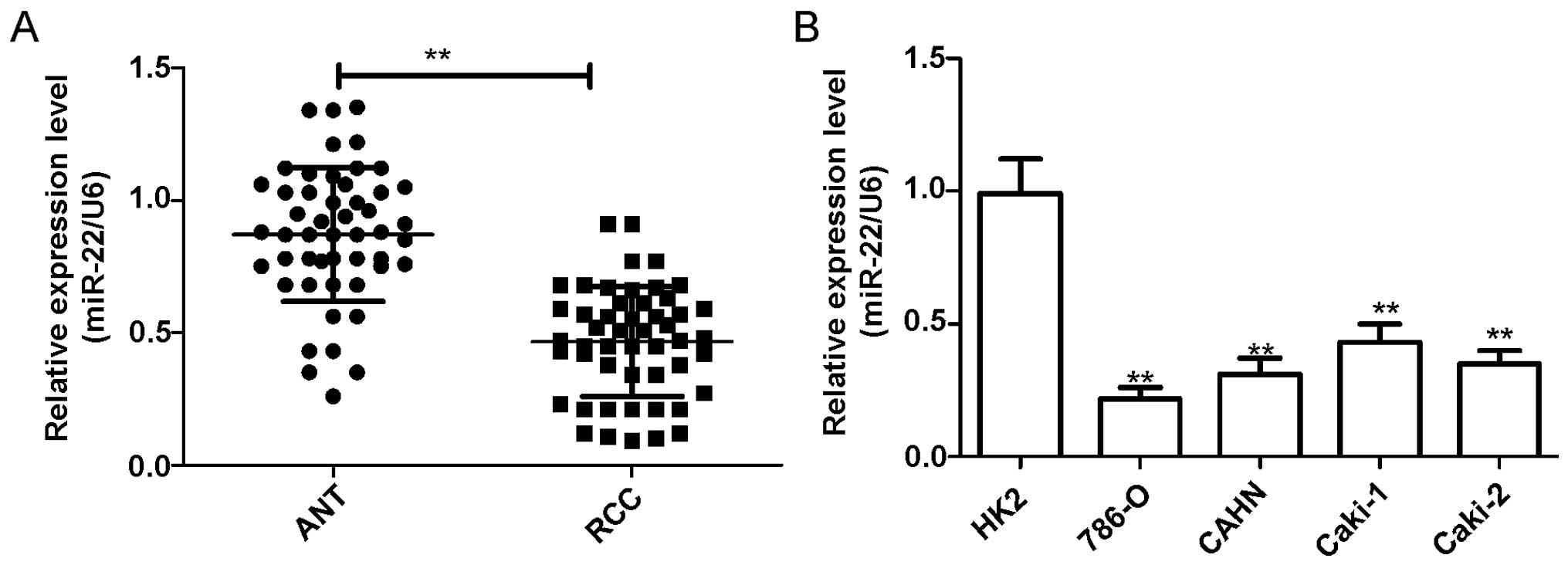
To investigate the expression levels of miR-22 in
renal cancer, the expression level of miR-22 in four RCC cell lines
was first analyzed by qRT-PCR. In comparison to immortalized normal
proximal tubule epithelial cell line HK-2, miR-22 expression was
downregulated in all four renal cancer cell lines (786-O, ACHN,
Caki-1, Caki-2) (P<0.05, Fig.
1A). The 786-O cell line, which possessed the lowest levels of
miR-22 expression among the four cell lines, was selected for
further study. In addition, we further assessed the expression
level of miR-22 in RCC tissues and adjacent normal tissues.
We found that the expression of miR-22 was
significantly decreased in RCC tissues as compared with the
corresponding adjacent tissues (P<0.05, Fig. 1B).
To investigate the association between miR-22
expression and clinicopathological features in the 50 RCC patients.
All RCC samples were divided into miR-22 low-expression group
(n=24) and high-expression group (n=26), median (0.467) was used as
cut-off. We found that miR-22 expression was significantly
correlated with histological grade, tumor stage and lymph node
metastasis (P<0.05), but was not significant associated with
patient gender, age or tumor size (Table I). These results indicate that
miR-22 is downregulated in renal cancer, suggesting miR-22 may play
a key role in the development and progression of renal cancer.
