Introduction
Breast cancer is the most common cancer among women
in many countries. Surgery, chemotherapy, radiotherapy, endocrine
therapy and targeted therapy significantly improve the disease-free
survival and overall survival of patients with breast cancer
(1). Molecular targeted treatment
bring breast cancer treatment into the molecular therapy era, with
the classification of breast cancer molecular subtypes and the
detection of genetic mutations (2).
However, local recurrence and distant metastasis of breast cancer
are the most important factors in breast cancer treatment failure.
The mechanisms of breast cancer metastasis and the molecular
mechanisms of this progression are still not understood. Therefore,
it is necessary to uncover new molecular targets in breast
cancer.
Extracellular sulfatases, especially heparan
endosulfatases (Sulfs), have attracted the attention of cancer
researchers because of accumulating evidence that they may play
important roles in cancer progression by modifying the sulfate
patterns of HSPGs located on the surface of most animal cells.
HSPGs can be released into the extracellular matrix and detected in
serum (3–6). HSPGs carry out many structural and
signaling functions through their ability to bind to diverse
protein ligands, including growth factors such as vascular
endothelial growth factor (VEGF), fibroblast growth factor-1
(FGF-1) and stromal cell-derived factor 1 (SDF-1) (7,8). The
progression of breast cancer growth and metastasis involves many
factors, including matrix metalloproteinases, adhesion molecules
and growth factors. Khurana et al (9) showed that Sulfs were involved in tumor
invasion and metastasis by removing the N-3-O and 6-O sulfate amino
glucose sulfuric acid from extracellular matrix HSPGs, thereby
forming a 'common receptor'. We hypothesized Sulf2 promoted breast
cancer progression and would possibly be a new target in breast
cancer treatment.
The Sulfs family includes Sulf1 and Sulf2, two
structurally similar, endogenous sulfatases with different
functions. They have highly conserved heparin-binding domains with
64% homology (10). At present,
most studies have shown that Sulf1 is a tumor suppressor protein,
but the role of Sulf2 in cancer progression is not uniform
(10–12). Sulf2 is dysregulated in many
cancers. It upregulates and promotes tumorigenesis in human
hepatocellular (13), pancreatic
(14), breast (15) and non-small cell lung carcinoma
(16). Based on these studies,
Sulf2 is considered a bona fide candidate cancer-causing agent in
multiple cancer types. It could therefore be a therapeutic target
for the treatment of non-small cell lung cancer (NSCLC) and other
cancers (17), but other studies
have shown conflicting results. Peterson et al (12) reported that Sulf2 overexpression in
MDA-MB-231 cells inhibited breast cancer cell proliferation,
invasion and metastasis in vitro, and the results were
further confirmed in vivo. In the present study, we
investigated the role of Sulf2 in breast cancer progression. We
detected endogenous expression of Sulf2 in several breast cancer
cell lines using western blot analysis and selected two human
breast cancer cell lines MCF-7 and MDA-MB-231, which respectively
express Sulf2 or not, to further study the role of Sulf2 in breast
cancer. Two breast cancer cell lines were created for Sulf2
function studies with stable knockdown or overexpression of Sulf2
(MCF-7 shSulf2 and MDA-MB-231 Sulf2, respectively).
Materials and methods
Cell lines
Human breast cancer cell lines (MCF-7, MDA-MB-231,
MDA-MB-468 and BT-549) and mammary epithelial cell line HBL-100
were obtained from the Cell Bank of the Chinese Academy of Sciences
(Shanghai, China). The HEK293T cells used for lentivirus packaging
were stocked in our own laboratory. Cells were cultured in
Dulbecco's modified Eagle's medium (DMEM; HyClone Laboratories,
Logan, UT, USA) with 10% fetal bovine serum (FBS; Gibco, Grand
Island, NY, USA) and penicillin-streptomycin (Gibco) at 37°C and 5%
CO2.
Sulf2 shRNA and overexpression vector
constructs
To construct the short-hairpin Sulf2 vector, shSulf2
target sequences were determined using siRNA scales (http://gesteland.genetics.utah.edu/siRNA_scales/)
and amplified using the primer pair miR30 XhoI
(5′-CAGAAGGCTCGAGAAG GTATATTGCTGTTGACAGTGAGCG-3′) and miR30
EcoRI (5′-CTAAAGTAGCCCCTTGAATTCCGAGGCAGTAGG CA-3′) to
introduce the miR-30 sequence and XhoI and EcoRI (MBI
Fermentas, Vilnius, Lithuania) restriction sites flanking the
shSulf2 sequence. The amplified sequences were inserted into MLP
vectors (Transomics, Shanghai, China) following XhoI and
EcoRI digestion of both PCR fragments and MLP vectors. The
newly constructed vectors were named shSulf2-1 to 4 (Table I). The silencing efficiency of
shSulf2-1 to 4 was measured in MCF-7 breast cancer cells using
western blot analysis.
 | Table ISulf2 oligo sequences. |
Table I
Sulf2 oligo sequences.
| No. | Sulf2 siRNA
target | Sulf2 oligo
sequence |
|---|
| ShRNA-1 |
GGAAGTATCTTAATGAATA |
5′-TGCTGTTGACAGTGAGCGCCGGGAAGTATCTTAATGAATA
TAGTGAAGCCACA GATGTATATTCATTAAGATACTTCCCG
ATGCCTACTGCCTCGGA-3′ |
| ShRNA-2 |
GCGCCAACAATAACACGTA |
5′-TGCTGTTGACAGTGAGCGACAGCGCCAACAATAACACGTA
TAGTGAAGCCAC AGATGTATACGTGTTATTGTTGGCGCTG
GTGCCTACTGCCTCGGA-3′ |
| ShRNA-3 |
CCTTTGACATTTTGTAAAA |
5′-TGCTGTTGACAGTGAGCGAAACCTTTGACATTTTGTAAAA
TAGTGAAGCCACA GATGTATTTTACAAAATGTCAAAGGTT
CTGCCTACTGCCTCGGA-3′ |
| ShRNA-4 |
TGAAGCTGCATAAGTGCAA |
5′-TGCTGTTGACAGTGAGCGCGCTGAAGCTGCATAAGTGCAA
TAGTGAAGCCAC AGATGTATTGCACTTATGCAGCTTCAGC
TTGCCTACTGCCTCGGA-3′ |
To generate the Sulf2 overexpression construct,
Sulf2 cDNA was amplified from the cDNA of MCF-7 cells using the
forward primer 5′-CTAGCTAGCAAAAAAGAAGATG GGCCCCC-3′ and reverse
primer 5′-CGGGATCCTTAACC TTCCCAGCCTTCCC-3′. The amplified fragment
was cloned into the pCDH vector (System Biosciences, Mountain View,
CA, USA) to form the pCDH-Sulf2 overexpression vector construct.
The sequences of these two positive clones were identified using
enzyme digestion and gene sequencing detection (Shanghai Meiji,
Shanghai, China).
Lentivirus was packaged in HEK293T cells using
Lipofectamine 2000 (Life Technologies, Carlsbad, CA, USA), and
transfection was performed following the manufacturer's
instructions. The MCF-7 cell lines transfected with blank MLP
vectors and shSulf2-1 were named as MCF-7 NC and MCF shSulf2. The
MDA-MB-231 cell lines transfected with blank pCDH vector and
pCDH-Sulf2 were name as MDA-MB-231 vector and MDA-MB-231 Sulf2.
qRT-PCR analysis
Total RNA was isolated from cells using TRIzol
reagent (Invitrogen, Madison, WI, USA) and reverse transcribed into
cDNA using SuperScript III Reverse Transcriptase (Invitrogen). The
mRNA level was determined by 7900HT qRT-PCR (Applied Biosystems,
Foster City, CA, USA) using SYBR® Green Real-time PCR
Master Mix (Takara, Shiga, Japan). Primers for qRT-PCR are listed
in Table II.
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an
internal control. Relative mRNA levels were calculated using the
ΔΔCt method.
 | Table IIReal-time PCR primers. |
Table II
Real-time PCR primers.
| Gene | Primer
sequences | Length (bp) |
|---|
| GAPDH
(control) | Forward:
5′-GGGAAACTGTGGCGTGAT-3′ | |
| Reverse:
5′-GAGTGGGTGTCGCTGTTGA-3′ | 299 |
| Sulf2 | Forward:
5′-GGCAGGTTTCAGAGGGACC-3′ | |
| Reverse:
5′-GAAGGCGTTGATGAAGTGCG-3′ | 207 |
| CD82 | Forward:
5′-AGCAGAACCCGCAGAGTCC-3′ | |
| Reverse:
5′-GCTTCCTTCCACGAAACCA-3′ | 101 |
| FGFR4 | Forward:
5′-GTTCTGCTCGGCTTCTTGG-3′ | |
| Reverse:
5′-ACGCCATTTGCTCCTGTTT-3′ | 237 |
| IGF1 | Forward:
5′-TGGTGGATGCTCTTCAGTTCG-3′ | |
| Reverse:
5′-GCACTCCCTCTACTTGCGTTCT-3′ | 265 |
| NR4A3 | Forward:
5′-GCTCGGAATACACCACGGA-3′ | |
| Reverse:
5′-GAAGGCTTGAGTTCGTAGTTGC-3′ | 154 |
| FIGF | Forward:
5′-GCTAAGGAGTCCCTGGTTCA-3′ | |
| Reverse:
5′-ATTCACAAGAGTTGCGATTAGC-3′ | 235 |
| MAPKAPK3 | Forward:
5′-AGCAGGATTCAGGAGCGTG-3′ | |
| Reverse:
5′-CCTTAGCAAAGCCAAAATCG-3′ | 196 |
| PDGFC | Forward:
5′-TGTGGAAACTACCCTGCGATT-3′ | |
| Reverse:
5′-GCCCGAAGAGGCTCATTTG-3′ | 165 |
| PGF | Forward:
5′-GCCCTGCTACCTGTTCTTGG-3′ | |
| Reverse:
5′-AAGCAAATGGCAAAGTGTGAG-3′ | 266 |
| SH2D2A | Forward:
5′-AAGGCTGTGGGTAAGGCGA-3′ | |
| Reverse:
5′-GAAGGTGCTGAAGGTTGGGA-3′ | 206 |
Western blot analysis
Cell lysate was prepared as previously described and
equal amount of protein was used in western blot analysis (18). Cells were harvested in RIPA lysis
buffer (Beyotime Institute of Biotechnology, Shanghai, China),
resolved in SDS-PAGE, transferred to polyvinylidene fluoride (PVDF)
membrane and probed with antibodies: mouse anti-Sulf2 (2B4; Novus
Biologicals, Littleton, CO, USA), rabbit anti-GAPDH (Sigma, St.
