Introduction
Breast cancer (BC) remains a worldwide burden with
an estimated incidence of more than 1.5 million new cases and
approximately half a million deaths per year (1). Due to early detection, improvement in
treatment options and changes in lifestyle paradigms, the mortality
rates have been decreasing in developed countries. Conversely,
developing countries are witnessing an increase in BC incidence and
mortality rates, most probably due to the lack of awareness
campaigns and changes in daily habits such as sedentary lifestyle,
high consumption of sugars and fat that lead to overweight and
obesity, known risk factors of BC (1). Moreover the proportion of cases
diagnosed in less developed countries is meagre when compared to
developed regions thus leading to higher mortality rates (2).
Although the molecular mechanisms that underlie the
development of breast cancer have been well investigated, our
current knowledge is far from complete. BC is a heterogeneous
malignancy. Clinical diagnosis and prescribed therapy relie on the
TNM staging system based on tumor (T), node status (N) and
metastasis (M). Estrogen and progesterone receptor status, human
epidermal growth factor receptor 2 (HER2/neu) status and the Ki-67
proliferative index, in addition to tumor-infiltrating lymphocytes,
as well as the age of the patient, are used to classify BC into
various subtypes (2–4). Nonetheless, these conventional breast
cancer prognostic factors have intrinsic limitations, and their use
does not allow an accurate prediction of treatment resistance or
relapse. Defining new molecular prognostic factors to refine BC
classification could be useful in improving the therapeutic
schemes.
Recently, various microRNAs (miRNAs) have been
characterized and identified as regulators and/or biomarkers in
breast cancer development, including initiation, metastasis and
therapeutic resistance (5–11). miRNAs are small 18–22 nucleotide RNA
molecules that regulate protein expression by mostly binding to the
3′-UTR (3′-untranslated region) of mRNAs, thus inhibiting
translation through repression or degradation of mRNAs (12,13).
Due to their size, miRNAs are stable in human samples and can
easily be used as molecular signatures in cancer (14–20).
miR-203 was originally described as a
keratinocyte-specific miRNA (21)
but was soon shown to play an important role in bladder cancer
(22) and to be epigenetically
silenced in hematopoietic cancers (23). Several studies have shown an
association between miR-203 and chemotherapeutic resistance to
cisplatin (24), invasiveness
(25,26), proliferation (26–29)
and metastases (30,31), and as a biomarker (32,33).
Some miR-203 targets have been identified, such as BMI1,
SNAI2, SOCS3, BIRC5 and LASP1 (24–26,28,34),
but a complete picture of the expression of miR-203 in different
cohorts of BC, its mechanisms of action and the circuitry of its
effects remain to be fully clarified.
With the aim of contributing to a better
understanding of the role of miR-203a in breast cancer, this study
reports on the assessment of miR-203a expression and
clinicopathological features in a Portuguese population with breast
carcinoma.
Materials and methods
Sample collection and processing
Patients with breast carcinoma were recruited for
the study at the Hospital de São José, from Centro Hospitalar de
Lisboa Central, during 2013 and 2014. Each patient signed a written
informed consent form and this study was reviewed and approved by
the Ethics Committees of the NOVA Medical School and of the Centro
Hospitalar de Lisboa Central. All clinical information was gathered
by trained and specialized clinicians. All samples originated from
surgical sections (mastectomy or tumorectomy). A total of 109
formalin-fixed paraffin-embedded (FFPE) paired normal and tumor
tissue samples were collected. Normal tissue was adjacent to the
tumor and in all cases was confirmed by the pathology team as being
only normal mammary tissue. Diagnosis and common
immunohistochemical markers for breast cancer classification such
as estrogen and progesterone receptor (ER and PR), HER2
amplification status and Ki-67 proliferative index were evaluated
by two highly trained and independent pathologists. Staging was
performed by tumor (T), node (N) and metastasis (M) classification
(35). The procedure for
immunohistochemical detection was carried out according to the
recommendations of the American Society of Clinical
Oncology/College of American Pathologists (ASCO/CAP) guidelines
(36,37) and the International Ki-67 in Breast
Cancer Working Group (38) at the
time of sample collection. With the existing canons at the time,
the molecular classification of breast tumors was as follows:
luminal A: ER-positive or PR-positive and Ki-67 <13%; luminal B:
ER-positive or PR-positive and Ki-67 ≥13%; HER2-positive:
ER-negative and PR-negative, HER2-positive; triple-negative:
ER-negative, PR-negative and HER2-negative.
Total RNA was purified from FFPE tissues using
RecoverAll™ Total Nucleic Acid isolation kit (Ambion®)
and according to the manufacturer's protocol with slight
alterations. Briefly, the samples were deparaffinized with xylol
and digested with protease/digestion buffer. Microtube pestles were
used to macerate hard samples during digestion. Then, nucleic acid
was precipitated with a mixture of absolute ethanol/isolation
additive and the supernatant used in the filter cartridges to
proceed with DNA digestion using a DNase/DNase buffer mixture.
Total RNA containing miRNAs was eluted and preserved at −80°C for
further use.
miR-203a quantification by quantitative
RT-PCR (RT-qPCR)
Total RNA concentration was quantified using a
NanoDrop™ spectrophotometer. miR-203a expression levels were
quantified by RT-qPCR using cDNA Synthesis and
SYBR®-Green Master Mix kits (Exiqon, Denmark) on an ABI
7300 real-time PCR machine, according to the manufacturer's
instructions. For each cDNA synthesis experience a no-enzyme
control was used to ensure that there was no contamination during
cDNA synthesis, and for each sample a no-template control was used
to detect any potential amplification of genomic DNA. U6 snRNA was
used as internal control for data normalization. Relative
expression of miR-203a in each FFPE sample was determined by
2−ΔCt, where ΔCt is Ct (miR-203a) - median Ct (U6
snRNA). Fold-change was determined by 2−ΔΔCt, where ΔΔCt
is ΔCt (tumor) - ΔCt (normal). All samples were analyzed in
duplicate.
Statistical analysis
All statistical analyses were performed using the
SPSS statistical software package version 21.0 (SPSS Inc., Chicago,
IL, USA). Non-parametric Wilcoxon signed-rank test was used to
analyze the differences between matched samples (normal vs. tumor
tissues). The Mann-Whitney U test and Kruskal-Wallis test were used
to analyze the differences of miR-203a expression levels in the
tumor tissue according to the clinicopathological characteristics.
For nominal variables, the relationships between
clinicopathological characteristics and miR-203a status were
studied using the Chi-square test and Fisher's exact test.