miR-22 suppresses cell proliferation,
colony formation, and induces cell arrest and apoptosis in renal
cancer cells
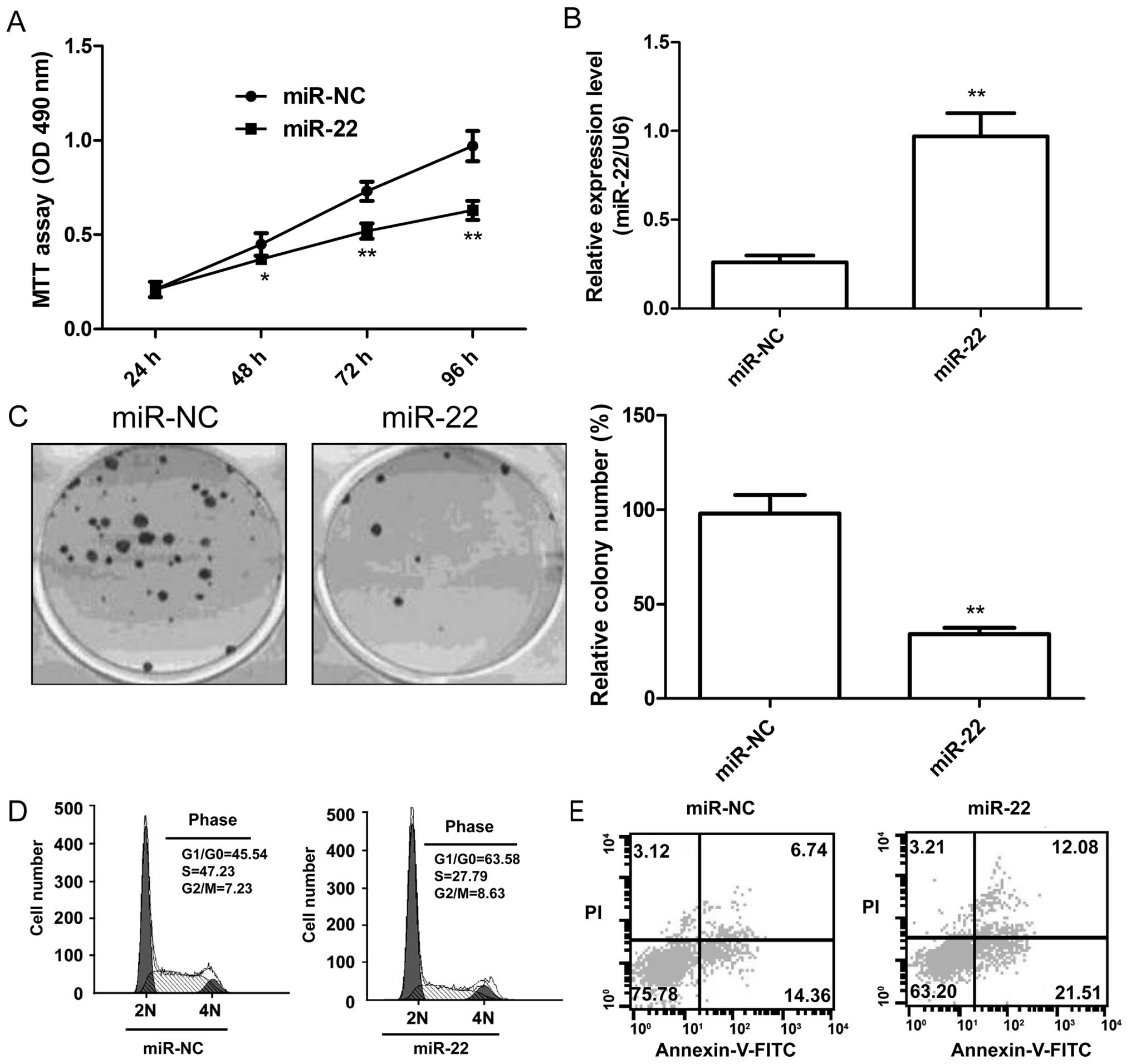
To investigate the effect of miR-22 on cell
proliferation, colony formation, cell cycle and apoptosis, we
performed overexpression experiments in the renal cancer cell line
786-O. Successful overexpression of miR-22 in the cells was
confirmed by qRT-PCR (Fig. 2A). MTT
assay and colony formation assay showed that overexpression of
miR-22 significantly inhibited cell proliferation (Fig. 2B) and colony formation (Fig. 2C). As the proliferation was directly
linked to cell cycle distribution, we investigated the effect of
miR-22 on RCC cell cycle progression, and found that overexpression
of miR-22 markedly induced G1 phase arrest and decreased S phase
arrest of renal cancer cell lines (Fig.
2D). Cell apoptosis assay showed that overexpression of miR-22
significantly induced apoptosis in RCC cells (Fig. 2E).
miR-22 inhibits the migration, invasion
and epithelial-to-mesenchymal transition (EMT) of renal cancer
cells
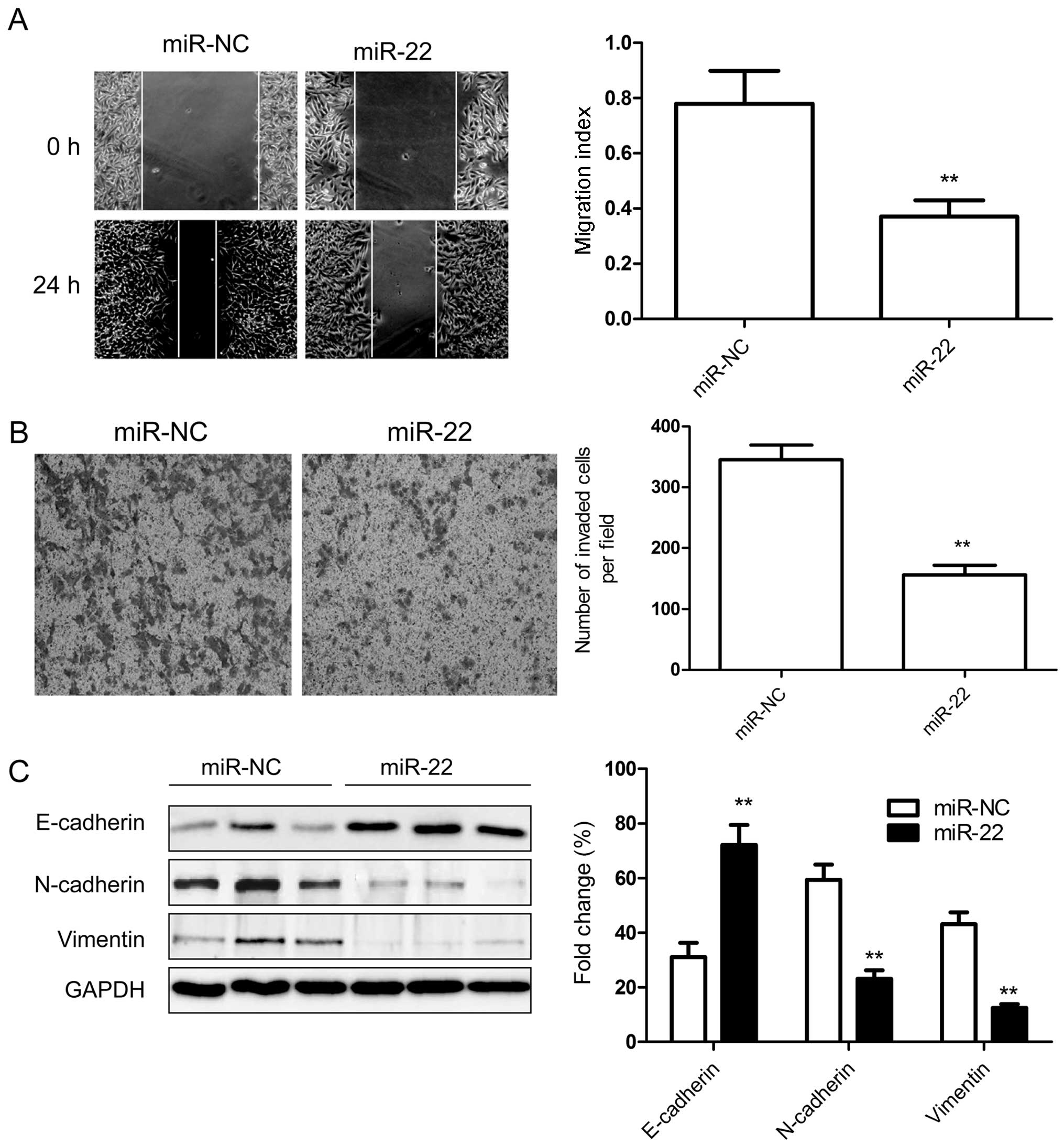