Louis, MO, USA); rabbit anti-FIGF (PAB4879; Abnova, Atlanta, GA,
USA), rabbit anti-PGF (PAB8004; Abnova), rabbit anti-CD82
(ab109529; Abnova), rabbit anti-NR43A (ab92777; Abnova), anti-mouse
HRP (Sigma) and anti-rabbit HRP (Sigma).
Cell proliferation assay
Four cell lines (MCF-7 NC, MCF-7 shSulf2, MDA-MB-231
vector and MDA-MB-231 Sulf2) were dissociated from cell flasks by
trypsin (Sigma) digestion and seeded into 24-well cell culture
plates (1×105 cells/well). Cells were dissociated from
wells with 0.25% trypsin and counted each day using a Bio-Rad cell
counter (Bio-Rad Laboratories, Hercules, CA, USA). Cell growth
curves were drawn from live cell numbers for seven days.
Apoptosis assay
Apoptosis was determined by dual staining using
Annexin V:FITC and propidium iodide (Invitrogen). Briefly, log
phase cells of these four cell lines were seeded into 24-well cell
culture plates (1×105 cells/well) and treated with 10
µg/ml cisplatin (Qilu Pharmaceutical Co., Shanghai, China)
for 24 h. Cells were dissociated from wells with 0.25% trypsin,
spun at 1,500 rpm for 5 min, resuspended in Annexin V binding
buffer, and stained with 1 µl Annexin V:FITC for 15 min and
1 µl propidium iodide for 1 min. Cells were analyzed using
the FACSCalibur System (BD Biosciences, San Jose, CA, USA). The
relative proportion of Annexin V-positive cells, representing
apoptotic cells, was determined using FlowJo software (FlowJo LLC,
Ashland, OR, USA).
Cell cycle assay
The cell cycle was determined by propidium iodide
staining and flow cytometric analysis. Log phase cells of these
four cell lines were seeded and harvested as described above. They
were then fixed in 70% ethanol overnight at −20°C and incubated in
RNase A at 37°C for 30 min. Propidium iodide was added and cells
were incubated in the dark for 30 min. Flow cytometry was used to
detect the cell cycle status. The proliferation index (PI) was
calculated using the formula: PI = (S+G2)/(S+G1+G2) × 100%.
Cell invasion assay
The migration of the four breast cancer cell lines
was measured in 12-well Boyden chamber plates with 8 µm pore
size polycarbonate membrane filter inserts (CoStar Group, Inc.,
Washington, DC, USA). For the cell invasion assay, the interior of
the Transwell insert was coated with diluted Matrigel (BD
Biosciences), which imitates the basement membrane. A total of
1×105 cells were seeded onto the upper chamber, the cell
suspension was also seeded onto the membrane in the upper chamber,
and the lower chamber was filled with 1 ml 10% FBS-DMEM. After
being incubated for 48 h, the non-migrating cells in the upper
chamber surface were removed by cotton swabs. The migrating cells
at the bottom of the membrane were fixed with formaldehyde for 1
min and stained with crystal violet. The stained membranes were cut
and placed onto a glass slide, and the numbers of invading cells at
the bottom surface of the membrane were counted three times under a
bright field light microscope.
Cell mobility assay
Four breast cancer cell lines were cultured in
6-well plates and incubated until 90% confluent. A scratch was made
using a sterile blade. Cell debris was then washed off, and the
cells continued to be cultured in low FBS medium for 18 h for the
MDA-MB-231 cells and 36 h for the MCF-7 cells. Then, the movement
of the breast cancer cells into the scratch area was monitored
under an inverted microscope to evaluate cell mobility in
vitro.
Cell adhesion assay
Four breast cancer cell lines were cultured in 10%
FBS-DMEM and seeded into 96-well plates with 30 µl/well
collagen I solution at 4°C for 12 h. After depriving cells of serum
for 8 h, cells were washed twice with DMEM and resuspended in DMEM
with 0.1% BSA to 2×105 cells/ml. Next, 100 µl
cell suspension was added into each collagen I-coated well at 37°C
for 20 min. Non-adherent cells were washed off and incubated at
37°C for 4 h. Then, 10 µl MTT and 100 µl DMSO were
added into each well. After 2 h the absorbance value of each well
was measured at 570 nm using a spectrophotometer.
Tumor metastasis and VEGF signaling PCR
array assay
Total mRNA was isolated and reverse transcribed as
above. Genetic screening was performed using the tumor metastasis
PCR array and the Human VEGF Signaling PCR array (Qiagen, Hilden,
Germany) in MCF-7 NC and MCF-7 shSulf2 cells following the
manufacturer's instructions.
Mouse mammary pad injection
Six-week-old, 18-g female NOD/SCID mice were
purchased from Shanghai Experimental Animal Center Chinese Academy
of Sciences (Shanghai, China). Four cultured breast cancer cell
lines were digested with 0.25% trypsin and resuspended in
HBSS/Matrigel (1:1 volume) to 107 cell/ml. Xenografts
were generated by injecting 0.2 ml cell suspension into the area of
the mammary fat pad. Growth of the xenografts was measured every
week, and the volumes of the xenografts were calculated using the
formula: volume = length × width × height × π/6. All experimental
protocols followed the instructions of the Chinese Council on
Animal Care and were approved by the Animal Experimental Ethical
Inspection of Shanghai Ninth People's Hospital affiliated to
Shanghai Jiaotong University, School of Medicine [permit no.
(20015) 25].
Xenograft invasion assay
Excised xenografts were fixed in formalin buffer and
embedded in paraffin. Five micron sections from formalin-fixed,
paraffin-embedded tissues were de-paraffinized with xylene and
rehydrated through a graded alcohol series. Heat induced epitope
retrieval (HIER) was performed by immersing the tissue sections at
98°C for 20 min in 8 mM EDTA (pH 8.0). Slides were counterstained
with hematoxylin (Thermo Fisher Scientific, Waltham, MA, USA), in
1% ammonium hydroxide and dehydrated.
Statistical analysis
Data are presented as the mean ± standard deviation
(SD) of at least three independent experiments with three or more
replicates. Continuous data were analyzed using a two-tailed
Student's t-test. P<0.05 was considered a significant
difference.
Results
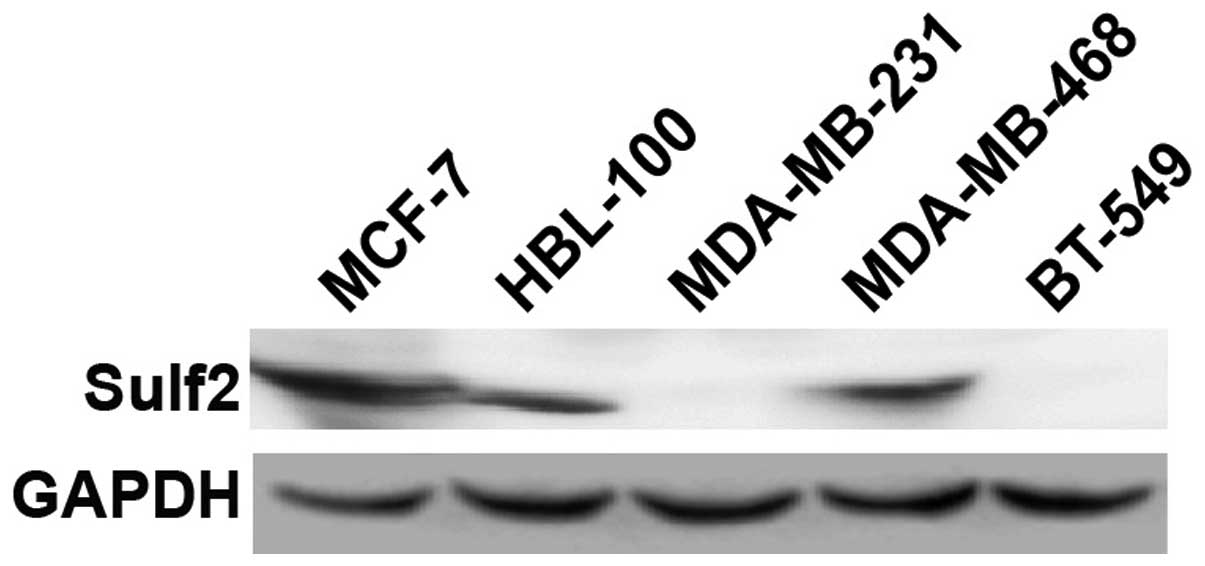
Sulf2 is differentially expressed in five
cell lines
To select breast cancer cell lines with different
Sulf2 expression levels for studying the biological function of
Sulf2, endogenous Sulf2 protein was detected using western blot
analysis in five cell lines: MCF-7, MDA-MB-468, HBL-100, MDA-MB-231
and BT-549. MCF-7, MDA-MB-468 and HBL-100 cells highly expressed
Sulf2 protein. Conversely, MDA-MB-231 and BT-549 cells did not
express Sulf2 protein (Fig. 1). The
MCF-7 and MDA-MB-231 breast cancer cell lines therefore were
selected to ascertain the role of Sulf2 in breast cancer.
Silencing efficiency of Sulf2 shRNA
vectors screened in MCF-7
To examine the efficiency of silencing constructs
shSulf2-1 to shSulf2-4, MCF-7 cells were transfected with these
constructs, and the Sulf2 protein levels were detected by western
blot analysis. Compared with the negative control (NC) transfect
with a blank vector, the Sulf2 protein expression was downregulated
in the MCF-7 shSulf2-1, shSulf2-2, shSulf2-3 and shSulf2-4 groups
by 86.23, 25.27, 28.66 and 79.79% respectively (Fig. 2). Therefore, shSulf2-1 was selected
to perform the Sulf2 silencing study.