Results
Study population description
Breast tumor and adjacent normal mammary tissues
were collected from 109 patients. The study population comprised
only Caucasian woman from the greater area of Lisbon and on the day
of diagnosis the median age was 62 years (range, 30–85). The median
age of menarche and menopause was 13 years (range, 8–17) and 50
years (range, 36–59), respectively, and ~66% of the population was
diagnosed with breast carcinoma in post menopause status.
Seventy-seven percent of the women had one or more pregnancies and
~73% had one or more children. Forty-five percent claimed to have
taken birth control pills. Approximately 50% were overweight or
obese. Regarding general tumor characteristics, the median tumor
size was 18.5 mm (range, 6–130), ~51% showed no invasion of the
lymph nodes, ~80% were ER-positive, 72.5% were PR-positive, and
13.8% showed high amplification of HER2 and 48% showed negative
Ki-67 proliferation index. The most common histological type was
invasive carcinoma NOS (83.5% of the cases), followed by invasive
lobular carcinoma (9.2%), ductal carcinoma in situ (6.4%)
and invasive lobular and ductal carcinoma (0.9%). The most common
molecular subtype was luminal A (47.4% of all cases). Luminal B
represented ~40% of the remaining cases. Triple-negative subtype
represented ~12% of the cases. The most frequent stage was II
(46.8%) followed by I (37.6%) and III (8.3%). All these data,
together with the stratification of several variables, is displayed
in Tables I–III.
 | Table IAssociation of miR-203a relative
expression with clinical characteristics of the breast cancer
cases. |
Table I
Association of miR-203a relative
expression with clinical characteristics of the breast cancer
cases.
| n (%) | Median relative
expression of miR-203
| Tumor tissue/normal
tissue | P-valuea |
|---|
| Normal tissue | Tumor tissue |
|---|
| No. of cases | 109 (100) | 0.07 | 0.12 | 1.7 |
0.002b |
| Age at diagnosis
(years), n (%) |
| 30–39 | 3 (2.8) | 0.04 | 0.07 | 1.75 | 0.593 |
| 40–49 | 17 (15.6) | 0.14 | 0.13 | 0.93 | 0.906 |
| 50–59 | 27 (24.8) | 0.14 | 0.15 | 1.07 | 0.657 |
| >60 | 54 (49.5) | 0.04 | 0.11 | 2.75 | 0.001 |
| Missing | 8 (7.3) | | | | |
| Age at menarche
(years), n (%) |
| ≤13 | 65 (59.6) | 0.06 | 0.12 | 2.00 | 0.066 |
| >13 | 35 (32.1) | 0.06 | 0.14 | 2.33 | 0.003 |
| Missing | 9 (8.3) | | | | |
| Age at menopause
(years), n (%) |
| ≤50 | 44 (52.4) | 0.04 | 0.14 | 3.50 |
<0.001 |
| >50 | 29 (34.5) | 0.05 | 0.10 | 2.00 | 0.539 |
| Missing | 11 (13.1) | | | | |
| Menopausal status,
n (%) |
| Pre | 25 (22.9) | 0.14 | 0.12 | 0.86 | 0.607 |
| Post | 72 (66.1) | 0.05 | 0.11 | 2.20 | 0.003 |
| Peri | 1 (0.9) | | | | |
| Missing | 11 (10.1) | | | | |
| No. of pregnancies,
n (%) |
| 0 | 15 (13.8) | 0.07 | 0.15 | 2.14 | 0.972 |
| 1–2 | 42 (38.5) | 0.06 | 0.11 | 1.83 | 0.046 |
| 3–4 | 27 (24.8) | 0.08 | 0.11 | 1.38 | 0.416 |
| >4 | 15 (13.8) | 0.04 | 0.16 | 4.00 | 0.001 |
| Missing | 10 (9.2) | | | | |
| No. of children, n
(%) |
| 0 | 21 (19.3) | 0.04 | 0.12 | 3.00 | 0.295 |
| 1–2 | 64 (58.7) | 0.08 | 0.11 | 1.38 | 0.012 |
| 3–4 | 12 (11.0) | 0.08 | 0.12 | 1.50 | 0.657 |
| >4 | 4 (3.7) | 0.03 | 0.21 | 7.00 | 0.068 |
| Missing | 8 (7.3) | | | | |
| Age at first birth
(years), n (%) |
| <20 | 13 (11.9) | 0.05 | 0.17 | 3.40 | 0.033 |
| 20–30 | 45 (41.3) | 0.09 | 0.12 | 1.33 | 0.074 |
| >30 | 12 (11.0) | 0.04 | 0.09 | 2.25 | 0.182 |
| Missing | 39 (35.8) | | | | |
| Breastfeeding, n
(%) |
| No | 32 (29.4) | 0.04 | 0.11 | 2.75 | 0.170 |
| Yes | 67 (61.5) | 0.08 | 0.13 | 1.62 | 0.002 |
| Missing | 10 (9.2) | | | | |
| Oral contraceptive,
n (%) |
| No | 48 (44.0) | 0.04 | 0.11 | 2.75 | 0.130 |
| Yes | 49 (45.0) | 0.08 | 0.12 | 1.50 | 0.004 |
| Missing | 12 (11.0) | | | | |
 | Table IIIAssociation of miR-203a relative
expression with the pathological characteristics of the breast
cancer patients. |
Table III
Association of miR-203a relative
expression with the pathological characteristics of the breast
cancer patients.