The above results showed that miR-22 expression is
inversely correlated with the metastatic ability of RCC, therefore,
to test whether miR-22 overexpression suppresses tumor cell
migration and invasion, the migration and invasion of 786-O cells
transfected with miR-22 or miR-NC were determined by wound-healing
assay and Transwell assay, respectively. We found that
overexpression of miR-22 significantly suppressed migration
(Fig. 3A) and invasion (Fig. 3B) in renal cancer cells.
To further investigate whether the inhibitory effect
of miR-22 on migration and invasion was mediated by
epithelial-mesenchymal transition (ETM) since EMT plays a key role
in the invasion of various cancer cells by the transformation of
polarized and adherent epithelial cells into motile and invasive
mesenchymal cells, we examined the ETM-related to protein
expression. As shown in Fig. 3C,
overexpression of miR-22 in RCC cells dramatically upregulated
E-cadherin protein expression, an epithelial marker, and
downregulated N-cadherin and vimentin protein expression,
mesenchymal marker, which contribute to suppression of ETM and
inhibition of migration and invasion.
miR-22 targets 3′UTR of SIRT1 and
suppresses the expression of SIRT1
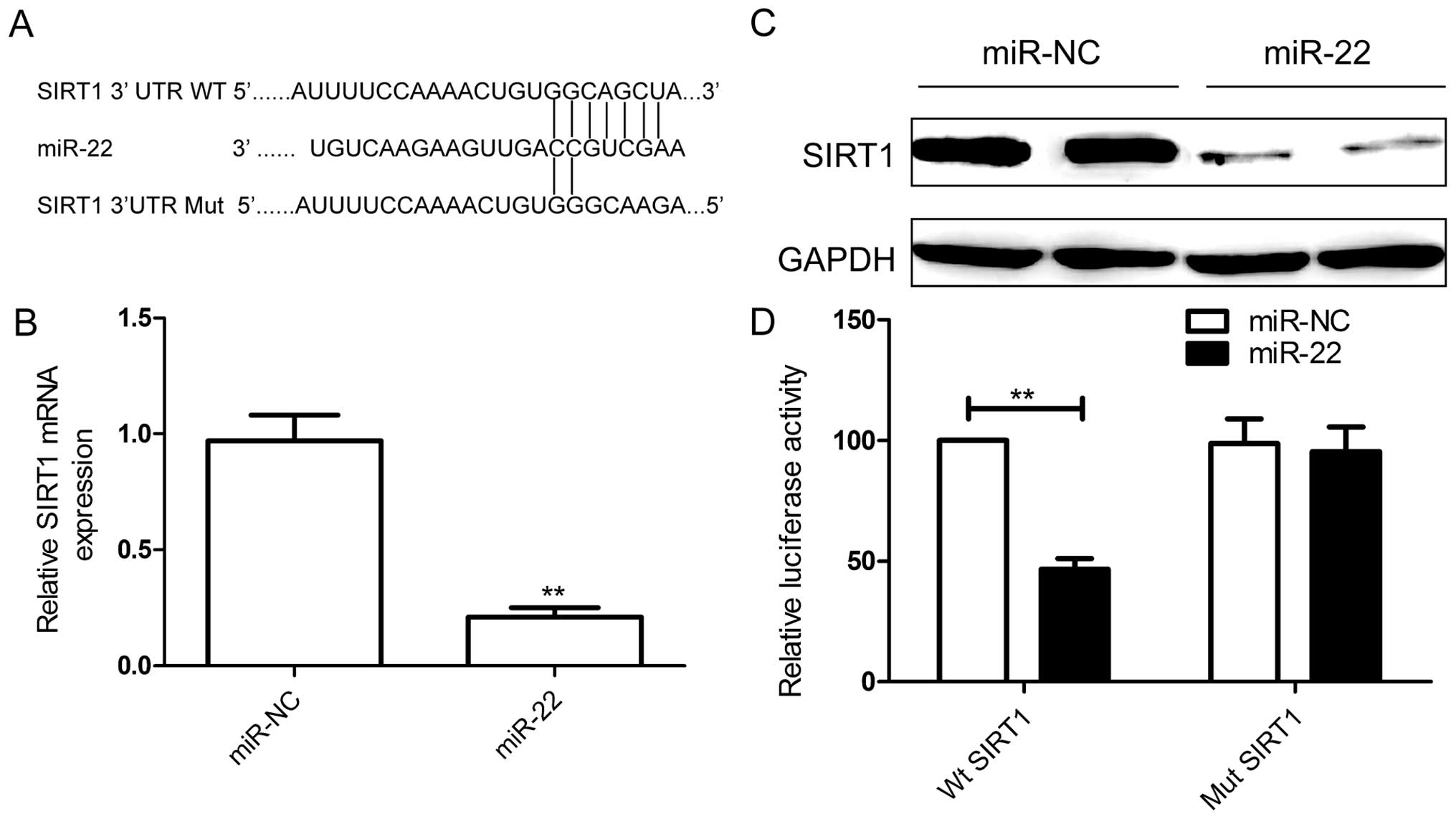
To explore the mechanism of the antitumor effects of
miR-22 on RCC cells, bioinformatic database (TargetScan, PicTar,
and miRanda) analyses were performed to predict putative miR-22
targets. The results revealed that SIRT1 was a potential target of
miR-22 (Fig. 4A). As expected, the
qRT-PCR and western blotting results confirmed that miR-22
restoration caused downregulation of SIRT1 expression on mRNA level
(Fig. 4B) and protein level
(Fig. 4C). To further prove the
direct interaction between miR-22 and its targets, we cloned 3′-UTR
sequences that contained the predicted target sites (wild type, WT)
or mutated sequences (mutant type) of SIRT1 into the psiCHECK-2
vectors. A luciferase reporter assay demonstrated that 786-O cells
co-transfected with miR-22 mimic and WT 3′-UTR SIRT1 significantly
suppressed the luciferase activity (Fig. 4D), whereas no obvious effect was
observed in their negative and mutant controls. These results
demonstrated that miR-22 negatively regulates SIRT1 expression by
