Introduction
Glioblastomas are the most frequent and aggressive
primary brain cancers in adults (1)
with a high recurrence and mortality rate (2). Glioblastoma prognosis is poor and
there are limited therapeutic options. In recent years, advances
have been made in multimodality including surgery, radiotherapy,
chemotherapy and biotherapy, but the overall 5-year survival rate
is still <3% for patients with glioblastoma (3). Thus, we try to identify prognostic
gene expression (upregulation or downregulation) that may
contribute to evaluate a more effective treatment to improve
patient survival and to address more precisely the use of
comprehensive therapy.
Benzyl isothiocyanate (BITC), one of the
isothiocyanates, is present in cruciferous plants, it acts against
carcinogenesis (4,5) and induces cell death through the
induction of apoptosis and cell cycle arrest in various human
cancer cells (6–10). In human prostate cancer cells, BITC
promoted the phosphorylation of Bcl-xL with simultaneous cell cycle
arrest and subsequent apoptosis (11). In our previous studies we have
demonstrated that BITC inhibited migration and invasion in human
colon (12) and gastric (13) cancer cells in vitro. There is
no available information to show whether BITC affects human brain
tumor cells, in particular regarding the effects of BITC on gene
expression in human glioblastoma cells.
In cell survival, to maintain the integrity of
genomic and mitochondrial DNA is critically important. It was
reported that damage to nuclear and mitochondrial DNA can increase
the accumulation of defective cellular components leading to impact
unfavorably on physiological functions, increasing entropy
(14). If an agent induces DNA
damage, the cell in order to respond to the DNA damage, activates
the cell cycle checkpoints (G1, S and G2/M) to stop cell cycle
progression in order to allow time for repair, thereby preventing
transmission of damaged or incompletely replicated chromosomes
(15). Thus, the associated gene
expression regarding cell cycle progression, cell apoptosis and DNA
damage in cells are important for cancer cell therapy. There is no
previous study showing the anticancer properties of BITC at the
genetic level of human glioblastoma. We investigated the effects of
BITC on gene expression in human brain cancer glioblastoma
multiforme (GBM 8401) in vitro.
Materials and methods
Chemicals and reagents
BITC, dimethyl sulfoxide (DMSO),
penicillin-streptomycin and trypsin-EDTA were obtained from Sigma
Chemical Co. (St. Louis, MO, USA). RPMI-1640 culture medium and
fetal bovine serum (FBS) were purchased from Gibco-BRL/Invitrogen
(Carlsbad, CA, USA). Tissue culture flasks and plates were obtained
from Gibco-BRL/Invitrogen.
Cell culture
Human brain glioblastoma GBM 8401 cells were
purchased from the Food Industry Research and Development Institute
(Hsinchu, Taiwan) and cultured following the supplier's
instructions. Cells were grown in 75 cm2 culture flasks
with RPMI-1640 medium supplemented with 10% FBS, 100 U/ml
penicillin and 100 µg/ml streptomycin and maintained in an
atmosphere of 5% CO2 and 95% air at 37°C. The medium was
changed every 2 days (16).
Cell morphological changes and viability
assays
GBM 8401 cells (8×104 cells/ml) were
seeded into a 12-well plate containing RPMI-1640 medium for 24 h.
In addition, BITC was added to wells at the final concentration of
6 µM for 0, 12, 24 and 48 h. After treatment, cells were
examined and photographed using contrast-phase microscopy at a
magnification of ×400 and then harvested for measuring the total
percentage of viable cells using flow cytometric assay (16).
Annexin V/PI staining for cell
apoptosis
GBM 8401 cells (8×104 cells/ml) were
seeded into a 12-well plate for 24 h and then treated with BITC (0
and 6 µM) for 0, 12, 24 and 48 h, and the cells were
collected, washed with phosphate-buffered saline (PBS) and stained
with Annexin V/propidium iodide (PI) staining kit (BD Biosciences,
San Diego, CA, USA) (17). All
samples were then immediately analyzed by flow cytometry.
cDNA microarray assay for gene
expression
GBM 8401 cells (2.4×106 cells/dish) were
maintained in a 10 cm dish for 24 h. Cells were treated with 6
µM BITC or DMSO for 48 h then cells were collected, and
washed twice with PBS. All samples were further isolated in total
RNA using Qiagen RNeasy Mini kit (Qiagen, Valencia, CA, USA). The
isolated RNA was further conducted for cDNA synthesis, labeling and
microarray hybridization. The fluorescent-labeled cDNA
hybridization (Affymetrix GeneChip Human Gene 1.0 ST array;
Affymetrix, Santa Clara, CA, USA) on the chip was conducted
(18), and the fluorescence from
each sample was measured by Asia BioInnovations Corporation
(Taipei, Taiwan). Expression Console software (Affymetrix) with
default RNA parameters (19,20)
was used to analyze the data. BITC affecting gene expression with
at least a 2-fold-change was considered significant and
recorded.