| n (%) | Median relative
expression of miR-203
| Tumor tissue/normal
tissue | P-valuea |
|---|
| Normal tissue | Tumor tissue |
|---|
| Size of the tumor
(mm), n (%) |
| ≤18.5 | 54 (49.5) | 0.08 | 0.12 | 1.50 | 0.019 |
| >18.5 | 54 (49.5) | 0.06 | 0.12 | 2.00 | 0.076 |
| Missing | 1 (1.0) | | | | |
| Lymph node
invasion, n (%) |
| No | 52 (51.4) | 0.05 | 0.12 | 2.40 | 0.013 |
| Yes | 53 (47.7) | 0.09 | 0.11 | 1.22 | 0.137 |
| Missing | 4 (0.9) | | | | |
| Estrogen receptor
status, n (%) |
| Negative | 16 (14.7) | 0.07 | 0.15 | 2.14 | 0.074 |
| Positive | 87 (79.8) | 0.07 | 0.12 | 1.71 | 0.042 |
| Missing | 6 (5.5) | | | | |
| Progesterone
receptor status, n (%) |
| Negative | 22 (20.2) | 0.04 | 0.12 | 3.00 | 0.091 |
| Positive | 79 (72.5) | 0.08 | 0.12 | 1.50 | 0.046 |
| Missing | 8 (7.3) | | | | |
| HER2 status, n
(%) |
| Negative | 87 (79.8) | 0.08 | 0.12 | 1.50 | 0.016 |
| Positive | 15 (13.8) | 0.72 | 0.73 | 1.01 | 0.609 |
| Missing | 7 (6.4) | | | | |
| Ki-67 index status,
n (%) |
| Negative | 53 (48.6) | 0.05 | 0.13 | 2.60 | 0.024 |
| Positive | 48 (44.0) | 0.08 | 0.10 | 1.25 | 0.253 |
| Missing | 8 (7.3) | | | | |
| Histological type,
n (%) |
| Ductal carcinoma
in situ | 7 (6.4) | 0.05 | 0.11 | 2.20 | 0.028 |
| Invasive carcinoma
NOS | 91 (83.5) | 0.07 | 0.12 | 1.71 | 0.009 |
| Invasive lobular
carcinoma | 10 (9.2) | 0.13 | 0.11 | 0.84 | 0.575 |
| Invasive lobular
and ductal carcinoma | 1 (0.9) | | | | |
| Molecular type, n
(%) |
| Luminal A | 48 (47.5) | 0.07 | 0.13 | 1.86 | 0.054 |
| Luminal B
(HER2-) | 27 (26.7) | 0.08 | 0.11 | 1.38 | 0.527 |
| Luminal B
(HER2+) | 13 (12.9) | 0.07 | 0.08 | 1.14 | 0.221 |
|
Triple-negative | 12 (11.9) | 0.06 | 0.15 | 2.50 | 0.139 |
|
HER2+ | 1 (1.0) | | | | |
| Stage, n (%) |
| 0 | 7 (6.4) | 0.05 | 0.11 | 2.20 | 0.028 |
| I | 41 (37.6) | 0.08 | 0.12 | 1.50 | 0.126 |
| II | 51 (46.8) | 0.06 | 0.13 | 2.17 | 0.009 |
| III | 9 (8.3) | 0.10 | 0.11 | 1.10 | 0.678 |
| Missing | 1 (0.9) | | | | |
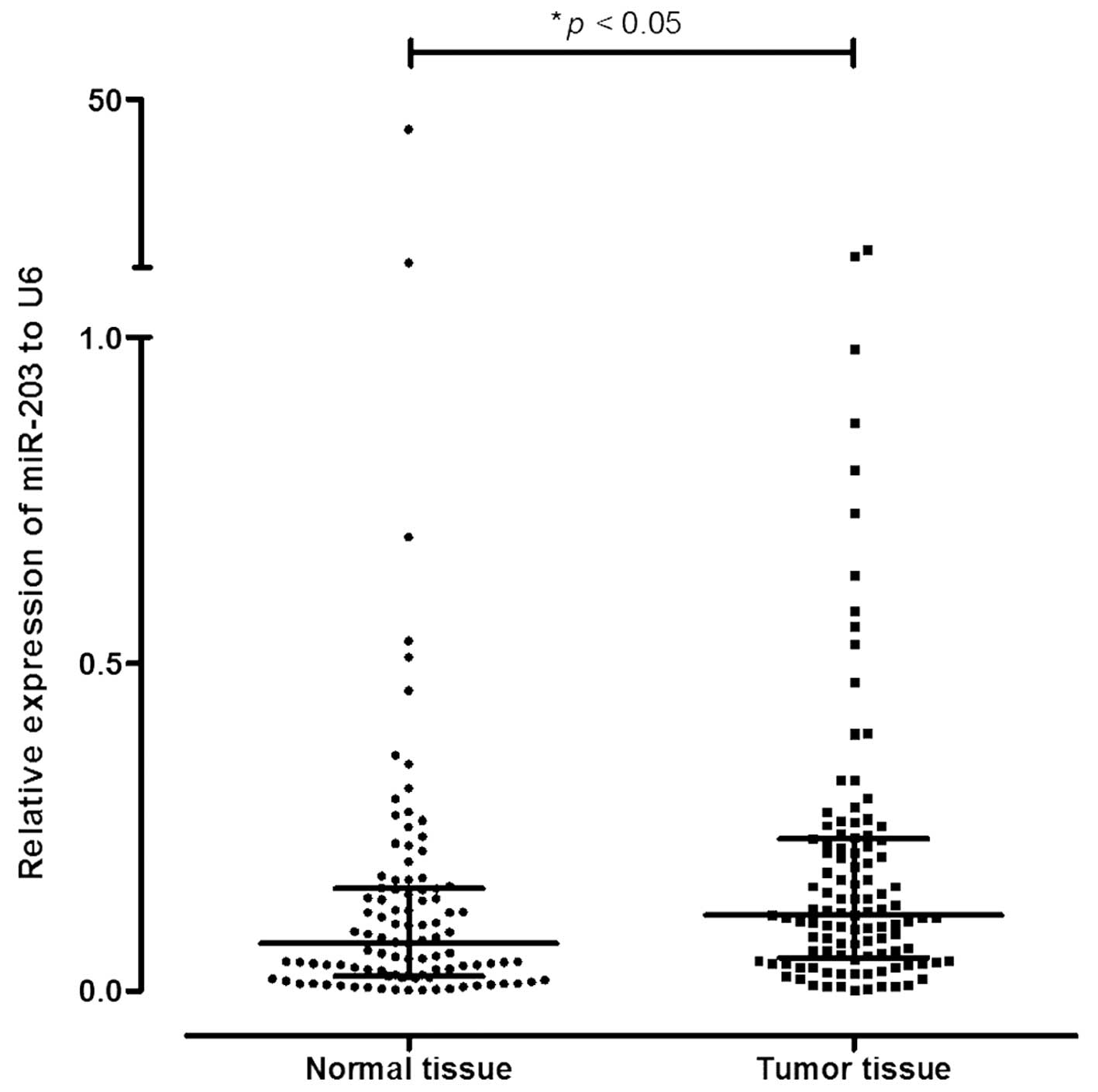
miR-203a is overexpressed in tumor
tissues compared to normal tissues
There was a significant overexpression of miR-203a
in the tumor tissues (1.7-fold higher) compared to the normal
adjacent tissues from the 109 patients (p=0.003; Wilcoxon
signed-rank test to matched samples) (Table I and Fig. 1).