directly binding to the 3′-UTR of SIRT1.
miR-22 activates the p53 signal pathway
of renal cancer cells
It has been reported that increased expression of
SIRT1 in normal or cancer cells decreases the ability of p53 by
deacetylating p53, leading to decreasing cellular apoptosis
(19,20). Therefore, we examined the effect of
miR-22 on acetylation of p53. Transfecting miR-22 mimic into 786-O
cells decreases SIRT1 (Fig. 4B and
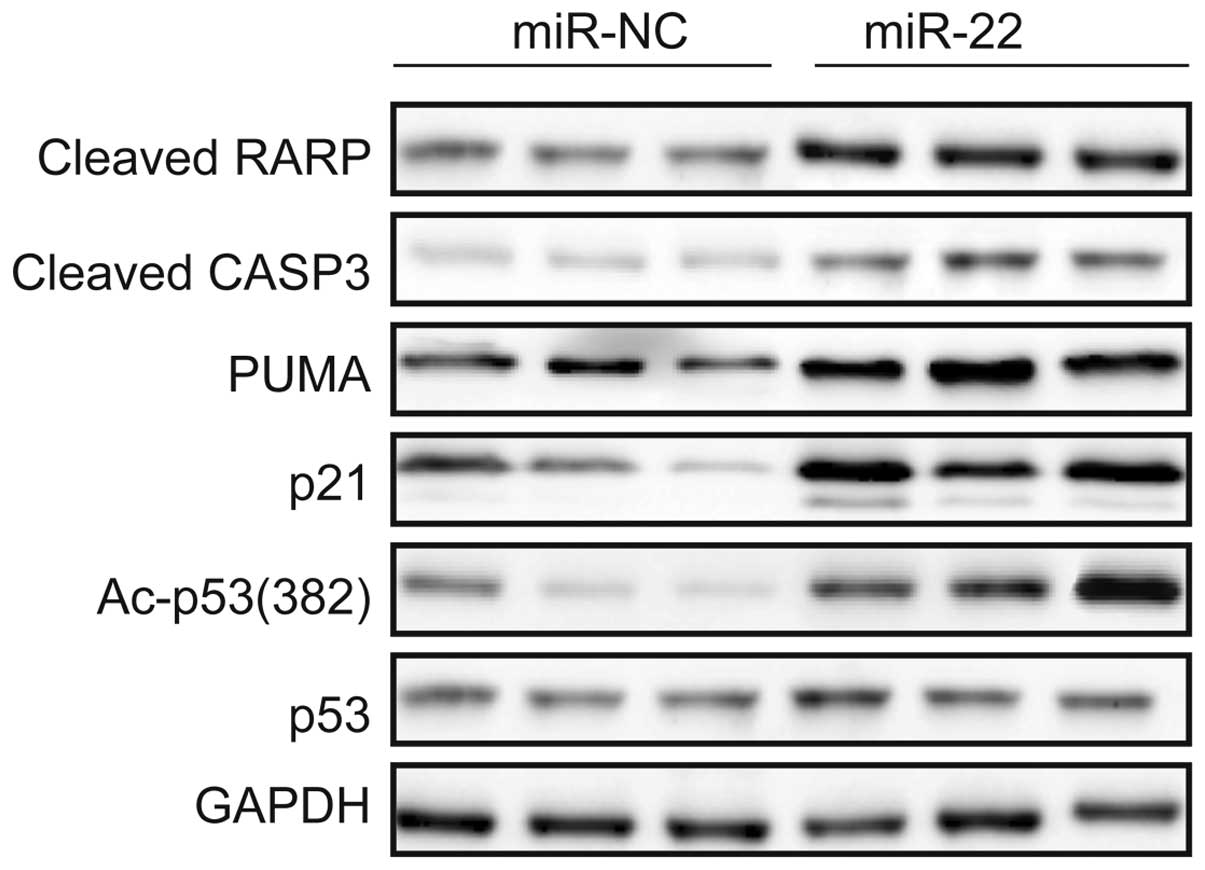
C) and increases acetylated p53 (Fig. 5). In addition, we detected p21 and
PUMA, both transcriptional targets of p53, and the apoptosis
markers cleaved CASP3 and PARP expression in 786-O cells
transfected with miR-22 mimic or miR-NC. The results showed that
overexpression of miR-22 in 786-O cells increased expression of
p21, PUMA, cleaved CASP3 and PARP (Fig.
5). These results suggest that miR-22 indirectly regulates p53
through SIRT1.
miR-22 inhibits tumor growth of RCC in
vivo
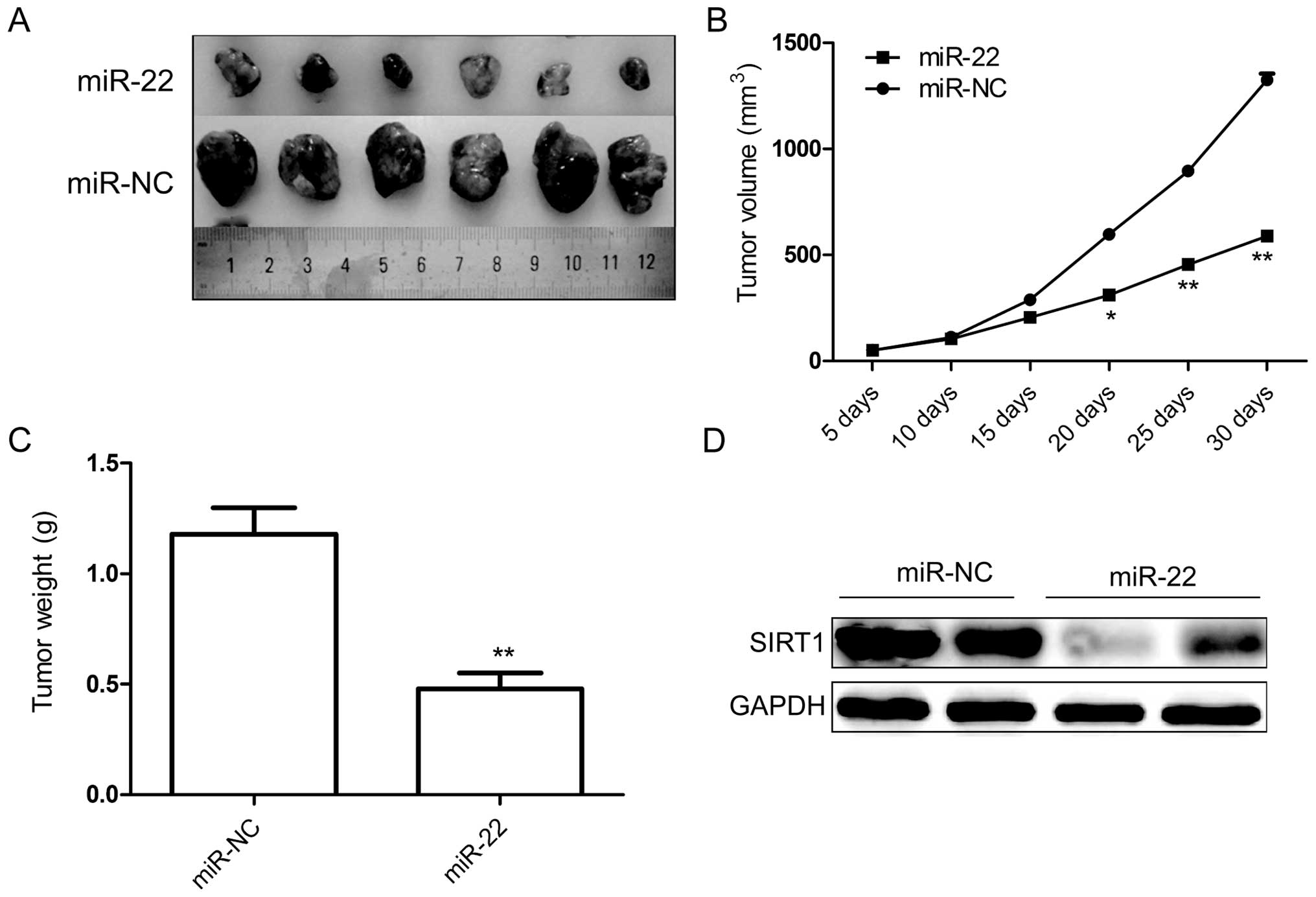
Finally, we tested whether ectopic expression of
miR-22 could influence the growth of breast tumors in vivo.