Results
Cytotoxic effects of BITC in GBM 8401
cells
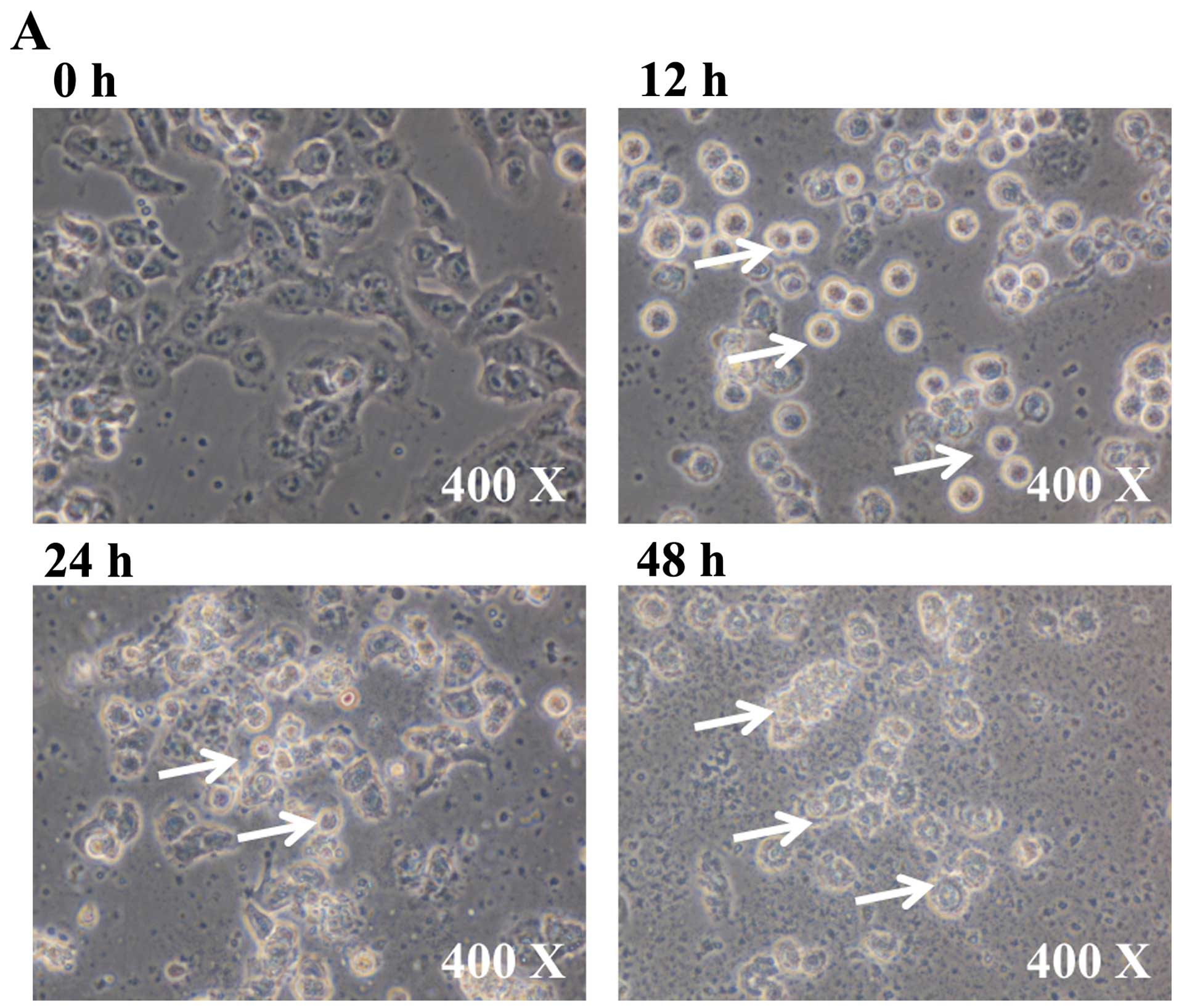
To investigate the cytotoxic effects of BITC in GBM
8401 cells, after treatment of cells with 6 µM BITC for 0,
12, 24 and 48 h, the cell morphological changes and percentage of
viable cells were measured and results are presented in Fig. 1A and B, respectively. BITC induced
cell morphological changes and decreased cell viability in GBM 8401
cells and these effects are time-dependent (Fig. 1A and B).
Induction of cell apoptosis in GBM 8401
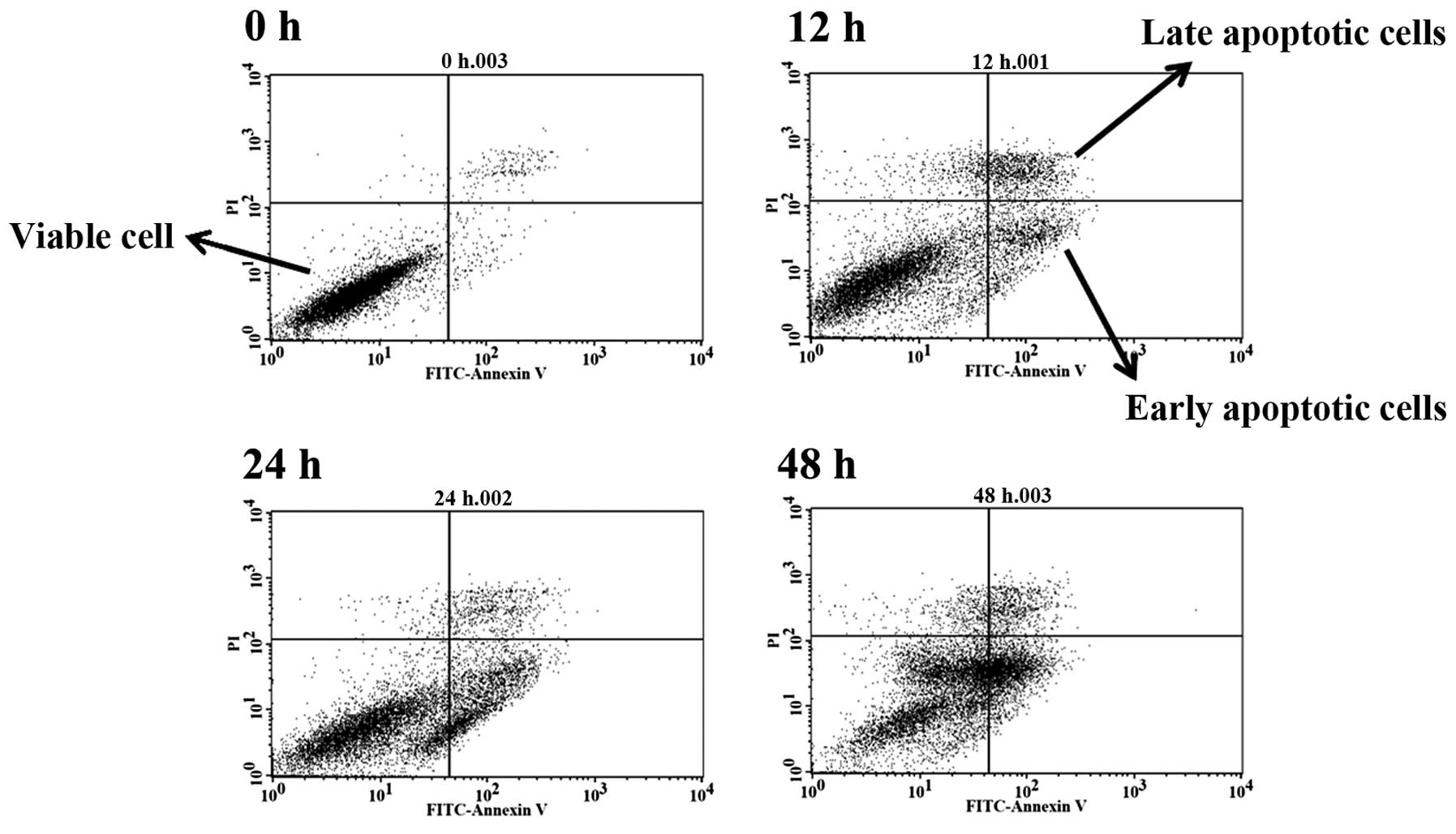
cells after exposure to BITC
In order to further examine whether cell death was
induced by BITC and through the induction of cell apoptosis, the
cells after treatment with 6 µM BITC were harvested and
apoptotic cells were measured by Annexin V/PI staining, and the
results are presented in Fig. 2.
Based on the data in Fig. 2,
BITC-induced apoptotic cell death and these effects are
time-dependent. The treatment of cells with BITC increased the
total apoptotic cell death to 36.81% at 48 h (Table I). The result is consistent with the
morphology and examination of total viable cells.
 | Table IBITC-induced apoptosis of GBM 8401
cells. |
Table I
BITC-induced apoptosis of GBM 8401
cells.
| Hours | Viable cells
(%) | Early apoptotic
cells (%) | Late apoptotic
cells (%) |
|---|
| 0 | 95.69±1.24 | 1.87±0.47 | 2.33±0.77 |
| 12 | 70.34±1.04b | 11.48±1.90b | 15.51±3.91b |
| 24 | 66.15±3.44b | 24.62±1.80b | 7.87±1.41a |
| 48 | 59.35±2.80b | 28.88±1.32b | 7.93±0.67a |
BITC alters the regulations of gene
expression in GBM 8401 cells
GBM 8401 cells were treated with or without 6