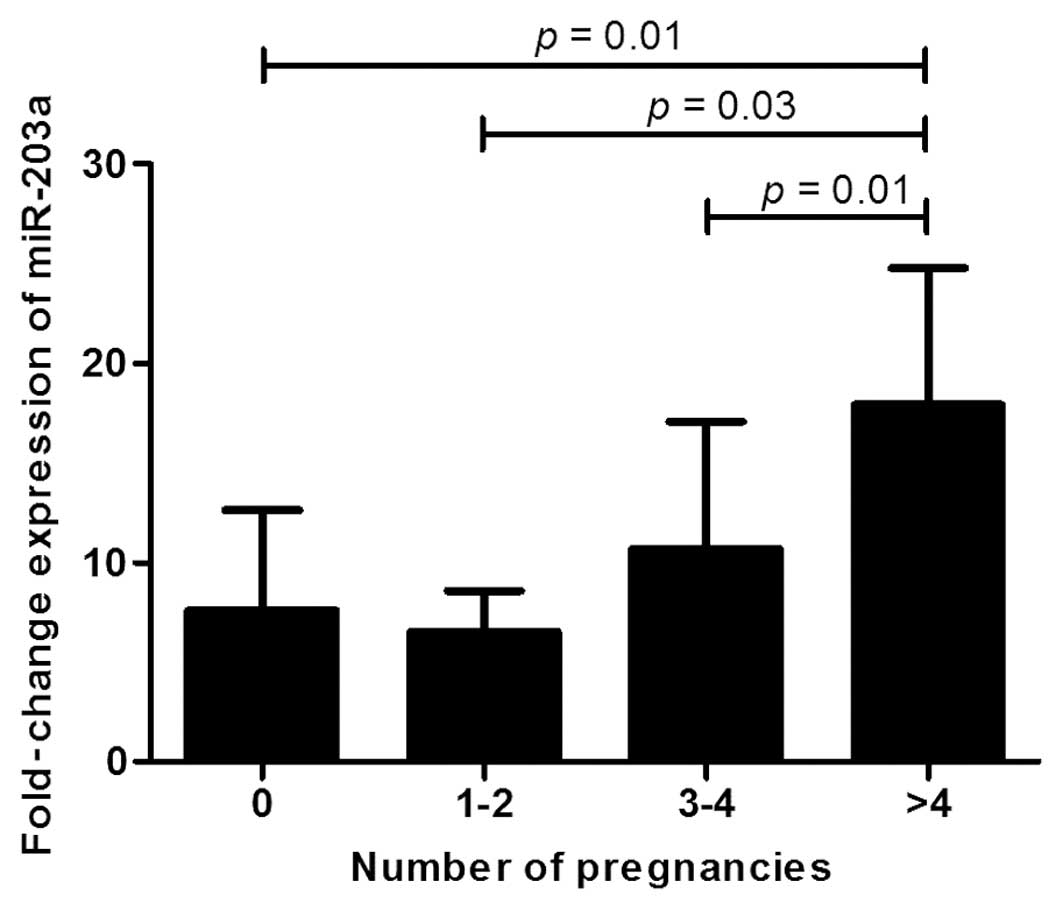
Association between miR-203a expression
and reproductive characteristics
The evaluation of clinical variables (Table I) revealed a significantly different
distribution in the fold-change of expression of miR-203a when
considering the number of pregnancies (Kruskal-Wallis p=0.006;
Fig. 2). Specifically, there was a
higher fold-change of expression in woman with four or more
pregnancies compared to the other classes (no pregnancies vs.
>4, p=0.01; 1–2 vs. >4, p=0.03; 3–4 vs. >4, p=0.01;
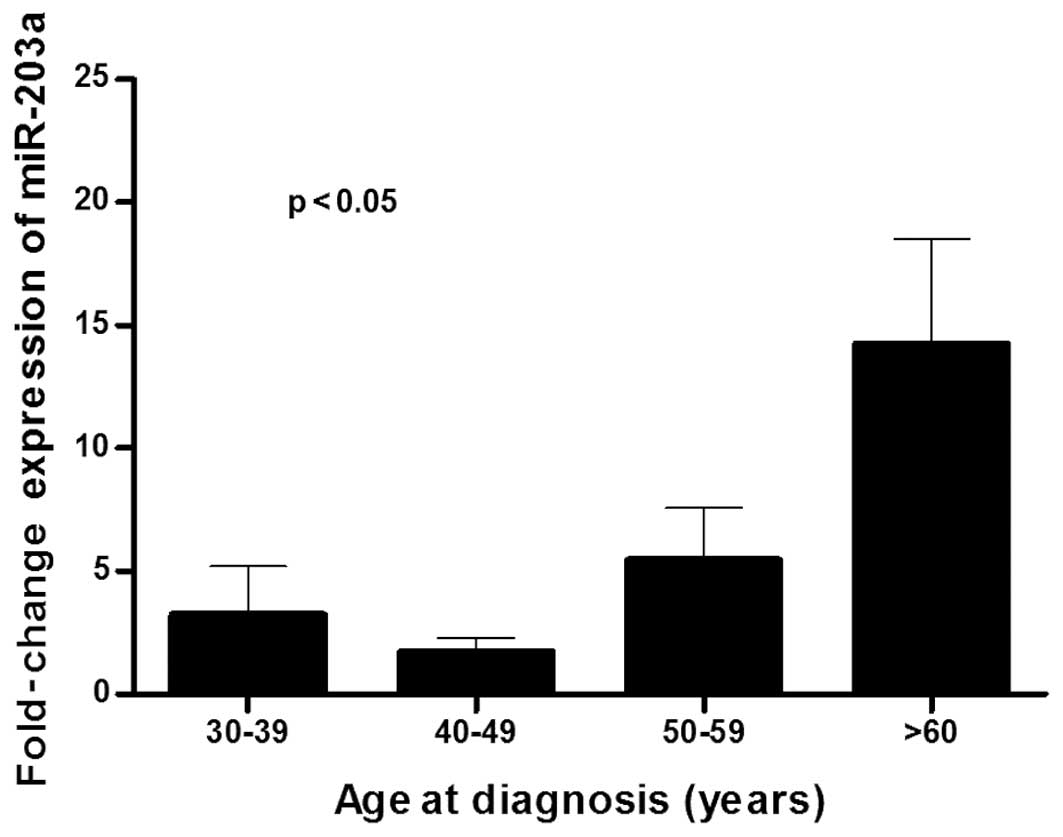
Fig. 2). Significant differences in
age were also found (Fig. 3).
Using Wilcoxon signed-rank test to matched samples
with the same variables, patients >60 years of age on the day of
diagnosis (fold-change, 2.75; p=0.001), with menarche age >13
years (fold-change, 2.33; p=0.003) and menopause <50 years
(fold-change, 3.50; p<0.001), showed a significant
overexpression of miR-203a when comparing tumor tissues with normal
adjacent tissues. Patients diagnosed in post-menopause status
(fold-change, 2.20; p=0.003) and who had <40 years of fertile
status (<30 years: fold-change, 3.75; p=0.041; 30–40 years:
fold-change, 2.40; p=0.005) also presented increased expression of
miR-203a in tumor tissues compared to adjacent normal tissues.