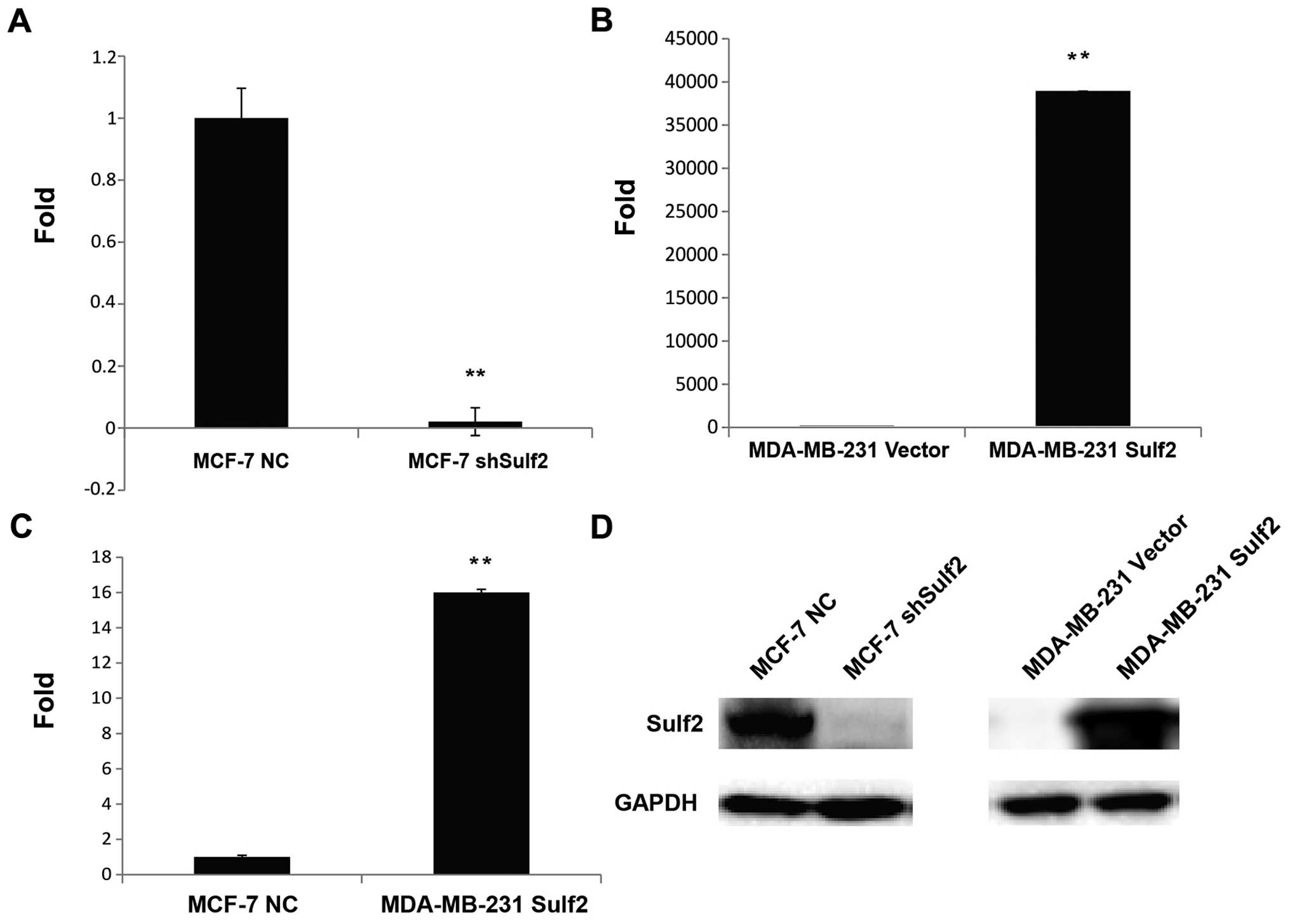
Sulf2 expression is altered in breast
cancer cell lines
The Sulf2 mRNA levels in MCF-7 NC, MCF-7 shSulf2,
MDA-MB-231 vector and MDA-MB-231 Sulf2 cells were determined by
qRT-PCR. The Sulf2 mRNA level in MCF-7 shSulf2 cells was reduced
98% compared with MCF-7 NC cells (0.02±0.045 vs. 1.00, P<0.01;
Fig. 3A). Conversely, the Sulf2
mRNA levels in MDA-MB-231 Sulf2 cells increased significantly
compared with MDA-MB-231 vector cells (38,906.96±0.061 vs. 1.00,
P<0.01; Fig. 3B). Moreover, the
Sulf2 expression in MDA-MB-231 Sulf2 cells was 16-fold higher than
that in MCF-7 NC cells (16.90±0.061 vs. 1.00, P<0.01; Fig. 3C). These data reflected significant
differences in Sulf2 mRNA levels in different breast cancer cell
lines. The changes in Sulf2 protein levels in these four cell lines
were also confirmed by western blot analysis (Fig. 3D).
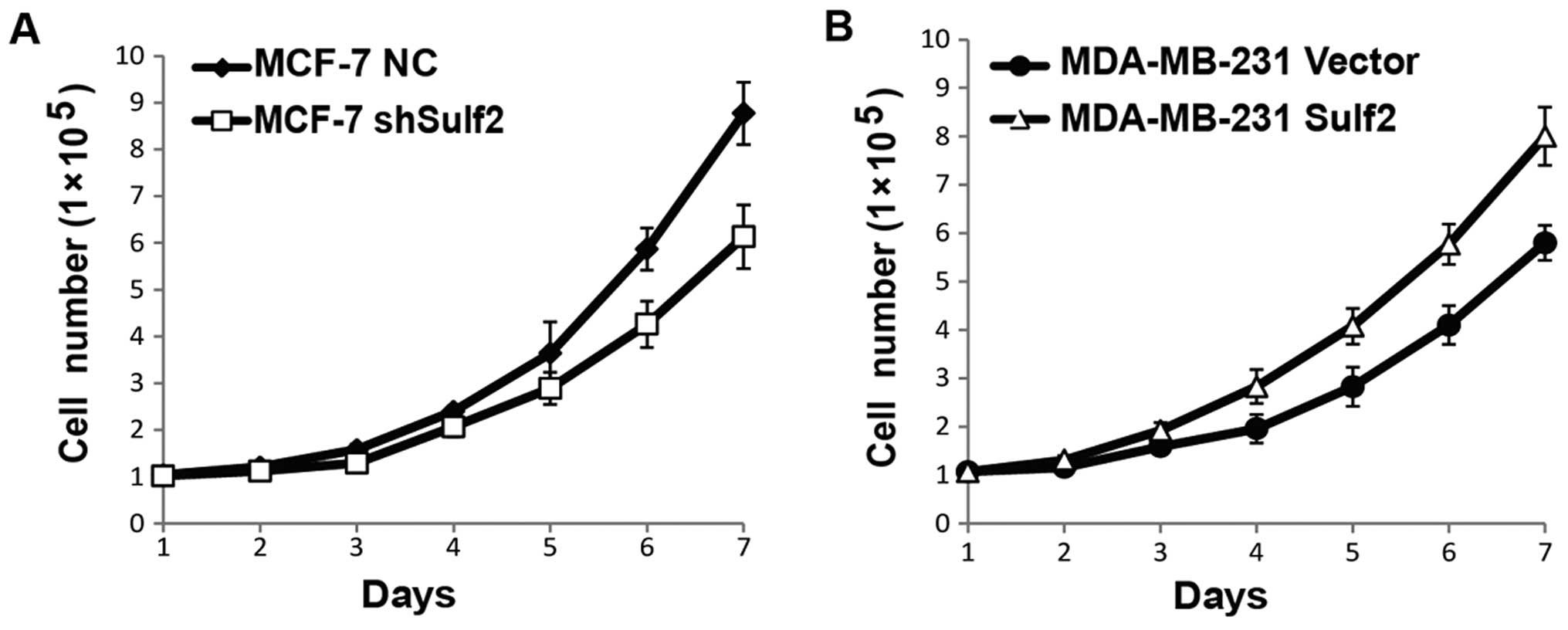
Sulf2 promotes cell proliferation
To evaluate the effects of Sulf2 on breast cancer
proliferation, cell numbers were counted every day for seven days
using the four cell lines above, and live cell numbers were used
for cell growth curves. The Sulf2 knockdown MCF-7 shSulf2 cells
showed a consistently lower growth rate than MCF-7 NC. The
difference was significant after the sixth day (P<0.05; Fig. 4A). Similarly, MDA-MB-231 Sulf2 also
showed a significantly higher growth rate than MDA-MB-231 vector.
The difference was significant after the fourth day (P<0.05;
Fig. 4B). Collectively, these data
indicated that Sulf2 promoted breast cancer proliferation.