µM BITC for 48 h and then harvested for total RNA
extraction. The expression of the top 10 up- and downregulated
genes was estimated by cDNA microarray analysis and the results are
presented in Tables II and
III. BITC induced 317 upregulated
genes and 182 downregulated genes of GBM 8401, respectively.
Fourty-six genes were upregulated in the range >3–<4-fold,
and 198 genes were upregulated >2–<3-fold. One gene was
downregulated >4-fold, and 11 genes were downregulated in the
range >3–<4-fold, and 170 genes were downregulated
>2–<3-fold (data not shown).
 | Table IIThe top 10 upregulated genes of GBM
8401 cells by BITC treatment. |
Table II
The top 10 upregulated genes of GBM
8401 cells by BITC treatment.
| Fold-change | Gene symbol | mRNA
description |
|---|
| 11.04 | LPAR6 | Lysophosphatidic
acid receptor 6 |
| 9.11 |
LOC344887 | NmrA-like family
domain containing 1 pseudogene |
| 8.26 | EGR1 | Early growth
response 1 |
| 7.65 | CLU | Custerin |
| 7.56 | CTH | Cystathionase
(cystathionine γ-lyase) |
| 6.05 | SAT1 | Spermidine/spermine
N1-acetyltransferase 1 |
| 5.81 | SLC7A11 | Solute carrier
family 7 (anionic amino acid transporter light chain, xc-system),
member 11 |
| 5.09 | AKR1B10 | Aldo-keto reductase
family 1, member B10 (aldose reductase) |
| 4.90 |
HIST2H4B | Histone cluster 2,
H4b; H4a; histone cluster 4, H4; histone cluster 1, H4l; H4e; H4b;
H4h; H4c; H4j; H4k; H4f; H4d; H4a; H4i |
| 4.62 | SLCO1B7 | Solute carrier
organic anion transporter family, member 1B7 (non-functional) |
 | Table IIIThe top 10 genes of GBM 8401 cells
downregulated by BITC treatment. |
Table III
The top 10 genes of GBM 8401 cells
downregulated by BITC treatment.