Regarding the number of pregnancies, patients with >4
pregnancies showed a significantly increased expression of miR-203a
in tumor tissues (fold-change, 4.00; p=0.001). These results are in
accordance with the ones described above where Kruskal-Wallis test
was applied. In accordance, although not significantly, women with
>4 children showed an increased expression of miR-203a. Patients
with first childbirth before 20 years of age also showed an
increased expression of miR-203a (fold-change, 3.40; p=0.033).
Breastfeeding status and oral contraceptive consumption also showed
statistically significant results (Table I).
Association between miR-203a expression
and lifestyle characteristics
It is known that various lifestyle habits can be a
risk factor for cancer. In our series we included body mass index
and smoking and alcohol habits. Overweight patients showed an
increase in miR-203a expression in tumor tissues (fold-change,
2.75; p=0.006) and those who did not smoke (fold-change, 2.60;
p=0.001) or sporadically drank alcoholic beverages (fold-change,
3.00; p= 0.037) also showed an increased expression of miR-203a
(Table II).
 | Table IIAssociation of miR-203a relative
expression with the lifestyle habits of the breast cancer
cases. |
Table II
Association of miR-203a relative
expression with the lifestyle habits of the breast cancer
cases.
| n (%) | Median relative
expression of miR-203
| Tumor tissue/normal
tissue | P-valuea |
|---|
| Normal tissue | Tumor tissue |
|---|
| Body mass index, n
(%) |
| Underweight | 2 (1.8) | 0.22 | 0.31 | 1.41 | 0.655 |
| Normal | 42 (38.5) | 0.08 | 0.13 | 1.63 | 0.252 |
| Overweight | 30 (27.5) | 0.04 | 0.11 | 2.75 | 0.006 |
| Obese | 23 (21.1) | 0.06 | 0.11 | 1.83 | 0.073 |
| Missing | 12 (11.1) | | | | |
| Smoking habit, n
(%) |
| No | 73 (67.0) | 0.05 | 0.13 | 2.60 | 0.001 |
| Yes | 22 (20.2) | 0.09 | 0.11 | 1.22 | 0.465 |
| Missing | 14 (12.8) | | | | |
| Alcohol habits, n
(%) |
| No | 54 (49.5) | 0.07 | 0.13 | 1.86 | 0.059 |
| Sporadically | 24 (22.0) | 0.04 | 0.12 | 3.00 | 0.037 |
| Daily | 17 (15.6) | 0.14 | 0.12 | 0.86 | 0.607 |
| Missing | 14 (12.8) | | | | |
Association between miR-203a expression
and clinicopathological characteristics
Several clinicopathological characteristics showed
an association with miR-203a expression (Table III). Tumors with diameter ≤18.5
mm, showed significant difference, albeit with a slight fold-change
of 1.5 compared with adjacent normal tissue (p=0.019), together
with tumors positive for ER (fold-change, 1.71; p=0.042), PR
(fold-change, 1.50; p=0.046), negative for HER2 (fold-change, 1.50;
p=0.016) and Ki-67 index (fold-change, 2.60; p=0.024). Tumors that
did not invade the lymph nodes also presented higher expression of
miR-203a (fold-change, 2.40; p=0.013). With regard to histological
classification, ductal carcinomas in situ (fold-change,
2.20; p=0.028) and invasive carcinoma NOS (fold-change, 1.71;
p=0.009) showed a significantly higher expression of miR-203a.
Stage 0 and II also showed significantly increased expression
(fold-change, 2.20; p=0.028; fold-change, 2.17; p=0.009,
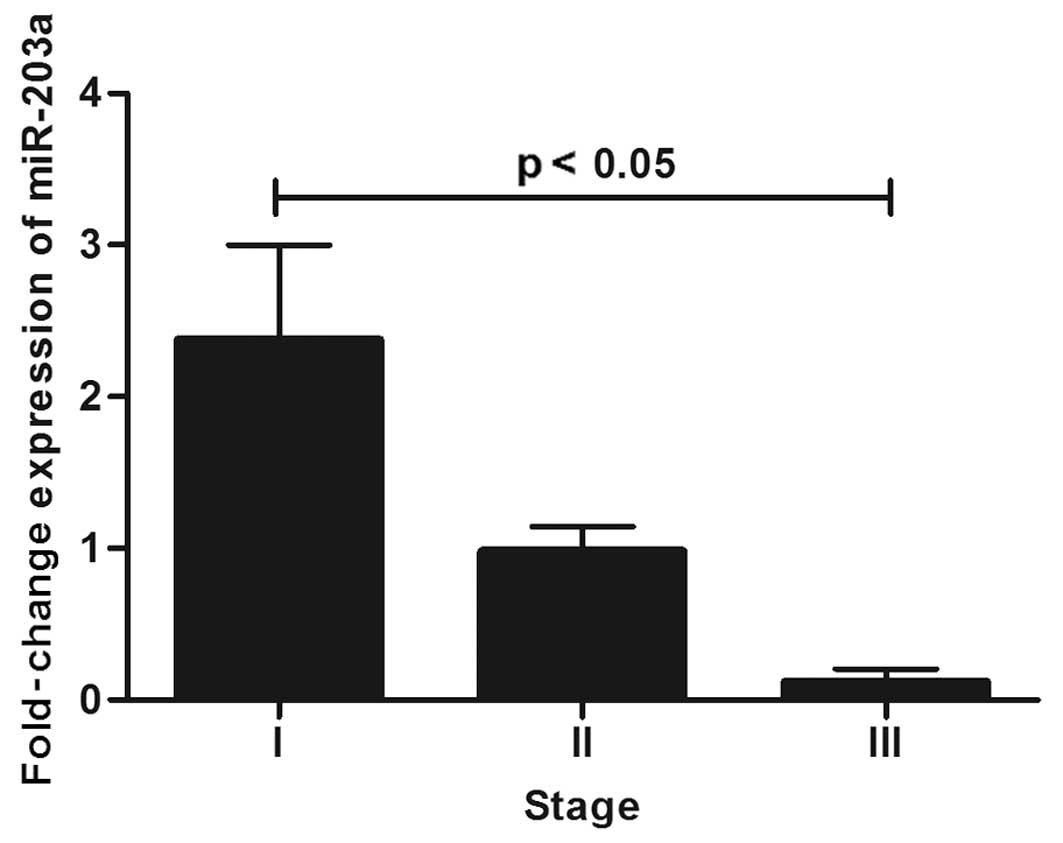
respectively). When considering only invasive lobular tumors
significant differences were found in staging, mainly when
comparing stage III with stage I (Fig.
4).