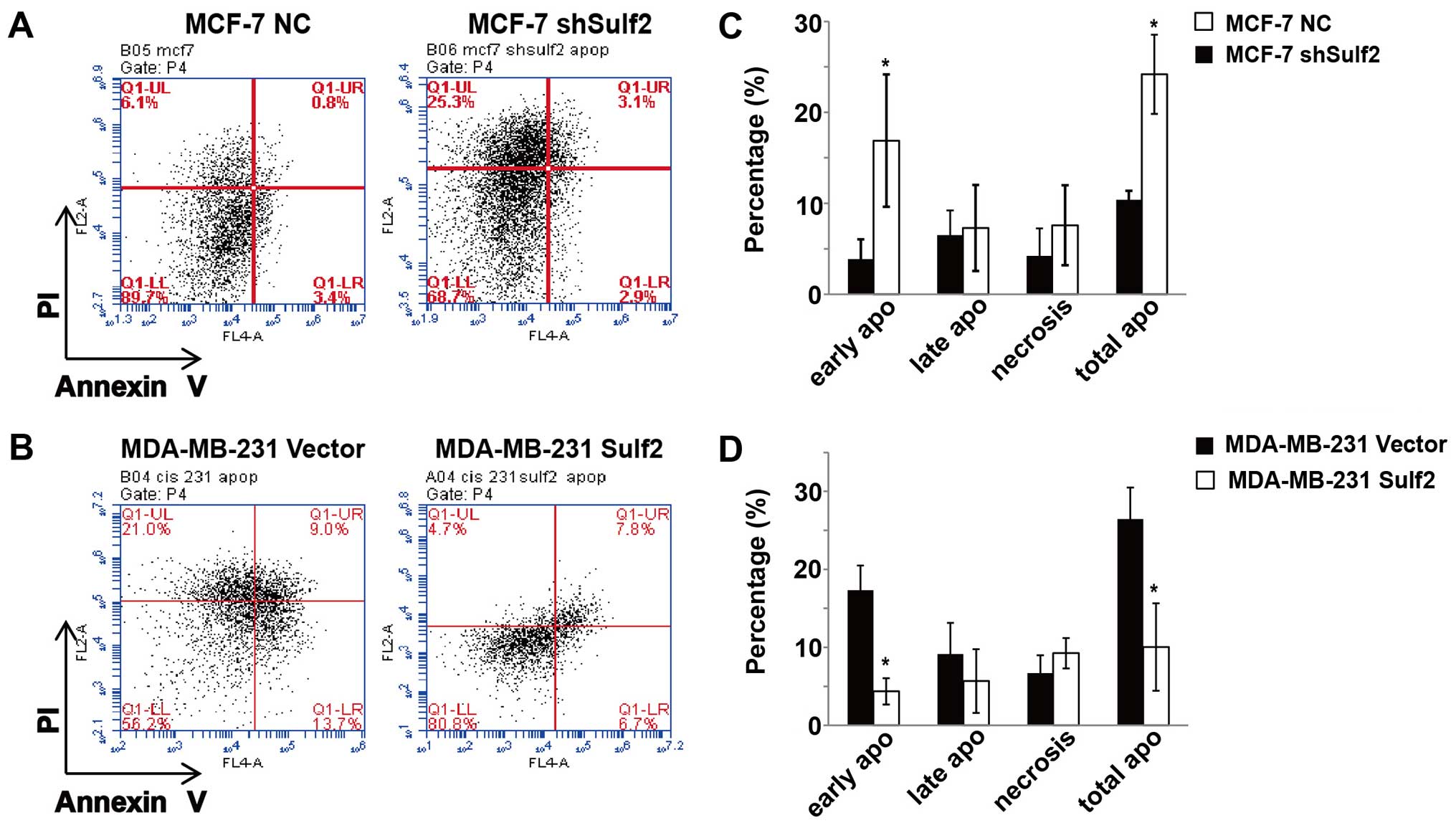
Sulf2 inhibits breast cancer
apoptosis
Previous studies show that the MCF-7 breast cancer
cell line is not sensitive to cisplatin-based chemotherapy and can
resist cisplatin induced cell apoptosis and necrosis (data not
shown). Conversely, the MDA-MB-231 breast cancer cell line is more
sensitive to cisplatin-based chemotherapy and cisplatin induced
MDA-MB-231 cell apoptosis and necrosis. Compared with MCF-7 NC, the
MCF-7 shSulf2 cell line had a significantly decreased live cell
percentage (68.2±4.76 vs. 85.4±3.89, P<0.05) and significantly
increased total apoptosis (24.19±4.35 vs. 10.39±0.99, P<0.05). A
closer look at the different stages of apoptosis showed that the
most significant difference between these two cell lines occurred
in early apoptosis (16.89±7.28 vs. 3.88±2.16, P<0.05; Fig. 5). Moreover, compared with MDA-MB-231
Sulf2, the MDA-MB-231 Sulf2 cell line showed a significant increase
in the live cell percentage (85.4±5.51 vs. 66.83±9.23, P<0.05)
and a significant decrease in total apoptosis (8.83±2.47 vs.
26.44±7.17, P<0.05), which was stronger in the early stage of
apoptosis (4.27±0.64 vs. 17.31±3.19, P<0.05; Fig. 5). Our studies showed that Sulf2
expression in breast cancer inhibited cisplatin-based breast cancer
cells apoptosis, especially at early stage.
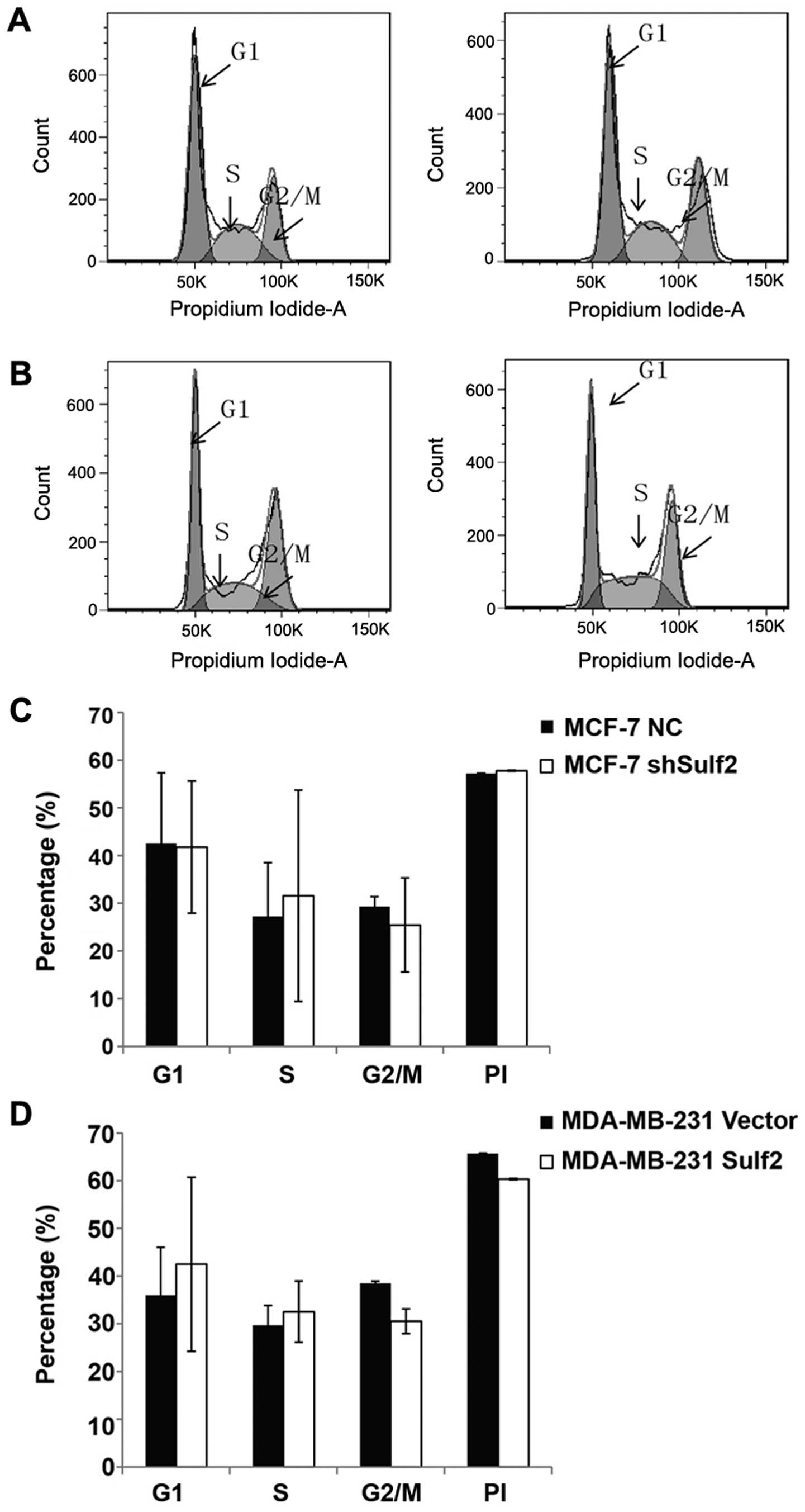
Sulf2 has no significant effect on the
cell cycle in breast cancer with cisplatin pre-treatment
Compared with MCF-7 NC, MCF-7 shSulf2 did not show
significant difference in G1 phase (41.79±13.87 vs. 42.51±14.84,
P>0.05), S phase (31.56±22.14 vs. 27.19±11.31, P<0.05), and
G2/M phase (25.43±9.85 vs. 29.28±2.09, P>0.05). The PI index
(0.58±0.13 vs. 0.57±0.14, P>0.05) also had no difference between
these cells (Fig. 6A and C).
Similarly, MDA-MB-231 Sulf2 did not show significant difference in
G1 phase (42.95±18.25 vs. 35.98±1.10, P>0.05), S phase
(32.54±6.41 vs. 29.69±4.51, P>0.05), G2/M phase (30.53±2.58 vs.
38.46±0.46, P>0.05), and the PI index (60.34±0.14 vs.
65.69%±0.08, P>0.05) (Fig. 6B and
D) when compared with MDA-MB-231 vector. These data indicated
that Sulf2 expression differences had no significant effect on the
cell cycles in breast cancer cell lines with cisplatin pretreatment
in vitro.