| Fold-change | Gene symbol | mRNA
description |
|---|
| −3.11 |
LOC730755 | Keratin associated
protein 2-4-like; 2-1; 2-4 |
| −3.12 | DKC1 | Dyskeratosis
congenita 1, dyskerin; small nucleolar RNA, H/ACA box 56 |
| −3.18 |
HIST1H1D | Histone cluster 1,
H1d |
| −3.24 | ACTC1 | Actin, α, cardiac
muscle 1 |
| −3.28 | EDN1 | Endothelin 1 |
| −3.71 |
HIST1H2AB | Histone cluster 1,
H2ab; histone cluster 1, H2ae |
| −3.74 |
HIST1H2BI | Histone cluster 1,
H2bi; H2bc; H2be; H2bf; H2bg |
| −3.76 | TRNAU2 | Transfer RNA
selenocysteine 2 (anticodon UCA) |
| −3.77 | KRT81 | Keratin 81 |
| −4.43 |
VTRNA1-1 | Vault RNA 1-1 |
Alterations in gene expression scored in
GBM 8401 cells after exposure to BITC
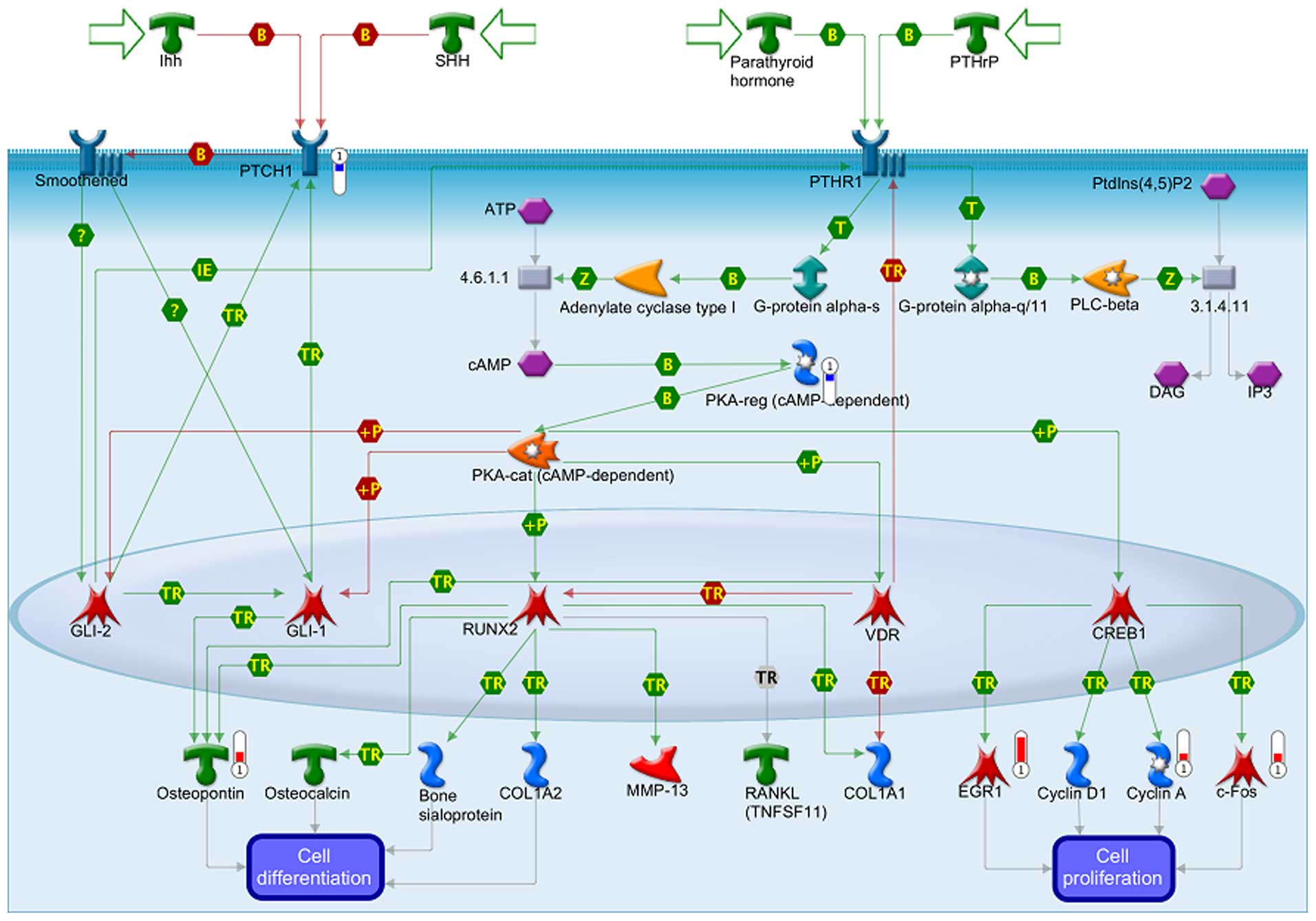
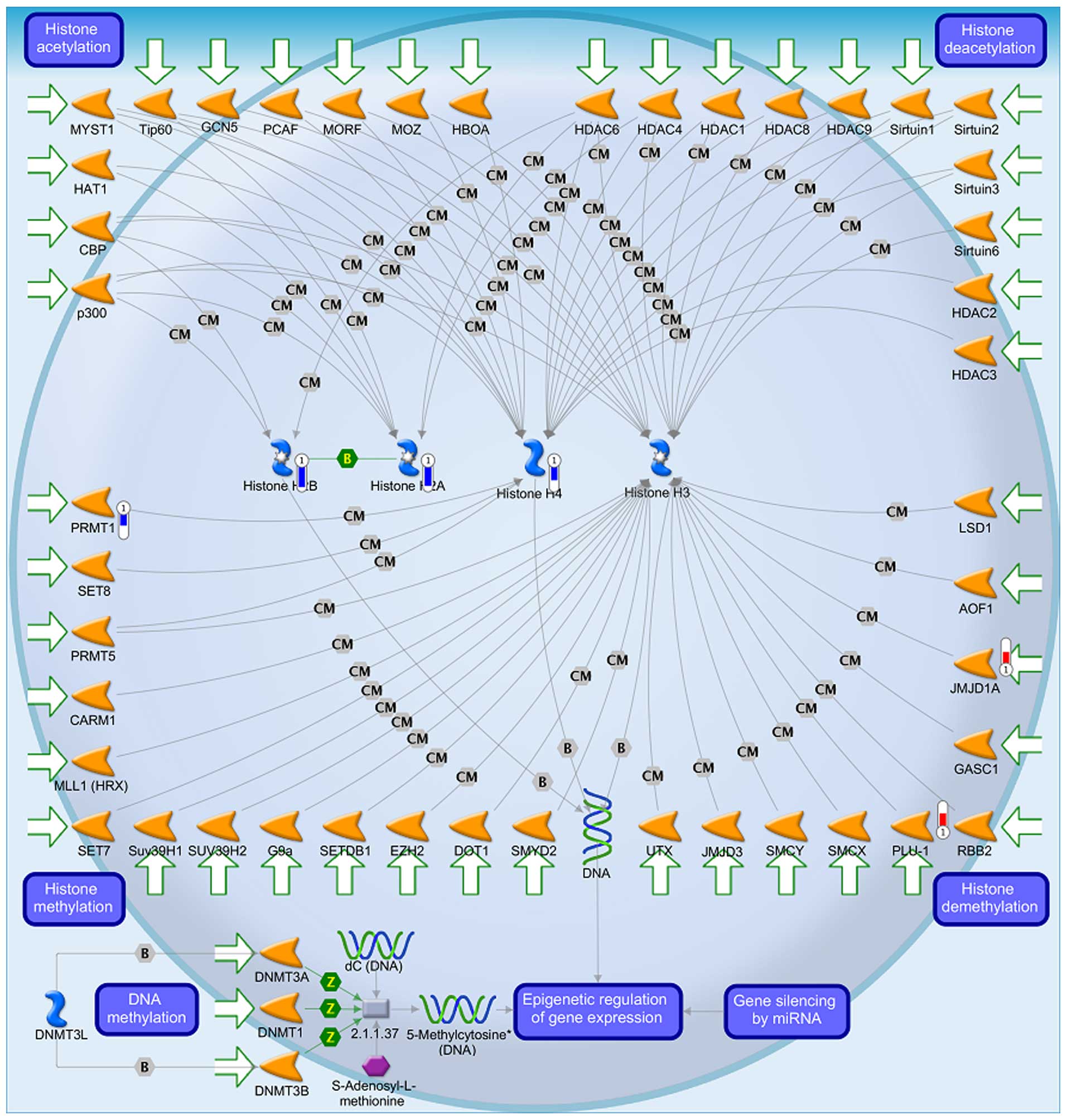
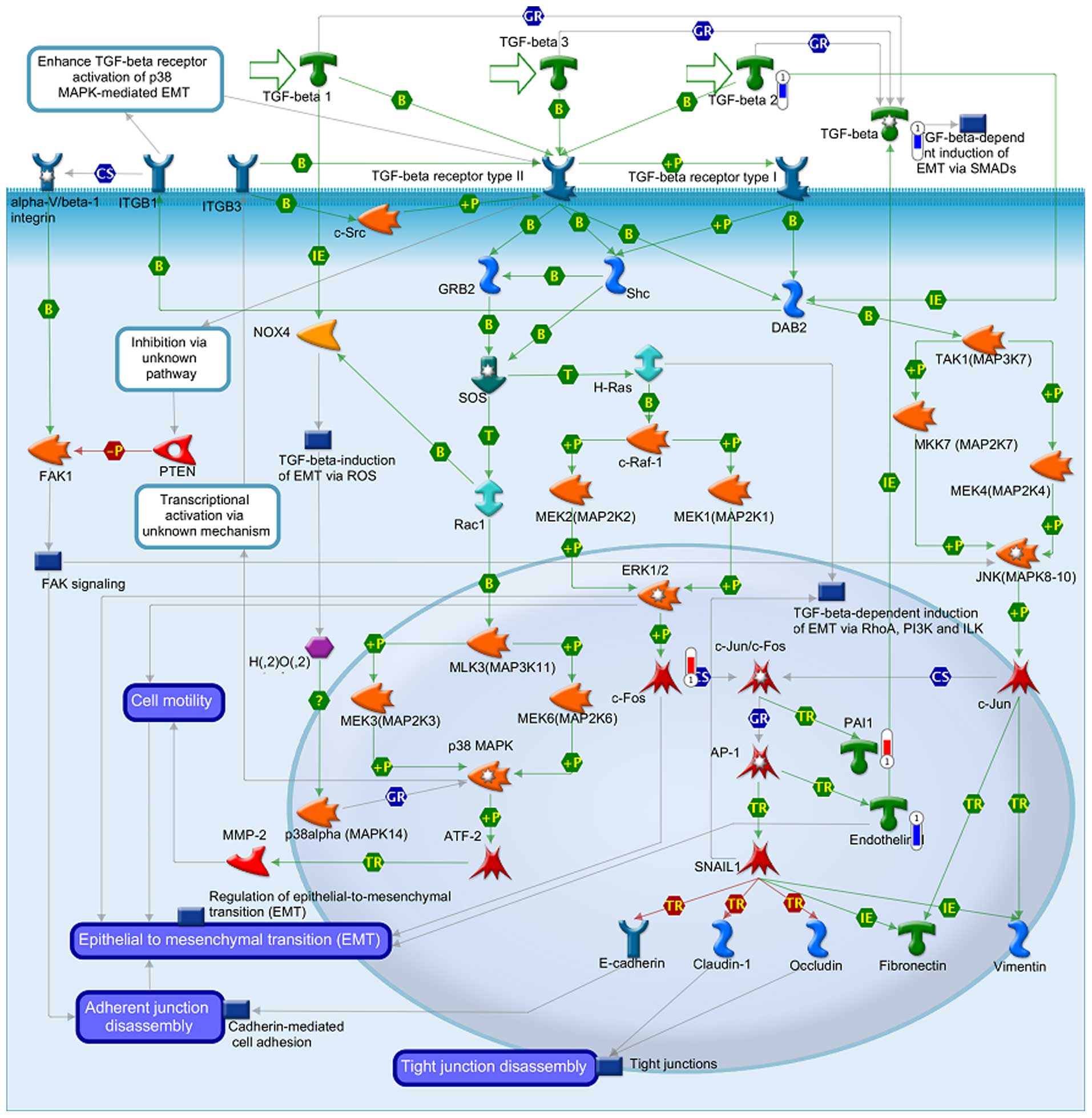
The data from GeneGo analysis were mapped and are
shown as upward thermometers in red color and indicate upregulated
signals and downward (blue) ones indicate downregulated expression
levels of the genes as presented in Figs. 3Figure 4–5. Fig. 3
shows the Development_Hedgehog and PTH signaling pathways in bone
and cartilage development. Fig. 4
shows the transcription and epigenetic regulation of gene
expression and Fig. 5 shows the
Development_TGF-β-dependent induction of EMT via MAPK.
Discussion
Numerous studies have shown that BITC present
biological activities including anticancer function in
vitro. In the present study, BITC-induced cell morphological
changes (Fig. 1A) and decreased the
percentage of viable GBM 8401 cells and these effects are
time-dependent (Fig. 1B). We also
used Annexin V/PI staining to show that BITC-induced cell death
through the induction of cell apoptosis in GBM 8401 cells (Fig. 2 and Table I) these effects are time-dependent.
In order to further examine whether or not BITC affects gene