Discussion
Several studies have established that specific miRNA
expression patterns can be correlated with biological and clinical
features. Studies of miRNA expression patterns in different
populations are of utmost importance in order to unveil the
significance of these molecules in the diagnosis and prognosis of
breast cancer. In the present study, we showed that miR-203a was
overexpressed in tumor tissues when compared to adjacent normal
tissues in a Portuguese cohort. To our knowledge this is the first
study to report miR-203a expression in a Portuguese breast cancer
population. Our results are in accordance with another study by Ru
et al (24). However, they
did not compare adjacent normal tissues with tumor tissues but an
independent disease-free population. The same pattern of
overexpression was also observed in ovarian cancer (39), cervical cancer (40), kidney and bladder cancers (22), colon adenocarcinoma (41) and head and neck squamous cell
carcinoma (42). Conversely,
miR-203 expression levels were decreased in hepatocellular
carcinoma (43). Altogether these
data support the notion that miR-203a plays an important role in
the development of cancer in a tissue-specific manner.
In the present study, we compared miR-203a
expression levels in ductal carcinoma in situ (n=7),
invasive carcinoma NOS (n=91) and invasive lobular carcinoma
(n=10). Our sample population also comprised one mixed tumor
(invasive ductal and lobular carcinoma) but this was not considered
in the statistical analysis of the histological subtypes. Comparing
matched samples, we observed significant differences between
miR-203a levels in the tumor and adjacent normal tissues in the
ductal carcinoma in situ and invasive carcinoma NOS.
However, there were no significant differences between the two
groups. Due to the fact that two different types of breast tumors
were presented, ductal and lobular, we analyzed ductal carcinoma
in situ and invasive carcinoma NOS separately. However, we
did not find significant differences either. Nevertheless, we
highlight the fact that there was a decrease in the miR-203a
expression level in invasive carcinoma NOS when compared with
ductal carcinoma in situ. These results suggest that during
tumor development, miR-203a may be downregulated, thus suggesting
that miR-203a may be implicated in early stages of tumor
development. Indeed, this involvement of miR-203a in invasiveness
through inhibition of the Polycomb group gene BMI1 has
alreadybeen reported in melanoma (34) and non-small cell lung cancer
(26), in which miR-203a expression
levels are inversely correlated with BMI1 expression levels
according to the cell type. Zhang et al (25) also reported an increased expression
of miR-203a in breast tumors compared to matched adjacent normal
tissue, even though their cohort was smaller. Additionally, the
authors determined miR-203a expression levels in several
non-tumorigenic, non-metastatic and metastatic breast cell lines
and showed an increased expression of miR-203a in non-metastatic
compared to non-tumorigenic and metastatic lines. These results led
the authors to speculate that miR-203a is overexpressed in a
protective mechanism to deal with cell proliferation and
invasiveness, and thereafter, most probably through epigenetic
mechanisms, the tumor cells repress miR-203a expression to enable
proliferation, invasion and metastasis through increased expression
of the pro-metastatic gene SNAI2. In fact, our data are in
accordance with this report, since when we stratified the tumors
according to lymph node invasion, the tumors that metastasized had
a decreased expression of miR-203a (fold-change expression, 1.22;
n=53; Table III) compared to
those that did not metastasize to the lymph node (fold-change
expression, 2.40; n=52; Table
III), although the result was not statistically different.
Additionally, we also found that miR-203a had decreased expression
in tumors positive for HER2 and a high level for the proliferation
index Ki-67 (Table III).
Altogether these data are in accordance with the fact that miR-203a
may act as a tumor suppressor, and in early stages of cancer
development miR-203a may play a protective role. Yet, throughout
tumor development, miR-203a may be repressed in order to enable
tumor cells to proliferate, invade and metastasize.
Notably, miR-203a expression decreased from stage 0
to stage I, then increased in stage II, and again decreased in
stage III. This upregulation and downregulation across stages was
unexpected, since as a putative tumor suppressor, miR-203a
expression should decrease with increasing stages. Petrovic et
al (44) observed a similar
pattern in invasive breast carcinomas but with miR-21. miRNA levels
are dependent on cell differentiation, thus displaying differently
expression levels according to stage. Analyzing only invasive
lobular carcinoma tumors, there were significant differences
between stage I (n=2), II (n=6) and III (n=2). Although the number
of samples was small, we observed a pronounced decrease in miR-203a
expression within the stages (Fig.
4). Thus, miR-203a expression levels in invasive lobular
carcinoma can be used as a marker to distinguish different
stages.
Breast cancer risk increases with age. However,
individual risk depends on other factors, including reproductive
history, lifestyle habits and family history, among others. Our
data showed significant differences with age stratification.
Indeed, women over 60 years at diagnosis presented an increased
expression of miR-203a when tumor tissue was compared with adjacent
normal tissue. As estimated by The Surveillance, Epidemiology, and
End Results (SEER) program of the National Cancer Institute
(45), there is a risk increment
for developing breast cancer with age. Our stratification was
carried out using the same criteria, and we observed a higher
expression of miR-203a in patients above 60 years of age. Regarding
age at menarche and age at menopause, known players for breast
cancer, the expression levels were higher for matched samples for
age at menarche >13 years and for age at menopause <50 years.
Although we observed significant differences in matched samples for
the classes referred, there were no differences for age at menarche
and menopause classes indicating that miR-203a expression levels
were not influenced by these factors.
Furthermore, it is known that female hormones, such
as 17-β-estradiol (E2), regulate gene expression by
binding to estrogen receptors (46). Indeed, Yu et al (27), showed that E2 can
regulate miRNA expression and thus control cell proliferation. The
authors showed that miR-16, miR-143 and miR-203a expression was
suppressed after E2 stimulation hence upregulating
bcl-2, cyclin D1 and survivin. Thus, the authors proposed a
mechanism whereby cells that undergo stimulation by E2
have increased proliferation by inhibiting tumor suppressor miRNAs
involved in cell proliferation and survival. Additionally, the
authors ascertained the expression levels of these miRNAs in
triple-positive and triple-negative breast tumors and showed that
they had increased levels of expression in triple-positive tumors,
indicating that these miRNAs may function as tumor suppressors in
triple-positive breast tumors. In contrast, our data showed that
triple-positive samples had lower expression of miR-203a than
triple-negative tumors (data not shown). Indeed, when we stratified
our data according to hormone receptor status, individually, we
obtained always an increased expression level of miR-203a in tumors
with negative receptor status. When sample matching was analyzed we
found significant differences between tumor tissues and adjacent
normal tissues with positive status. To confirm these data, when we
analyzed the samples by stratifying them by molecular subtype, we
observed that basal-like tumors had higher expression of miR-203a.