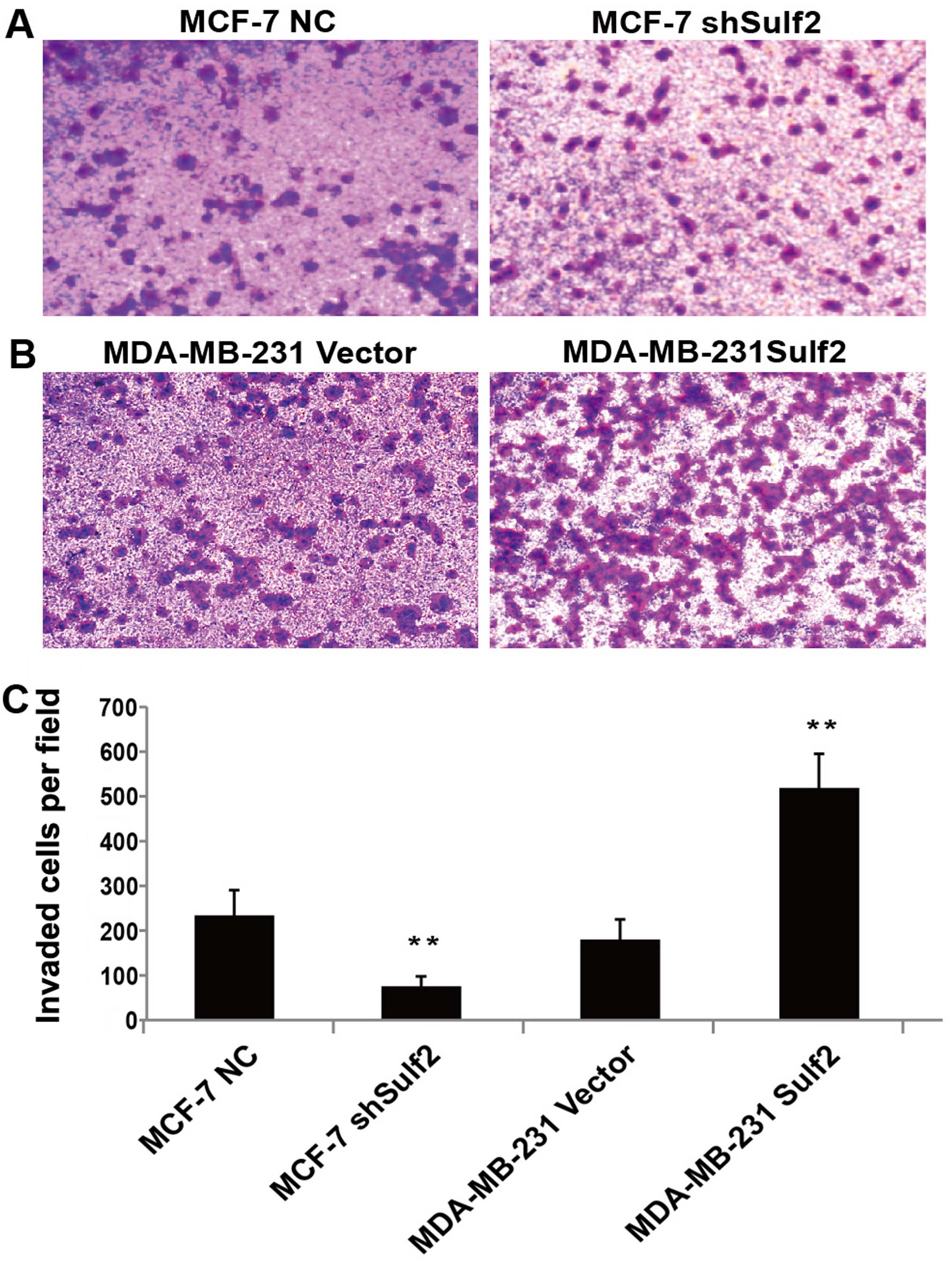
Sulf2 enhances breast cancer
invasion
Sulf2 expression was also positively associated with
higher invasion in Transwell chamber assay. Compared with MCF-7 NC,
the downregulation of Sulf2 in the MCF-7 shSulf2 cell line
significantly reduced the average number of cells that migrated
though the chamber membrane (74.50±22.15 vs. 232.83±56.76,
P<0.01; Fig. 7). The opposite
observation was made when the overexpression of Sulf2 in MDA-MB-231
Sulf2 caused a significant increase (517.50±77.02 vs. 179.67±44.81,
P<0.01; Fig. 7) in the number of
cells that migrated to the lower chamber, compared with MDA-MB-231
vector. These observations clearly suggested that Sulf2 expression
improved breast cancer invasion.
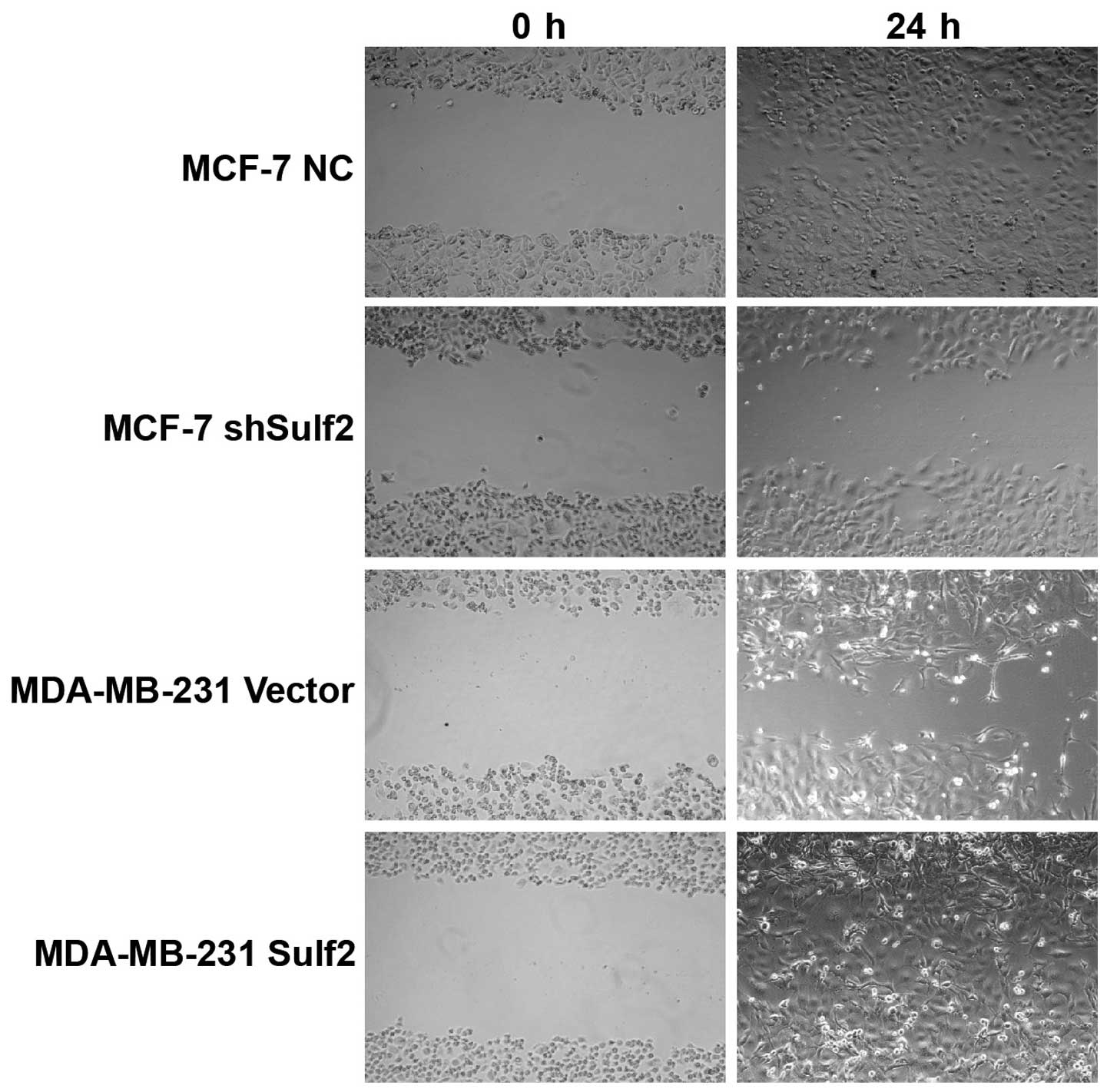
Sulf2 promotes breast cancer
migration
Higher Sulf2 expression was positively associated
with cell migration, as demonstrated by scratch wound healing
assays using the same four cell lines. Compared with MCF-7 NC, the
MCF-7 shSulf2 cell line showed markedly slower healing of the
scratch after 36 h (Fig. 8).
Overexpression of Sulf2 in the MDA-MB-231 Sulf2 cell line markedly
increased the scratch wound healing ability (Fig. 8) compared with MDA-MB-231 vector at
18 h. These observations suggested that Sulf2 expression was
associated with breast cancer cell migration.