expression of GBM 8401 cells, we treated cells with 6 µM of
BITC for 24 h before cells were harvested, total RNA was extracted
for cDNA microarray and underwent further analysis for gene
expression and the results are shown in Tables II and III.
It is well documented that after cells are exposed
to anticancer agents, it may cause DNA damage or induce cell cycle
arrest for causing cell death (21–23).
We found that BITC decreased total viable cell number (Fig. 1B) based on cells incubated with BITC
and then harvesting and staining by PI and examination by flow
cytometric assay as previously described (24,25).
We also confirmed cell apoptosis by Annexin V/PI staining and
evaluation by flow cytometry and results indicated that BITC
significantly induced cell death in GBM 8401 cells in vitro
(Fig. 2).
Table II indicates
that expression of 317 genes was promoted, and among them two genes
associated with DNA damage in GBM 8401 cells, the
DNA-damage-inducible transcript 3 (DDIT3) was increased 3.66-fold,
and the growth arrest and DNA-damage-inducible α (GADD45A) was
increased 2.34-fold. Based on these observations, BITC induced DNA
damage as shown previously (9), our
results indicated that BITC-induced DNA damage was associated with
gene expression. Table II
indicates that BITC also promoted four heat protein gene
expression, the heat shock protein 70 kDa family member 13
(HSPA13), which was increased 2.16-fold, the heat shock protein 70
kDa protein 1A, 1B (HSPA1A) increased 2.13-fold [heat shock protein
90 kDa β (Grp94), membrane 2, pseudogene (HSP90B2P)] and increased
2.03-fold. It was reported that heat shock proteins (HSPs) have
anti-apoptotic properties and they are often elevated in many human
cancers; furthermore, the overexpression of HSPs is associated with
poor survival and response to therapy (26–28).