Stably expressing 786-O cells either with miR-22 or miR-NC were
injected subcutaneously into nude mice, and the tumor formation was
monitored. Cells transfected with miR-NC showed progressive growth,
while cells transfected with miR-22 mimic retarded tumor growth
compared to miR-NC group (Fig. 6A and
B). Thirty days after injection, the nude mice were sacrificed,
and the tumors weighed. Our results showed that in miR-22
overexpressing group the weights were significantly lower than in
the miR-NC group (Fig. 6C). In
addition, the result of western blot analysis showed that SIRT1
protein expression was decreased in miR-22 group compared to the
miR-NC group (Fig. 6D). These
findings suggested that miR-22 suppressed renal cancer
tumorigenicity in vivo by targeting SIRT1.
Discussion
miRNAs, as a novel class of small, single-stranded,
non-coding RNA regulatory molecules, have been demonstrated to play
critical roles as either oncogenes or tumor suppressors in various
human cancers including renal cancer (21). For example, miR-1 functions as a
tumor suppressor and inhibits RCC progression by targeting CDK4,
CDK6, Caprin1 and Slug (22).
MicroRNA-429 acts as a tumor suppressor, and inhibits cell
proliferation, epithelial-mesenchymal transition, and metastasis by
direct targeting of BMI1 and E2F3 in renal cell carcinoma (23). MicroRNA-21 (miR-21) functions as an
oncogene, and downregulates tumor suppressor PDCD4 and promotes
cell transformation, proliferation, and metastasis in renal cell
carcinoma (24). The present study
first showed that miR-22 could suppress RCC tumor growth in
vitro and in vivo by targeting SIRT1, suggesting that
miR-22 functions as a tumor suppressor in renal cell carcinoma.
miR-22, located at a fragile cancer-relevant genomic
region in chromosome 17 (17p13.3), is mapped to an exon of the
C17orf91 gene (25). microRNA-22
(miR-22) has been shown to be upregulated in prostate cancer
(26), and downregulated in breast
cancer (12), osteosarcoma
(13), esophageal squamous cell
carcinoma (14), gastric cancer
(15) and colorectal cancer
(16). Recent studies have shown
that miR-22 is involved in various cellular processes related to
carcinogenesis, including cell growth, apoptosis, motility, and the
cell cycle (17). For example,
Xiong et al (27) reported
that miR-22 was also downregulated in breast cancer, and it
suppressed breast cancer development through directly targeting
estrogen receptor α (ERα) and downstream signaling (12). Guo et al reported that miR-22
inhibits osteosarcoma cell proliferation and migration by targeting
HMGB1 and inhibiting HMGB1-mediated autophagy (13). Yang et al demonstrated that
miR-22 expression is decreased in human ESCC tissues and cell lines
compared with matching adjacent normal tissues and cell lines and
restoration of miR-22 in Eca109 and Kyse410 cells significantly
inhibited cellular migration and invasion (14). However, miR-22 expression was found
to be upregulated in prostate cancer, and its upregulation
increased phosphatidylinositol 3-kinase-Akt (PI3K/AKT) signal
pathway activation (28). These
controversial results of miR-22 in cancer development suggested
miR-22 play different roles in different types of cancers. Here, we
demonstrated that miR-22 expression is decreased in human RCC
tissues and cell lines compared with matching adjacent normal
tissues and cell lines, which is consistent with findings from a
previous study (18). Restoration
of miR-22 in 786-O cells inhibited proliferation, migration and
invasion, and induced cell apoptosis in vitro, and
suppressed tumor growth in vivo by targeting SIRT1. These
results suggest that miR-22 as a tumor suppressor plays a role in
the metastasis and progression of RCC.
SIRT1, a class III histone deacetylase, is highly
conserved from bacteria to humans and is homologous to silent
information regulator 2 (Sir2) in mammals (28). SIRT1 involvement has been shown in
regulating various physiological processes, including gene
transcription, energy metabolism, cell senescence, glucose
metabolism, lipid metabolism, and insulin secretion (29). SIRT1 overexpression is found in
prostate (30), ovarian (31), renal (32) colorectal (33) and liver cancers (34), suggesting that SIRT1 may play a role
in tumorigenesis. It has been demonstrated that SIRT1 plays an
important role in the longevity and cellular senescence of most
organisms through directly modulating the p53 signal pathway
(35). In addition, recent studies
have suggested that SIRT1 expression in cancer cells is regulated
by miR-34a (34), miR-126 (36), miR-217 (37), miR-449 (38) and miR-494 (39), and miRNAs regulating SIRT1 are
involved in many cellular pathways, including cancer cell
proliferation, cycle distribution, migration and invasion (35–39).