Although the terms basal-like tumor and triple-negative tumors are
not used interchangeably (3), in
this case we can consider that all basal-like were triple-negative
tumors. Interestingly, women who had used oral contraceptives had
lower expression of miR-203a in these tissues. Thus, miR-203a
expression might be influenced by estrogen and progesterone
(27).
In summary, miR-203a appears to be involved in
breast cancer development, mainly in the early stages of
development. Early-stage tumor cells might upregulate miR-203a in a
self-protective manner in order to manage the augmented cell
proliferation and then, most probably, through epigenetic
mechanisms or E2 mediated suppression, miR-203a might be
downregulated and its targets upregulated. Accordingly, miR-203a
could represent a potential marker for invasiveness. In the present
study, we also showed that miR-203a may be a potential marker to
discriminate stages in invasive lobular carcinoma. Further studies
with larger populations of invasive lobular carcinoma cases must be
performed in order to validate these results.
Acknowledgments
The present study was scientifically supported in
part by the grant PEst-OE/SAU/UI0009/2014 from Fundação de Ciência
e Tecnologia (FCT) and was funded through the Centre for
Toxicogenomics and Human Health - UID/BIM/00009/2013 (FCT). B.C.G.
was supported by a fellowship (SFRH/BD/64131/2009) from FCT.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
De Abreu FB, Schwartz GN, Wells WA and
Tsongalis GJ: Personalized therapy for breast cancer. Clin Genet.
86:62–67. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Badve S, Dabbs DJ, Schnitt SJ, Baehner FL,
Decker T, Eusebi V, Fox SB, Ichihara S, Jacquemier J, Lakhani SR,
et al: Basal-like and triple-negative breast cancers: A critical
review with an emphasis on the implications for pathologists and
oncologists. Mod Pathol. 24:157–167. 2011. View Article : Google Scholar
|
|
4
|
Győrffy B, Hatzis C, Sanft T, Hofstatter
E, Aktas B and Pusztai L: Multigene prognostic tests in breast
cancer: Past, present, future. Breast Cancer Res. 17:112015.
View Article : Google Scholar
|
|
5
|
Vasudevan S, Tong Y and Steitz JA:
Switching from repression to activation: microRNAs can up-regulate
translation. Science. 318:1931–1934. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
He L, Thomson JM, Hemann MT,
Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe
SW, Hannon GJ, et al: A microRNA polycistron as a potential human
oncogene. Nature. 435:828–833. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Takamizawa J, Konishi H, Yanagisawa K,
Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y,
et al: Reduced expression of the let-7 microRNAs in human lung
cancers in association with shortened postoperative survival.
Cancer Res. 64:3753–3756. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K,
Guo J, Zhang Y, Chen J, Guo X, et al: Characterization of microRNAs
in serum: A novel class of biomarkers for diagnosis of cancer and
other diseases. Cell Res. 18:997–1006. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ma L, Reinhardt F, Pan E, Soutschek J,
Bhat B, Marcusson EG, Teruya-Feldstein J, Bell GW and Weinberg RA:
Therapeutic silencing of miR-10b inhibits metastasis in a mouse
mammary tumor model. Nat Biotechnol. 28:341–347. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cheng CJ, Bahal R, Babar IA, Pincus Z,
Barrera F, Liu C, Svoronos A, Braddock DT, Glazer PM, Engelman DM,
et al: MicroRNA silencing for cancer therapy targeted to the tumour
microenvironment. Nature. 518:107–110. 2015. View Article : Google Scholar
|
|
11
|
Rueff J and Rodrigues AS: Cancer drug
resistance: A brief overview from a genetic viewpoint. Methods Mol
Biol. 1395:1–18. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lin S and Gregory RI: MicroRNA biogenesis
pathways in cancer. Nat Rev Cancer. 15:321–333. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Andreasen D, Fog JU, Biggs W, Salomon J,
Dahslveen IK, Baker A and Mouritzen P: Improved microRNA
quantification in total RNA from clinical samples. Methods.
50:S6–S9. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hui A, How C, Ito E and Liu FF: Micro-RNAs
as diagnostic or prognostic markers in human epithelial
malignancies. BMC Cancer. 11:5002011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Graveel CR, Calderone HM, Westerhuis JJ,
Winn ME and Sempere LF: Critical analysis of the potential for
microRNA biomarkers in breast cancer management. Breast Cancer
(Dove Med Press). 7:59–79. 2015.
|
|
17
|
Azam AT, Bahador R, Hesarikia H, Shakeri M
and Yeganeh A: Downregulation of microRNA-217 and microRNA-646 acts
as potential predictor biomarkers in progression, metastasis, and
unfavorable prognosis of human osteosarcoma. Tumour Biol. Jul
31–2015.Epub ahead of print. PubMed/NCBI
|
|
18
|
Sadeghian Y, Kamyabi-Moghaddam Z, Nodushan
SM, Khoshbakht S, Pedram B, Yahaghi E, Mokarizadeh A and Mohebbi M:
Profiles of tissue microRNAs; miR-148b and miR-25 serve as
potential prognostic biomarkers for hepatocellular carcinoma.
Tumour Biol. Jul 25–2015.Epub ahead of print. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang S, Li H, Wang J, Wang D, Yao A and Li
Q: Prognostic and biological significance of microRNA-127
expression in human breast cancer. Dis Markers. 2014:4019862014.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gomes BC, Rueff J and Rodrigues AS:
MicroRNAs and cancer drug resistance. Methods Mol Biol.
1395:137–162. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sonkoly E, Wei T, Janson PC, Sääf A,
Lundeberg L, Tengvall-Linder M, Norstedt G, Alenius H, Homey B,
Scheynius A, et al: MicroRNAs: Novel regulators involved in the
pathogenesis of psoriasis? PLoS One. 2:e6102007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gottardo F, Liu CG, Ferracin M, Calin GA,
Fassan M, Bassi P, Sevignani C, Byrne D, Negrini M, Pagano F, et
al: Micro-RNA profiling in kidney and bladder cancers. Urol Oncol.
25:387–392. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bueno MJ, Pérez de Castro I, Gómez de
Cedrón M, Santos J, Calin GA, Cigudosa JC, Croce CM,
Fernández-Piqueras J and Malumbres M: Genetic and epigenetic
silencing of microRNA-203 enhances ABL1 and BCR-ABL1 oncogene
expression. Cancer Cell. 13:496–506. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ru P, Steele R, Hsueh EC and Ray RB:
Anti-miR-203 upregulates SOCS3 expression in breast cancer cells
and enhances cisplatin chemosensitivity. Genes Cancer. 2:720–727.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang Z, Zhang B, Li W, Fu L, Fu L, Zhu Z
and Dong JT: Epigenetic silencing of miR-203 upregulates SNAI2 and
contributes to the invasiveness of malignant breast cancer cells.