HSP expression in selected brain tumor cell lines (27,29)
have been reported using mainly immunohistochemistry (29–31).
Table II indicates that BITC also
promoted expression of seven genes associated with cell cycle such
as CLK (CDC-like kinase 4), which was increased 3.29-fold, CCNG2
(cyclin G2) was increased 3.19-fold, cyclin A1 (CCNA1) increased
2.30-fold, cyclin Y-like 1 (CCNYL1) increased 2.20-fold,
cyclin-dependent kinase-like 5 (CDKL5) increased 2.19-fold, cyclin
D binding myb-like transcription factor 1 (DMTF1) increased
2.04-fold and cell cycle progression 1 (CCPG1) was increased
2.01-fold in GBM 8401 cells.
Table III
indicates that it suppressed expression of 182 genes in GBM 8401
cells, and among them a gene associated with receptor for cell
responses to stimuli, the EGF containing fibulin-like extracellular
matrix protein 1 (EFEMP1) was inhibited 2.01-fold, and the TNF
receptor-associated protein 1 (TRAP1) was inhibited 2.08-fold. Both
receptors are associated with cell sensitivity for stimuli agents
(32,33). Furthermore, BITC inhibited
mitochondria ribosomal genes such as mitochondrial ribosomal
protein; tumor protein D52 (MRPS28) was inhibited 2.06-fold,
mitochondria ribosomal protein S2 (MRPS2) decreased 2.07-fold,
mitochondria ribosomal protein L23 (MRPL23) decreased 2.08-fold,
mitochondria ribosomal protein S2 (MRPS2) decreased 2.07-fold,
mitochondria ribosomal protein S12 (MRPS12) decreased 2.08-fold,
mitochondria ribosomal protein L12 (MRPL12) decreased 2.25-fold and
mitochondria ribosomal protein S34 (MRPS34) was decreased 2.30-fold
in GBM 8401 cells. It is well documented that agents inducing
cancer cell apoptosis are involved in the mitochondria (34,35),
thus, in the present study, we found that BITC-induced cell death
may be through the induction of DNA damage and affects mitochondria
ribosomal gene expression in GBM 8401 cells.