Here, we confirmed that SIRT1 is a target of miR-22 by luciferase
assay, and that upregulation of miR-22 decreased the expression of
SIRT1 on mRNA and protein levels. Overexpression of miR-22 actived
the p53 signal pathway and its downstream protein. These finding
may suggest that miR-22 inhibited RCC growth and metastasis by
targeting SIRT1.
In summary, the results presented here demonstrate
that miR-22 expression level was downregulated in RCC tissue and
cell lines, and its expression level was significantly associated
with histological grade, tumor stage and lymph node metastasis.
miR-22 functions as a tumor suppressor and suppresses RCC tumor
growth in vitro and in vivo by targeting SIRT1.
Moreover, miR-22 activated the p53 signal pathway and its
downstream proteins, and was able to inhibit ETM contributing to
inhibition of tumor growth and metastasis. These results suggested
that miR-22 may be a novel tumor suppressor that blocks the growth
of RCC through p53 signaling pathways by targeting SIRT1.
Therefore, miR-22 may be a therapeutic target for the treatment of
RCC.
References
|
1
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
DeSantis CE, Lin CC, Mariotto AB, Siegel
RL, Stein KD, Kramer JL, Alteri R, Robbins AS and Jemal A: Cancer
treatment and survivorship statistics, 2014. CA Cancer J Clin.
64:252–271. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chow WH, Dong LM and Devesa SS:
Epidemiology and risk factors for kidney cancer. Nat Rev Urol.
7:245–257. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pantuck AJ, Zisman A and Belldegrun AS:
The changing natural history of renal cell carcinoma. J Urol.
166:1611–1623. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
DiBiase SJ, Valicenti RK, Schultz D, Xie
Y, Gomella LG and Corn BW: Palliative irradiation for focally
symptomatic metastatic renal cell carcinoma: Support for dose
escalation based on a biological model. J Urol. 158:746–749. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fabian MR, Sonenberg N and Filipowicz W:
Regulation of mRNA translation and stability by microRNAs. Annu Rev
Biochem. 79:351–379. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Guo H, Ingolia NT, Weissman JS and Bartel
DP: Mammalian microRNAs predominantly act to decrease target mRNA
levels. Nature. 466:835–840. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
McManus MT: MicroRNAs and cancer. Semin
Cancer Biol. 13:253–258. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Farazi TA, Spitzer JI, Morozov P and
Tuschl T: miRNAs in human cancer. J Pathol. 223:102–115. 2011.
View Article : Google Scholar :
|
|
11
|
Garzon R and Marcucci G: Potential of
microRNAs for cancer diagnostics, prognostication and therapy. Curr
Opin Oncol. 24:655–659. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kong LM, Liao CG, Zhang Y, Xu J, Li Y,
Huang W, Zhang Y, Bian H and Chen ZN: A regulatory loop involving
miR-22, Sp1, and c-Myc modulates CD147 expression in breast cancer
invasion and metastasis. Cancer Res. 74:3764–3778. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Guo S, Bai R, Liu W, Zhao A, Zhao Z, Wang
Y, Wang Y, Zhao W and Wang W: miR-22 inhibits osteosarcoma cell
proliferation and migration by targeting HMGB1 and inhibiting
HMGB1-mediated autophagy. Tumour Biol. 35:7025–7034. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yang C, Ning S, Li Z, Qin X and Xu W:
miR-22 is down-regulated in esophageal squamous cell carcinoma and
inhibits cell migration and invasion. Cancer Cell Int. 14:1382014.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang X, Yu H, Lu X, Zhang P, Wang M and Hu
Y: miR-22 suppresses the proliferation and invasion of gastric
cancer cells by inhibiting CD151. Biochem Biophys Res Commun.
445:175–179. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang G, Xia S, Tian H, Liu Z and Zhou T:
Clinical significance of miR-22 expression in patients with
colorectal cancer. Med Oncol. 29:3108–3112. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Song SJ and Pandolfi PP: miR-22 in
tumorigenesis. Cell Cycle. 13:11–12. 2014. View Article : Google Scholar :
|
|
18
|
He H, Wang L, Zhou W, Zhang Z, Wang L, Xu
S, Wang D, Dong J, Tang C, Tang H, et al: MicroRNA expression
profiling in clear cell renal cell carcinoma: Identification and
functional validation of key miRNAs. PLoS One. 10:e01256722015.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Vaziri H, Dessain SK, Ng Eaton E, Imai SI,
Frye RA, Pandita TK, Guarente L and Weinberg RA: hSIR2(SIRT1)
functions as an NAD-dependent p53 deacetylase. Cell. 107:149–159.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Langley E, Pearson M, Faretta M, Bauer UM,
Frye RA, Minucci S, Pelicci PG and Kouzarides T: Human SIR2
deacetylates p53 and antagonizes PML/p53-induced cellular
senescence. EMBO J. 21:2383–2396. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li JY, Yong TY, Michael MZ and Gleadle JM:
Review: The role of microRNAs in kidney disease. Nephrology
(Carlton). 15:599–608. 2010. View Article : Google Scholar
|
|
22
|
Xiao H, Zeng J, Li H, Chen K, Yu G, Hu J,
Tang K, Zhou H, Huang Q, Li A, et al: miR-1 downregulation
correlates with poor survival in clear cell renal cell carcinoma
where it interferes with cell cycle regulation and metastasis.