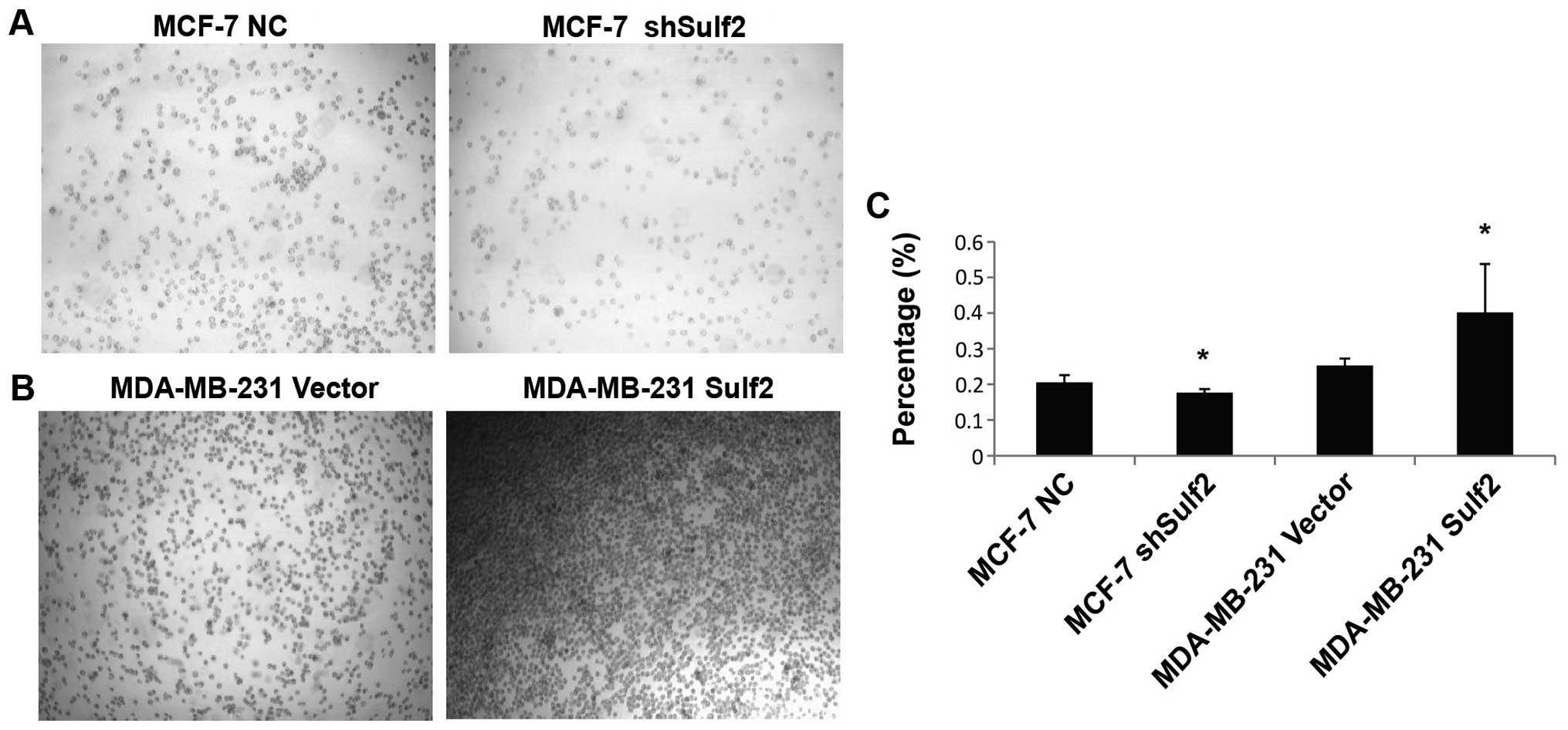
Sulf2 improves breast cancer
adhesion
Compared with MCF-7 NC, the absorbance value in
MCF-7 shSulf2 was significantly reduced (0.17±0.02 vs. 0.21±0.02,
P<0.05). This result suggested that Sulf2 silencing reduced the
adhesion of breast cancer cells with the extracellular matrix.
Compared with MDA-MB-231 vector, MDA-MB-231 Sulf2 cells cultured in
a fibronectin (FN)-coated 96-well plate showed increased cell
adhesion. The absorbance value in MDA-MB-231 Sulf2 cells was
significantly higher than that in MDA-MB-231 vector cells
(0.40±0.13 vs. 0.25±0.02, P<0.05), indicating that Sulf2 was
associated with breast cancer adhesion in vitro (Fig. 9).
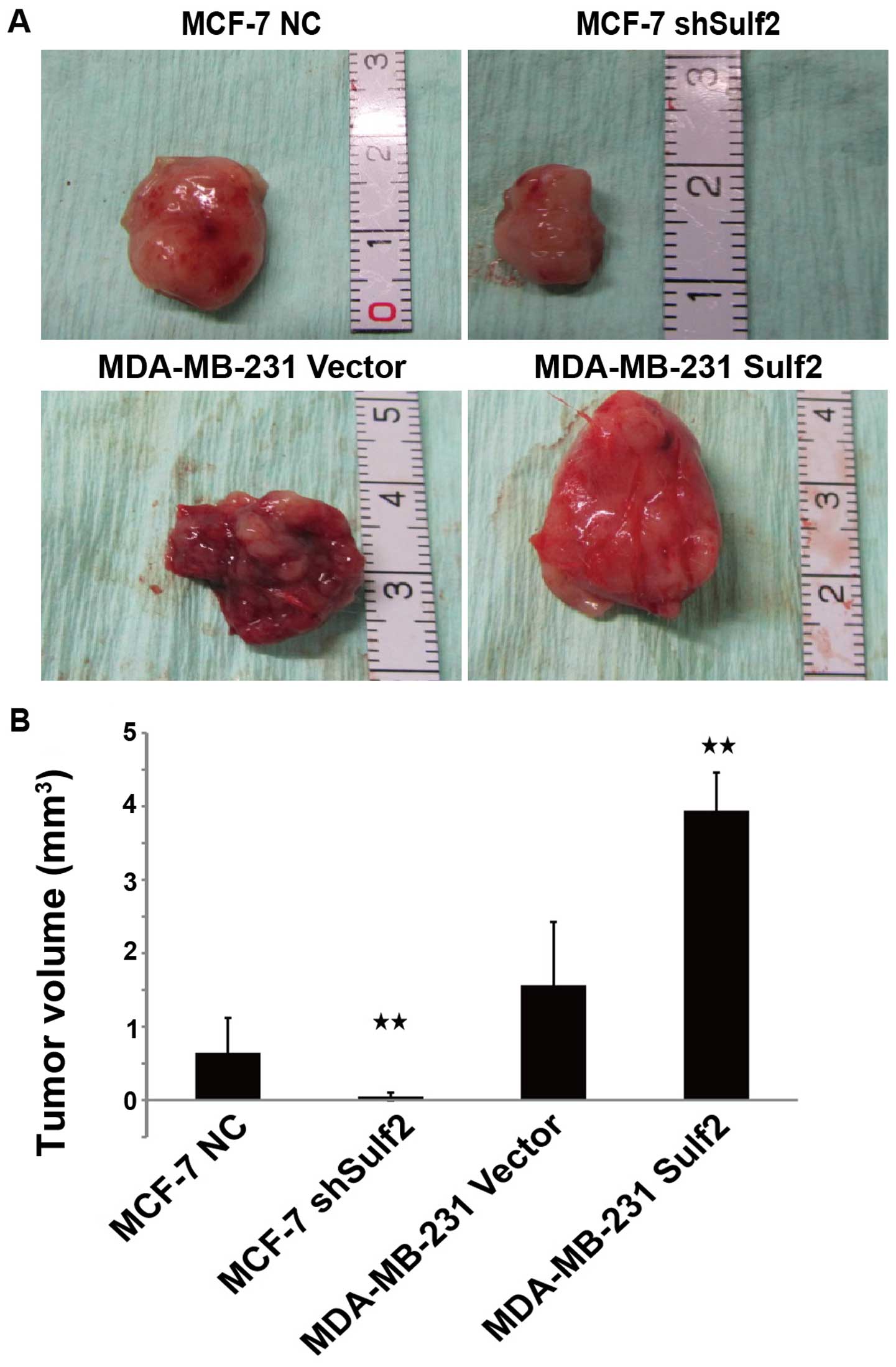
Sulf2 promotes tumorigenicity of breast
cancer cells in vivo
To determine whether the Sulf2 expression level
affects tumorigenicity of breast cancer cells in vivo, we
injected the four cell lines into the mammary pads of nude mice and
began measurement of tumor formation from day 7. There was no
significant difference in the time required for tumor formation
between MDA-MB-231 Sulf2 and MDA-MB-231 vector (7.17±0.98 vs.
6.83±0.41, P>0.05). However, MCF-7 shSulf2 formed tumor later
than MCF-7 NC (12.38±1.52 vs. 7.17±0.98, P<0.05). After 35 days,
100% (6 out of 6) of the nude mice injected with MCF-7 NC,
MDA-MB-231 vector, and MDA-MB-231 Sulf2 developed tumors, while 50%
(3 out of 6) (P<0.05) of the nude mice injected with MCF-7
shSulf2 formed tumors. The sizes of these tumors were measured and
calculated every week. The tumors derived from MCF-7 shSulf2 and
MDA-MB-231 vector were significantly smaller than those from MCF-7
NC and MDA-MB-231 Sulf2, respectively. Xenografts were measured
each week using a Vernier caliper, and the length and the width
were recorded to evaluate the effect of Sulf2 on xenograft growth.
Compared with MCF-7 NC, the volume of MCF-7 shSulf2 tumors was
significantly smaller (0.05±0.05 vs. 0.64±0.48, P<0.05), which
suggested that Sulf2 downregulated MCF-7 proliferation. Conversely,
the MDA-MB-231 Sulf2 tumor volume was significantly larger than
that of the MDA-MB-231 vector tumor (3.94±0.51 vs. 1.56±0.86,
P<0.05). These data suggested that Sulf2 promoted breast cancer
xenograft growth (Fig. 10).