In conclusion, we found that many genes are
associated with DNA damage and cell cycle regulation and various
genes that associated with the mitochondria were affected by BITC
in GBM 8401 cells. These changes of gene expression in GBM 8401
cells, after exposure to BITC, provide further knowledge on the
effects of BITC at the genetic level, and for future development of
potential biomarkers for glioblastoma therapy.
Acknowledgments
The present study was supported by grant no.
CMU103-ASIA-01 from the China Medical University, Taichung,
Taiwan.
References
|
1
|
Di Cristofori A, Ferrero S, Bertolini I,
Gaudioso G, Russo MV, Berno V, Vanini M, Locatelli M, Zavanone M,
Rampini P, et al: The vacuolar H+ ATPase is a novel
therapeutic target for glioblastoma. Oncotarget. 6:17514–17531.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wen PY and Kesari S: Malignant gliomas in
adults. N Engl J Med. 359:492–507. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Stupp R, Mason WP, van den Bent MJ, Weller
M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn
U, et al European Organisation for Research and Treatment of Cancer
Brain Tumor and Radiotherapy Groups; National Cancer Institute of
Canada Clinical Trials Group: Radiotherapy plus concomitant and
adjuvant temozolomide for glioblastoma. N Engl J Med. 352:987–996.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nakamura Y and Miyoshi N: Cell death
induction by isothiocyanates and their underlying molecular
mechanisms. Biofactors. 26:123–134. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sehrawat A and Singh SV: Benzyl
isothiocyanate inhibits epithelial-mesenchymal transition in
cultured and xenografted human breast cancer cells. Cancer Prev
Res. 4:1107–1117. 2011. View Article : Google Scholar
|
|
6
|
Huang SH, Wu LW, Huang AC, Yu CC, Lien JC,
Huang YP, Yang JS, Yang JH, Hsiao YP, Wood WG, et al: Benzyl
isothiocyanate (BITC) induces G2/M phase arrest and apoptosis in
human melanoma A375. S2 cells through reactive oxygen species (ROS)
and both mitochondria-dependent and death receptor-mediated
multiple signaling pathways. J Agric Food Chem. 60:665–675. 2012.
View Article : Google Scholar
|
|
7
|
Singh SV, Srivastava SK, Choi S, Lew KL,
Antosiewicz J, Xiao D, Zeng Y, Watkins SC, Johnson CS, Trump DL, et
al: Sulforaphane-induced cell death in human prostate cancer cells
is initiated by reactive oxygen species. J Biol Chem.
280:19911–19924. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wicker CA, Sahu RP, Kulkarni-Datar K,
Srivastava SK and Brown TL: BITC sensitizes pancreatic
adenocarcinomas to TRAIL-induced apoptosis. Cancer Growth
Metastasis. 2009:45–55. 2010.PubMed/NCBI
|
|
9
|
Wu CL, Huang AC, Yang JS, Liao CL, Lu HF,
Chou ST, Ma CY, Hsia TC, Ko YC and Chung JG: Benzyl isothiocyanate
(BITC) and phenethyl isothiocyanate (PEITC)-mediated generation of
reactive oxygen species causes cell cycle arrest and induces
apoptosis via activation of caspase-3, mitochondria dysfunction and
nitric oxide (NO) in human osteogenic sarcoma U-2 OS cells. J
Orthop Res. 29:1199–1209. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xiao D, Powolny AA and Singh SV: Benzyl
isothiocyanate targets mitochondrial respiratory chain to trigger
reactive oxygen species-dependent apoptosis in human breast cancer
cells. J Biol Chem. 283:30151–30163. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Basu A and Haldar S: Dietary
isothiocyanate mediated apoptosis of human cancer cells is
associated with Bcl-xL phosphorylation. Int J Oncol. 33:657–663.
2008.PubMed/NCBI
|
|
12
|
Lai KC, Huang AC, Hsu SC, Kuo CL, Yang JS,
Wu SH and Chung JG: Benzyl isothiocyanate (BITC) inhibits migration
and invasion of human colon cancer HT29 cells by inhibiting matrix
metalloproteinase-2/-9 and urokinase plasminogen (uPA) through PKC
and MAPK signaling pathway. J Agric Food Chem. 58:2935–2942. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ho CC, Lai KC, Hsu SC, Kuo CL, Ma CY, Lin
ML, Yang JS and Chung JG: Benzyl isothiocyanate (BITC) inhibits
migration and invasion of human gastric cancer AGS cells via
suppressing ERK signal pathways. Hum Exp Toxicol. 30:296–306. 2011.
View Article : Google Scholar
|
|
14
|
Salminen A and Kaarniranta K: Genetics vs
entropy: Longevity factors suppress the NF-kappaB-driven entropic
aging process. Ageing Res Rev. 9:298–314. 2010. View Article : Google Scholar
|
|
15
|
Erol A: Genotoxic stress-mediated cell
cycle activities for the decision of cellular fate. Cell Cycle.