Genes Cancer. 2:782–791. 2011. View Article : Google Scholar
|
|
26
|
Chen T, Xu C, Chen J, Ding C, Xu Z, Li C
and Zhao J: MicroRNA-203 inhibits cellular proliferation and
invasion by targeting Bmi1 in non-small cell lung cancer. Oncol
Lett. 9:2639–2646. 2015.PubMed/NCBI
|
|
27
|
Yu X, Zhang X, Dhakal IB, Beggs M,
Kadlubar S and Luo D: Induction of cell proliferation and survival
genes by estradiol-repressed microRNAs in breast cancer cells. BMC
Cancer. 12:292012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang C, Zheng X, Shen C and Shi Y:
MicroRNA-203 suppresses cell proliferation and migration by
targeting BIRC5 and LASP1 in human triple-negative breast cancer
cells. J Exp Clin Cancer Res. 31:582012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hailer A, Grunewald TG, Orth M, Reiss C,
Kneitz B, Spahn M and Butt E: Loss of tumor suppressor mir-203
mediates overexpression of LIM and SH3 protein 1 (LASP1) in
high-risk prostate cancer thereby increasing cell proliferation and
migration. Oncotarget. 5:4144–4153. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ding X, Park SI, McCauley LK and Wang CY:
Signaling between transforming growth factor β (TGF-β) and
transcription factor SNAI2 represses expression of microRNA miR-203
to promote epithelial-mesenchymal transition and tumor metastasis.
J Biol Chem. 288:10241–10253. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Taipaleenmäki H, Browne G, Akech J, Zustin
J, van Wijnen AJ, Stein JL, Hesse E, Stein GS and Lian JB:
Targeting of Runx2 by miR-135 and miR-203 impairs progression of
breast cancer and metastatic bone disease. Cancer Res.
75:1433–1444. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Madhavan D1, Zucknick M, Wallwiener M, Cuk
K, Modugno C, Scharpff M, Schott S, Heil J, Turchinovich A, Yang R,
et al: Circulating miRNAs as surrogate markers for circulating
tumor cells and prognostic markers in metastatic breast cancer.
Clin Cancer Res. 18:5972–5982. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Imaoka H1, Toiyama Y, Okigami M, Yasuda H,
Saigusa S, Ohi M, Tanaka K, Inoue Y, Mohri Y and Kusunoki M:
Circulating microRNA-203 predicts metastases, early recurrence, and
poor prognosis in human gastric cancer. Gastric Cancer. Aug
2–2015.Epub ahead of print. PubMed/NCBI
|
|
34
|
Chang X, Sun Y, Han S, Zhu W, Zhang H and
Lian S: miR-203 inhibits melanoma invasive and proliferative
abilities by targeting the polycomb group gene BMI1. Biochem
Biophys Res Commun. 456:361–366. 2015. View Article : Google Scholar
|
|
35
|
Singletary SE, Allred C, Ashley P, Bassett
LW, Berry D, Bland KI, Borgen PI, Clark G, Edge SB, Hayes DF, et
al: Revision of the American Joint Committee on Cancer staging
system for breast cancer. J Clin Oncol. 20:3628–3636. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hammond ME, Hayes DF, Dowsett M, Allred
DC, Hagerty KL, Badve S, Fitzgibbons PL, Francis G, Goldstein NS,
Hayes M, et al: American Society of Clinical Oncology/College Of
American Pathologists guideline recommendations for
immuno-histochemical testing of estrogen and progesterone receptors
in breast cancer. J Clin Oncol. 28:2784–2795. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wolff AC, Hammond ME, Hicks DG, Dowsett M,
McShane LM, Allison KH, Allred DC, Bartlett JM, Bilous M,
Fitzgibbons P, et al American Society of Clinical Oncology; College
of American Pathologists: Recommendations for human epidermal
growth factor receptor 2 testing in breast cancer: American Society
of Clinical Oncology/College of American Pathologists clinical
practice guideline update. J Clin Oncol. 31:3997–4013. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Dowsett M, Nielsen TO, A'Hern R, Bartlett
J, Coombes RC, Cuzick J, Ellis M, Henry NL, Hugh JC, Lively T, et
al International Ki-67 in Breast Cancer Working Group: Assessment
of Ki67 in breast cancer: Recommendations from the International
Ki67 in Breast Cancer working group. J Natl Cancer Inst.
103:1656–1664. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Iorio MV, Visone R, Di Leva G, Donati V,
Petrocca F, Casalini P, Taccioli C, Volinia S, Liu CG, Alder H, et
al: MicroRNA signatures in human ovarian cancer. Cancer Res.
67:8699–8707. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Gocze K, Gombos K, Juhasz K, Kovacs K,
Kajtar B, Benczik M, Gocze P, Patczai B, Arany I and Ember I:
Unique microRNA expression profiles in cervical cancer. Anticancer
Res. 33:2561–2567. 2013.PubMed/NCBI
|
|
41
|
Schetter AJ, Leung SY, Sohn JJ, Zanetti
KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK, et
al: MicroRNA expression profiles associated with prognosis and
therapeutic outcome in colon adenocarcinoma. JAMA. 299:425–436.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
de Carvalho AC, Scapulatempo-Neto C, Maia
DC, Evangelista AF, Morini MA, Carvalho AL and Vettore AL: Accuracy
of microRNAs as markers for the detection of neck lymph node
metastases in patients with head and neck squamous cell carcinoma.
BMC Med. 13:1082015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Liu Y, Ren F, Rong M, Luo Y, Dang Y and
Chen G: Association between underexpression of microrna-203 and
clinicopathological significance in hepatocellular carcinoma
tissues. Cancer Cell Int. 15:622015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Petrović N, Mandušić V, Dimitrijević B,
Roganović J, Lukić S, Todorović L and Stanojević B: Higher miR-21
expression in invasive breast carcinomas is associated with
positive estrogen and progesterone receptor status in patients from
Serbia. Med Oncol. 31:9772014. View Article : Google Scholar
|
|
45
|
Howlader N, Noone AM, Krapcho M, Neyman N,
Aminou R, Waldron W, Altekruse SF, Kosary CL, Ruhl J, Tatalovich Z,
et al: SEER Cancer Statistics Review, 1975–2009 (Vintage 2009
Populations). National Cancer Institute; Bethesda, MD: http://seer.cancer.gov/csr/1975_2009_pops09/,
based on November 2011 SEER data submission, posted to the SEER web
site. April. 2012
|
|
46
|
Marino M, Galluzzo P and Ascenzi P:
Estrogen signaling multiple pathways to impact gene transcription.
Curr Genomics. 7:497–508. 2006. View Article : Google Scholar
|