Oncotarget. 6:13201–13215. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Qiu M, Liang Z, Chen L, Tan G, Wang K, Liu
L, Liu J and Chen H: MicroRNA-429 suppresses cell proliferation,
epithelial-mesenchymal transition, and metastasis by direct
targeting of BMI1 and E2F3 in renal cell carcinoma. Urol Oncol.
33:332.e9–e18. 2015. View Article : Google Scholar
|
|
24
|
Li X, Xin S, He Z, Che X, Wang J, Xiao X,
Chen J and Song X: MicroRNA-21 (miR-21) post-transcriptionally
downregulates tumor suppressor PDCD4 and promotes cell
transformation, proliferation, and metastasis in renal cell
carcinoma. Cell Physiol Biochem. 33:1631–1642. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gurha P, Abreu-Goodger C, Wang T, Ramirez
MO, Drumond AL, van Dongen S, Chen Y, Bartonicek N, Enright AJ, Lee
B, et al: Targeted deletion of microRNA-22 promotes stress-induced
cardiac dilation and contractile dysfunction. Circulation.
125:2751–2761. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pasqualini L, Bu H, Puhr M, Narisu N,
Rainer J, Schlick B, Schäfer G, Angelova M, Trajanoski Z, Börno ST,
et al: miR-22 and miR-29a are members of the androgen receptor
cistrome modu lating LAMC1 and Mcl-1 in prostate cancer. Mol
Endocrinol. 29:1037–1054. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xiong J1, Yu D, Wei N, Fu H, Cai T, Huang
Y, Wu C, Zheng X, Du Q, Lin D and Liang Z: An estrogen receptor
alpha suppressor, microRNA-22, is downregulated in estrogen
receptor alpha-positive human breast cancer cell lines and clinical
samples. FEBS J. 277:1684–1694. PubMed/NCBI
|
|
28
|
Poliseno L, Salmena L, Riccardi L, Fornari
A, Song MS, Hobbs RM, Sportoletti P, Varmeh S, Egia A, Fedele G, et
al: Identification of the miR-106b~25 microRNA cluster as a
proto-oncogenic PTEN-targeting intron that cooperates with its host
gene MCM7 in transformation. Sci Signal. 3:ra292010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Dong YJ, Liu N, Xiao Z, Sun T, Wu SH, Sun
WX, Xu ZG and Yuan H: Renal protective effect of sirtuin 1. J
Diabetes Res. 2014:8437862014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Huffman DM, Grizzle WE, Bamman MM, Kim JS,
Eltoum IA, Elgavish A and Nagy TR: SIRT1 is significantly elevated
in mouse and human prostate cancer. Cancer Res. 67:6612–6618. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Jang KY, Kim KS, Hwang SH, Kwon KS, Kim
KR, Park HS, Park BH, Chung MJ, Kang MJ, Lee DG, et al: Expression
and prognostic significance of SIRT1 in ovarian epithelial tumours.
Pathology. 41:366–371. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jung YJ, Lee JE, Lee AS, Kang KP, Lee S,
Park SK, Lee SY, Han MK, Kim DH and Kim W: SIRT1 overexpression
decreases cisplatin-induced acetylation of NF-κB p65 subunit and
cytotoxicity in renal proximal tubule cells. Biochem Biophys Res
Commun. 419:206–210. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Stünkel W, Peh BK, Tan YC, Nayagam VM,
Wang X, Salto-Tellez M, Ni B, Entzeroth M and Wood J: Function of
the SIRT1 protein deacetylase in cancer. Biotechnol J. 2:1360–1368.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chen J, Zhang B, Wong N, Lo AW, To KF,
Chan AW, Ng MH, Ho CY, Cheng SH, Lai PB, et al: Sirtuin 1 is
upregulated in a subset of hepatocellular carcinomas where it is
essential for telomere maintenance and tumor cell growth. Cancer
Res. 71:4138–4149. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yamakuchi M, Ferlito M and Lowenstein CJ:
miR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci
USA. 105:13421–13426. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xu JQ, Liu P, Si MJ and Ding XY:
MicroRNA-126 inhibits osteosarcoma cells proliferation by targeting
Sirt1. Tumour Biol. 34:3871–3877. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Deng S, Zhu S, Wang B, Li X, Liu Y, Qin Q,
Gong Q, Niu Y, Xiang C, Chen J, et al: Chronic pancreatitis and
pancreatic cancer demonstrate active epithelial-mesenchymal
transition profile, regulated by miR-217-SIRT1 pathway. Cancer
Lett. 355:184–191. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhang H, Feng Z, Huang R, Xia Z, Xiang G
and Zhang J: MicroRNA-449 suppresses proliferation of hepatoma cell
lines through blockade lipid metabolic pathway related to SIRT1.
Int J Oncol. 45:2143–2152. 2014.PubMed/NCBI
|
|
39
|
Liu Y, Li X, Zhu S, Zhang JG, Yang M, Qin
Q, Deng SC, Wang B, Tian K, Liu L, et al: Ectopic expression of
miR-494 inhibited the proliferation, invasion and chemoresistance
of pancreatic cancer by regulating SIRT1 and c-Myc. Gene Ther. Apr
28–2015.Epub ahead of print. View Article : Google Scholar
|