10:3239–3248. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang DY, Yeh CC, Lee JH, Hung CF and Chung
JG: Berberine inhibited arylamine N-acetyltransferase activity and
gene expression and DNA adduct formation in human malignant
astrocytoma (G9T/VGH) and brain glioblastoma multiforms (GBM 8401)
cells. Neurochem Res. 27:883–889. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kwon HY, Kim KS, An HK, Moon HI, Kim HJ
and Lee YC: Triptolide induces apoptosis through extrinsic and
intrinsic pathways in human osteosarcoma U2OS cells. Indian J
Biochem Biophys. 50:485–491. 2013.
|
|
18
|
Hsia TC, Yu CC, Hsu SC, Tang NY, Lu HF, Yu
CS, Wu SH, Lin JG and Chung JG: cDNA microarray analysis of the
effect of cantharidin on DNA damage, cell cycle and
apoptosis-associated gene expression in NCI-H460 human lung cancer
cells in vitro. Mol Med Rep. 12:1030–1042. 2015.PubMed/NCBI
|
|
19
|
Gardina PJ, Clark TA, Shimada B, Staples
MK, Yang Q, Veitch J, Schweitzer A, Awad T, Sugnet C, Dee S, et al:
Alternative splicing and differential gene expression in colon
cancer detected by a whole genome exon array. BMC Genomics.
7:3252006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lin JJ, Yu CC, Lu KW, Chang SJ, Yu FS,
Liao CL, Lin JG and Chung JG: α-Phellandrene alters expression of
genes associated with DNA damage, cell cycle, and apoptosis in
murine leukemia WEHI-3 cells. Anticancer Res. 34:4161–4180.
2014.PubMed/NCBI
|
|
21
|
Li X, Tian J, Bo Q, Li K, Wang H, Liu T
and Li J: Targeting DNA-PKcs increased anticancer drug sensitivity
by suppressing DNA damage repair in osteosarcoma cell line MG63.
Tumour Biol. 36:9365–9372. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Neumann J, Yang Y, Köhler R, Giaisi M,
Witzens-Harig M, Liu D, Krammer PH, Lin W and Li-Weber M: Mangrove
dolabrane-type of diterpenes tagalsins suppresses tumor growth via
ROS-mediated apoptosis and ATM/ATR-Chk1/Chk2-regulated cell cycle
arrest. Int J Cancer. 137:2739–2748. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang D, Tang B, Xie X, Xiao YF, Yang SM
and Zhang JW: The interplay between DNA repair and autophagy in
cancer therapy. Cancer Biol Ther. 16:1005–1013. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Anuchapreeda S, Tima S, Duangrat C and
Limtrakul P: Effect of pure curcumin, demethoxycurcumin, and
bisdemethoxycurcumin on WT1 gene expression in leukemic cell lines.
Cancer Chemother Pharmacol. 62:585–594. 2008. View Article : Google Scholar
|
|
25
|
Ji BC, Hsu WH, Yang JS, Hsia TC, Lu CC,
Chiang JH, Yang JL, Lin CH, Lin JJ, Suen LJ, et al: Gallic acid
induces apoptosis via caspase-3 and mitochondrion-dependent
pathways in vitro and suppresses lung xenograft tumor growth in
vivo. J Agric Food Chem. 57:7596–7604. 2009. View Article : Google Scholar
|
|
26
|
Ciocca DR and Calderwood SK: Heat shock
proteins in cancer: Diagnostic, prognostic, predictive, and
treatment implications. Cell Stress Chaperones. 10:86–103. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ciocca DR, Rozados VR, Cuello Carrión FD,
Gervasoni SI, Matar P and Scharovsky OG: Hsp25 and Hsp70 in rodent
tumors treated with doxorubicin and lovastatin. Cell Stress
Chaperones. 8:26–36. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gyrd-Hansen M, Nylandsted J and Jäättelä
M: Heat shock protein 70 promotes cancer cell viability by
safeguarding lysosomal integrity. Cell Cycle. 3:1484–1485. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Graner MW and Bigner DD: Chaperone
proteins and brain tumors: Potential targets and possible
therapeutics. Neuro Oncol. 7:260–278. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Alexiou GA, Vartholomatos G, Stefanaki K,
Patereli A, Dova L, Karamoutsios A, Lallas G, Sfakianos G, Moschovi
M and Prodromou N: Expression of heat shock proteins in
medulloblastoma. J Neurosurg Pediatr. 12:452–457. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Assimakopoulou M: Human meningiomas:
Immunohistochemical localization of progesterone receptor and heat
shock protein 27 and absence of estrogen receptor and PS2. Cancer
Detect Prev. 24:163–168. 2000.PubMed/NCBI
|
|
32
|
Kim YJ, Yoon HY, Kim SK, Kim YW, Kim EJ,
Kim IY and Kim WJ: EFEMP1 as a novel DNA methylation marker for
prostate cancer: Array-based DNA methylation and expression
profiling. Clin Cancer Res. 17:4523–4530. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ou Y, Liu L, Xue L, Zhou W, Zhao Z, Xu B,
Song Y and Zhan Q: TRAP1 shows clinical significance and promotes
cellular migration and invasion through STAT3/MMP2 pathway in human
esophageal squamous cell cancer. J Genet Genomics. 41:529–537.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ly JD, Grubb DR and Lawen A: The
mitochondrial membrane potential (Δψm) in apoptosis; An update.
Apoptosis. 8:115–128. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Rego AC and Oliveira CR: Mitochondrial
dysfunction and reactive oxygen species in excitotoxicity and
apoptosis: Implications for the pathogenesis of neurodegenerative
diseases. Neurochem Res. 28:1563–1574. 2003. View Article : Google Scholar : PubMed/NCBI
|