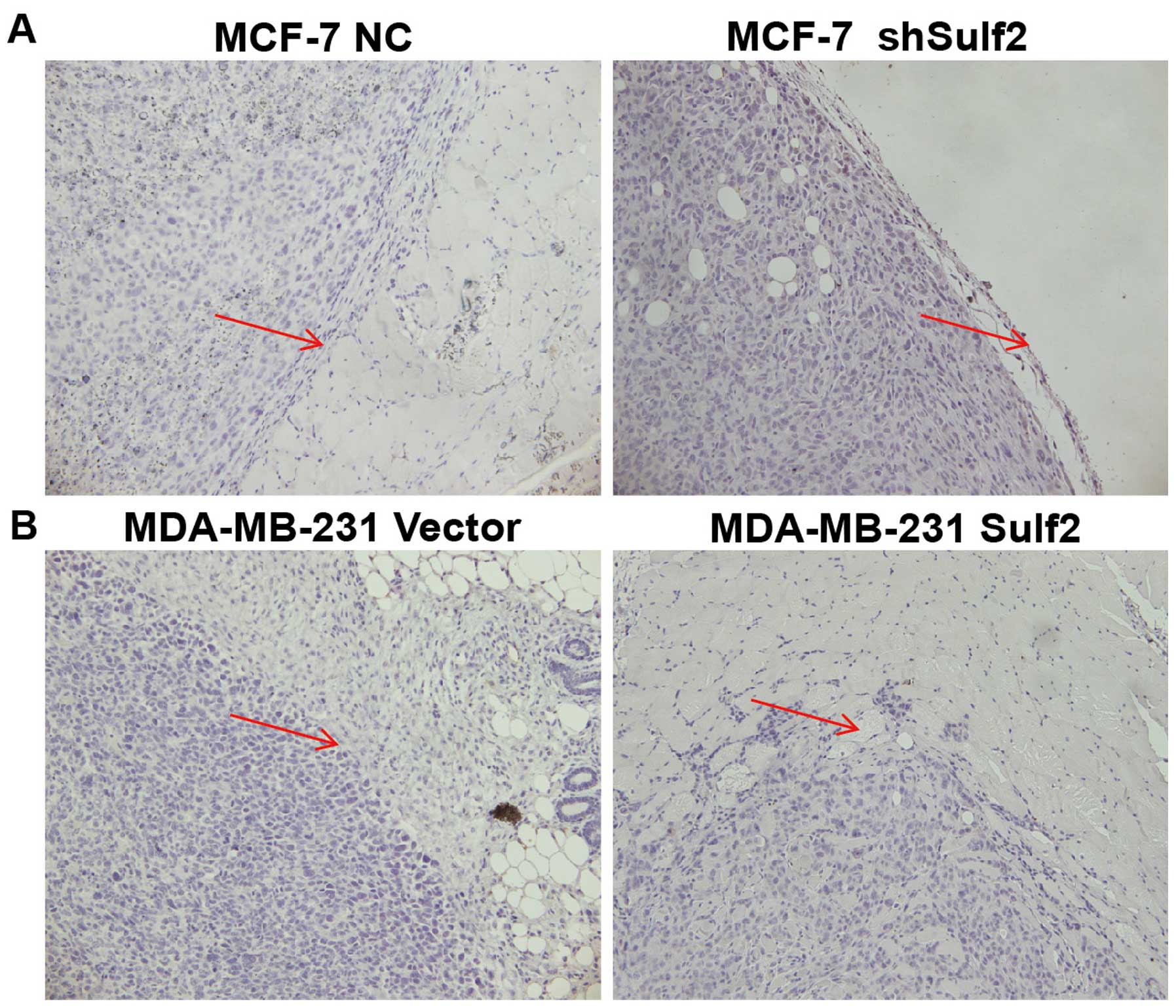
Sulf2 promotes breast cancer invasion in
vivo
The xenografts were examined by pathological
sections. The MDA-MB-231 Sulf2 tumor cells invaded the surrounding
muscle tissue, showing significant infiltration and invasion
ability. Their boundary was unclear, suggesting that Sulf2 can
promote breast cancer invasion and increase the malignancy of
breast cancer. The MCF-7 shSulf2 xenografts showed membrane
structures and no invasion. MCF-7 xenografts formed a capsule-like
structure within the surrounding muscle tissue. The boundary line
between the tumor and surrounding tissue was clear and showed fewer
infiltrating tumor cells (Fig.
11).
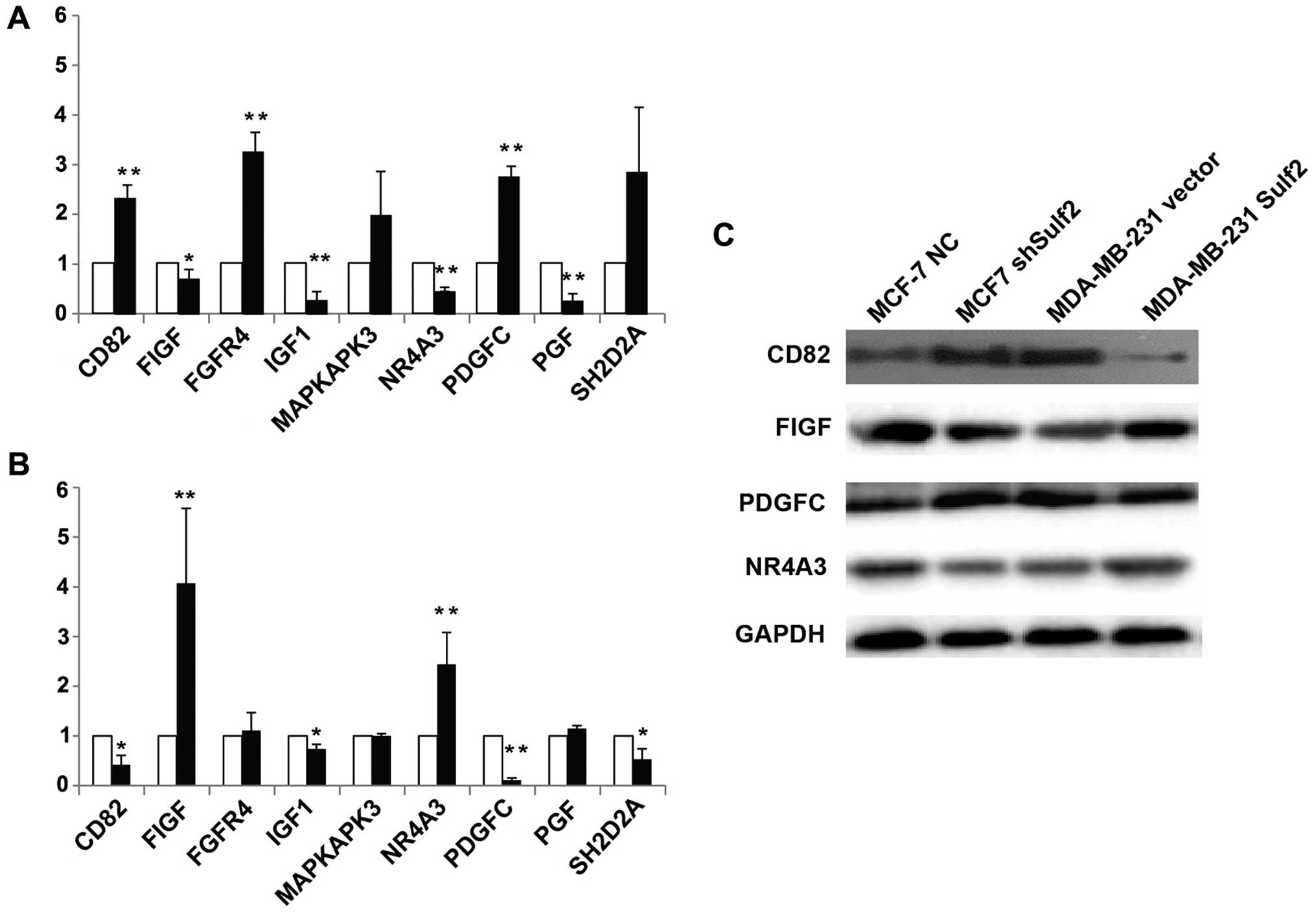
Sulf2 regulates growth factor expression
in breast cancer
Messenger RNA levels of a panel of tumor metastasis
and VEGF signaling genes were first analyzed by PCR microarray
followed by qRT-PCR and western blot verification. Compared with
MCF-7 NC, the genes significantly upregulated in MCF-7 shSulf2 were
CD82 (2.18±0.21 vs. 1.00, P<0.01), FGFR (3.05±0.33 vs. 1.00,
P<0.01) and PDGFC (2.58±0.16 vs. 1.00, P<0.01). The genes
significantly downregulated were FIGF (0.67±0.14 vs. 1.00,
P<0.05), IGF1 (0.27±0.12 vs. 1.00, P<0.01), NR4A3 (0.44±0.04
vs. 1.00, P<0.01) and PGF (0.26±0.1 vs. 1.00, P<0.01)
(Fig. 12A). Compared with
MDA-MB-231 vector, FIGF (4.07±1.51 vs. 1.00, P<0.01) and NR4A3
(2.44±0.64 vs. 1.00, P<0.01) were significantly upregulated in
MDA-MB-231 Sulf2, while CD82 (0.42±0.19 vs. 1.00, P<0.01), IGF1
(0.74±0.09 vs. 1.00, P<0.05), PDGFC (0.11±0.04 vs. 1.00,
P<0.01) and SH2D2A (0.53±0.21 vs. 1.00, P<0.05) were
significantly downregulated (Fig.
12A). Four genes (CD82, FIGF, NR4A3 and PDGFC) that showed
significant changes in mRNA level in response to altered Sulf2
expression were chosen for western blot verification. CD82 and
PDGFC protein expression were negatively correlated with Sulf2
expression, but FIGF and NR4A3 protein expression were positively
correlated with Sulf2 expression. Sulf2 may be the upstream gene
regulating these four genes.
Discussion
Sulf2 has been reported as an important mediator of
carcinogenesis in many cancer types. However, its role in breast
cancer was inconsistent in different reports, making it necessary
to clearly define the effects of Sulf2 in specific cancers,
especially breast cancer (19).
Morimoto et al (15)
reported that Sulf2 mRNA was upregulated in two mouse models of
mammary carcinoma compared with its expression in the normal
mammary gland. Although the mRNA was present in normal tissues,
Sulf2 protein was undetectable. However, Sulf2 can be detected in
some premalignant lesions and in tumors. Khurana et al
(20) found that Sulf2 silencing
promoted matrix detachment, induced MCF10DCIS cell death, promoted
apoptosis and maintained the acne-like structure. In their later
study, the Sulf2 specific inhibitor Bortezomib significantly
reduced tumor size, caused massive apoptosis and, more importantly,
reduced Sulf2 levels in vivo.
In the present study, we used two human breast
cancer cell lines, MCF-7 and MDA-MB-231, that respectively
expresses Sulf2 and lacks Sulf2 expression. We altered the Sulf2
expression in these two cell lines by knocking down or
overexpressing Sulf2 using shRNA and overexpression constructs to
study the biological effects of Sulf2 in breast cancer. We found
that Sulf2 silencing in MCF-7 cells significantly decreased cell
proliferation, invasion, mobility and adhesion, and increased
cisplatin-induced cell apoptosis, especially in early stage
apoptosis. Conversely, Sulf2 overexpression in MDA-MB-231 increased
cell proliferation and decreased cisplatin-induced cell apoptosis
and necrosis. Our experiments suggested that Sulf2 was positively
related to the apoptosis of breast cancer cells, and Sulf2 may be a
molecular target for breast cancer drug resistance. In the present
study, Sulf2 has no effect on breast cancer cell cycle after
cisplatin pre-treatment. We speculated that Sulf2 could not
completely salvage the cells from the damage caused by cisplatin
possibly because cisplatin is a non-specific platinum-containing
antitumor drug that could block cell cycles in all breast cancer
cells. Our data indicated that Sulf2 could influence the cell cycle
status of breast cancer without cisplatin-treatment which improved
cell proliferation in vitro and in vivo. Further
study is needed to verify this hypothesis.
Invasion of tumor cells of the surrounding tissue
and the distant metastasis are complicated procedures which involve
many mechanisms (21,22), including matrix metalloproteinases,
adhesion molecules and growth factors. Many of these factors are
regulated by HSPGs, such as growth factors, cytokines, chemotactic
factors, proteases, lipases, lipoproteins, matrix proteins and cell
adhesion molecules (23–25). Sulf2 can affect a large number of
protein ligands and their molecular biological activity by removing
the HSPG sulfate amino glucose sulfuric acid. Sulf2 can therefore
regulate these factors by forming a common receptor with HSPGs
(19,26).
In this study, we assessed the effects of Sulf2 on
breast cancer cell extracellular matrix adhesion, cell migration,
motility and invasion using a series of assays, including adhesion
assay, scratch assay and Transwell chamber assay. Our results
showed that invasion, mobility and adhesion were decreased when
Sulf2 was inhibited in MCF-7 cells, and we observed converse
results when Sulf2 was overexpressed in MDA-MB-231 cells.
Sulf2 increased breast cancer cell proliferation,
invasion and metastasis and decreased apoptosis in the present
study. To further evaluate the mechanism of action of Sulf2 in
breast cancer metastasis using the tumor metastasis PCR array.
Furthermore, because Sulf2 is closely related to angiogenesis, we
used the VEGF signal PCR array to screen its signaling pathways.
All results were confirmed by qRT-PCR and western blot analysis.
Sulf2 increased FIGF and NR4A3 expression and inhibited CD82 and
PDGFC expression. FIGF is also known as vascular endothelial growth
factor (VEGF-D) and is a member of the VEGF family. VEGF-D is a
ligand of the vascular endothelial growth factor receptors (VEGFRs)
which can activate the subsequent signaling pathways and active
angiogenesis, lymphangiogenesis and endothelial cell growth and
migration (26). Our previous
studies showed that VEGF-D was significantly elevated in the
clinical specimens of patients with lymph node-positive breast
cancer and closely related to the density of lymphatic vessels
around the tumor (27). In this
study, we found that the expression of VEGF-D in breast cancer
cells decreased after Sulf2 inhibition in MCF-7 and increase in
MDA-MB-231 after Sulf2 overexpression, suggesting that Sulf2 was
positively correlated to VEGF-D expression. Our next aim is to
understand the role of Sulf2 in the lymphangiogenesis of breast
cancer.
References
|
1
|
Joy AA, Ghosh M, Fernandes R and Clemons
MJ: Systemic treatment approaches in her2-negative advanced breast
cancer-guidance on the guidelines. Curr Oncol. 22(Suppl 1):
S29–S42. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fina E, Callari M, Reduzzi C, D'Aiuto F,
Mariani G, Generali D, Pierotti MA, Daidone MG and Cappelletti V:
Gene expression profiling of circulating tumor cells in breast
cancer. Clin Chem. 61:278–289. 2015. View Article : Google Scholar
|
|
3
|
Bishop JR, Schuksz M and Esko JD: Heparan
sulphate proteoglycans fine-tune mammalian physiology. Nature.
446:1030–1037. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Turnbull J, Powell A and Guimond S:
Heparan sulfate: Decoding a dynamic multifunctional cell regulator.
Trends Cell Biol. 11:75–82. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rosen SD and Lemjabbar-Alaoui H: Sulf-2:
An extracellular modulator of cell signaling and a cancer target
candidate. Expert Opin Ther Targets. 14:935–949. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Maltseva I, Chan M, Kalus I, Dierks T and
Rosen SD: The SULFs, extracellular sulfatases for heparan sulfate,
promote the migration of corneal epithelial cells during wound
repair. PLoS One. 8:e696422013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fujita K, Takechi E, Sakamoto N, Sumiyoshi
N, Izumi S, Miyamoto T, Matsuura S, Tsurugaya T, Akasaka K and
Yamamoto T: HpSulf, a heparan sulfate 6-O-endosulfatase, is
involved in the regulation of VEGF signaling during sea urchin
development. Mech Dev. 127:235–245. 2010. View Article : Google Scholar
|
|
8
|
Uchimura K, Morimoto-Tomita M, Bistrup A,
Li J, Lyon M, Gallagher J, Werb Z and Rosen SD: HSulf-2, an
extracellular endoglucosamine-6-sulfatase, selectively mobilizes
heparin-bound growth factors and chemokines: Effects on VEGF,
FGF-1, and SDF-1. BMC Biochem. 7:22006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Khurana A, Jung-Beom D, He X, Kim SH,
Busby RC, Lorenzon L, Villa M, Baldi A, Molina J, Goetz MP, et al:
Matrix detachment and proteasomal inhibitors diminish Sulf2
expression in breast cancer cell lines and mouse xenografts. Clin
Exp Metastasis. 30:407–415. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hammond E, Khurana A, Shridhar V and
Dredge K: The role of heparanase and sulfatases in the modification
of heparan sulfate proteoglycans within the tumor microenvironment
and opportunities for novel cancer therapeutics. Front Oncol.
4:1952014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Khurana A, Liu P, Mellone P, Lorenzon L,
Vincenzi B, Datta K, Yang B, Linhardt RJ, Lingle W, Chien J, et al:
HSulf-1 modulates FGF2- and hypoxia-mediated migration and invasion
of breast cancer cells. Cancer Res. 71:2152–2161. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Peterson SM, Iskenderian A, Cook L,
Romashko A, Tobin K, Jones M, Norton A, Gómez-Yafal A, Heartlein
MW, Concino MF, et al: Human sulfatase 2 inhibits in vivo tumor
growth of MDA-MB-231 human breast cancer xenografts. BMC Cancer.
10:4272010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lai JP, Sandhu DS, Yu C, Han T, Moser CD,
Jackson KK, Guerrero RB, Aderca I, Isomoto H, Garrity-Park MM, et
al: Sulfatase 2 up-regulates glypican 3, promotes fibroblast growth
factor signaling, and decreases survival in hepatocellular
carcinoma. Hepatology. 47:1211–1222. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nawroth R, van Zante A, Cervantes S,
McManus M, Hebrok M and Rosen SD: Extracellular sulfatases,
elements of the Wnt signaling pathway, positively regulate growth
and tumorigenicity of human pancreatic cancer cells. PLoS.
2:e3922007. View Article : Google Scholar
|
|
15
|
Morimoto-Tomita M, Uchimura K, Bistrup A,
Lum DH, Egeblad M, Boudreau N, Werb Z and Rosen SD: Sulf-2, a
proangiogenic heparan sulfate endosulfatase, is upregulated in
breast cancer. Neoplasia. 7:1001–1010. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lemjabbar-Alaoui H, van Zante A, Singer
MS, Xue Q, Wang YQ, Tsay D, He B, Jablons DM and Rosen SD: Sulf-2,
a heparan sulfate endosulfatase, promotes human lung
carcinogenesis. Oncogene. 29:635–646. 2010. View Article : Google Scholar :
|
|
17
|
Rosen SD and Lemjabbar-Alaoui H: Sulf-2:
An extracellular modulator of cell signaling and a cancer target
candidate. Expert Opin Ther Targets. 14:935–949. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhu C, Qi X, Chen Y, Sun B, Dai Y and Gu
Y: PI3K/Akt and MAPK/ERK1/2 signaling pathways are involved in
IGF-1-induced VEGF-C upregulation in breast cancer. J Cancer Res
Clin Oncol. 137:1587–1594. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Khurana A, Beleford D, He X, Chien J and
Shridhar V: Role of heparan sulfatases in ovarian and breast
cancer. Am J Cancer Res. 3:34–45. 2013.PubMed/NCBI
|
|
20
|
Khurana A, McKean H, Kim H, Kim SH,
McGuire J, Roberts LR, Goetz MP and Shridhar V: Silencing of
HSulf-2 expression in http://MCF10DCIS.comurisimpleMCF10DCIS.com cells
attenuate ductal carcinoma in situ progression to invasive ductal
carcinoma in vivo. Breast Cancer Res. 14:R432012. View Article : Google Scholar
|
|
21
|
Guo J, Huang Y, Yang L, Xie Z, Song S, Yin
J, Kuang L and Qin W: Association between abortion and breast
cancer: An updated systematic review and meta-analysis based on
prospective studies. Cancer Causes Control. 26:811–819. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Scully OJ, Bay BH, Yip G and Yu Y: Breast
cancer metastasis. Cancer Genomics Proteomics. 9:311–320.
2012.PubMed/NCBI
|
|
23
|
Sun DW, Zhang YY, Qi Y, Zhou XT and Lv GY:
Prognostic significance of MMP-7 expression in colorectal cancer: A
meta-analysis. Cancer Epidemiol. 39:135–142. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kasthuri RS, Taubman MB and Mackman N:
Role of tissue factor in cancer. J Clin Oncol. 27:4834–4838. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Eichhorn ME, Kleespies A, Angele MK, Jauch
KW and Bruns CJ: Angiogenesis in cancer: Molecular mechanisms,
clinical impact. Langenbecks Arch Surg. 392:371–379. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Harris NC, Davydova N, Roufail S,
Paquet-Fifield S, Paavonen K, Karnezis T, Zhang YF, Sato T,
Rothacker J, Nice EC, et al: The propeptides of VEGF-D determine
heparin binding, receptor heterodimerization, and effects on tumor
biology. J Biol Chem. 288:8176–8186. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gu Y, Qi X and Guo S: Lymphangiogenesis
induced by VEGF-C and VEGF-D promotes metastasis and a poor outcome
in breast carcinoma: A retrospective study of 61 cases. Clin Exp
Metastasis. 25:717–725. 2008. View Article : Google Scholar : PubMed/NCBI
|