Introduction
Colorectal cancer (CRC) is one of the most common
malignancies associated with high recurrence incidence and poor
prognosis (1). Although diagnostic
methods and comprehensive treatment have been developed over the
past decades, the mortality rate of CRC remains as the second and
third highest-ranked cancer in male and female patients,
respectively, in the USA (2) and is
ranked fifth in China (3). The
liver is the most common metastatic site of CRC. It is estimated
that 15–25% of patients with CRC have synchronous liver metastasis,
which is detected at the time of the initial diagnosis. Another 20%
of patients with CRC have heterochronous liver metastases, which
typically develops following resection of the primary tumor
(4). Liver metastasis is one of the
leading causes of cancer-associated mortalities in patients with
CRC. The 5-year overall survival rate of patients with liver
metastatic CRC is only 25–40%, which is markedly lower compared
with patients without liver metastasis (69.5–95.7%) (5). Since the morbidity and mortality of
liver metastatic CRC remains high, identifying the causes,
molecular mechanisms and molecular biomarkers for early prediction
and personalized therapy is important and urgently required.
MicroRNAs (miRNAs) are short (21–25 nucleotide)
non-coding RNAs involved in the translational and
post-transcriptional regulation of gene expression. miRNAs promote
the translational repression or degradation of their targets by
binding to the complementary sequences in the 3′-untranslated
region of target mRNAs, thus affecting multiple signaling pathways
(6). Recent studies have suggested
the critical role of miRNAs (miRs) in various physiological and
pathological processes in CRC, including metastasis (7). For example, miR-21, miR-31, miR-103,
miR-107 and miR-122 have been implicated in tumor local invasion;
and the miR-17-91 cluster (miR-17, miR-18a, miR-19a/b, miR-20a and
miR-92a), miR-21, miR-126 and miR-155 may be involved in tumor
intravasation, circulation and extravasation (7). Therefore, identifying diagnostic and
therapeutic miRNAs could additionally be of great clinical
significance.
The microarray technique is increasingly being used
for life science research purposes. Bioinformatics data-mining of
gene and non-coding RNA expression microarray data is a useful tool
for identifying novel significant genes and non-coding RNAs
involved in the pathogenesis of diseases, providing valuable
insights and a basis for further novel research (8). In the present study, 3 gene expression
profile datasets, GSE41258 (9),
GSE68468 (10) and
GSE81582-GPL15207 (11), and 4miRNA
expression profile datasets, GSE35834 (12), GSE44121, GSE72199 (13) and GSE81582-GPL16384 (11) were downloaded from the Gene
Expression Omnibus (GEO). The GEO2R online tool was used to
identify the differentially expressed (DE) genes (DEGs) and DE
miRNAs of primary CRC tumor tissues and liver metastatic CRC tumor
tissues. Gene Ontology (GO) function and Kyoto Encyclopedia of
Genes and Genomes (KEGG) pathways enrichment analyses were
conducted for the DEGs. A protein-protein interaction (PPI) network
of the DEGs was constructed and the hub genes with a higher degree
of connectivity were selected. Target genes of these DE miRNAs were
predicted using online datasets, and novel miRNA-mRNA regulatory
axes were constructed, which were expected to serve vital roles in
the liver metastasis of CRC, and to thus serve as novel biomarkers
for diagnosis and therapy of CRC.
In the present study, 3 DE miRNAs were identified,
namely miR-10b, miR-122 and miR-885. Among them, the function and
mechanism of miR-885 in tumorigenesis and progression remain
unclear. Previous studies revealed that miR-885 served as a tumor
suppressor by reducing matrix metalloproteinase-9 (MMP-9)
expression (14), repressing
cyclin-dependent kinase 2 and DNA replication licensing factor MCM5
expression (15) and inhibiting
Wnt/β-catenin signaling (16). A
recent study observed that miR-885 was overexpressed in CRC liver
metastasis and induced CRC metastasis by targeting cytoplasmic
polyadenylation element-binding protein 2 (CPEB2) (17). In the present study, the results
demonstrated that miR-885 promoted metastasis of CRC in
vitro, and that von Willebrand factor (vWF) and insulin-like
growth factor binding protein 5 (IGFBP5) are potential novel target
genes of miR-885, indicating that the miR-885/vWF and
miR-885/IGFBP5 axes may serve roles in the metastasis of CRC.
Materials and methods
Microarray data
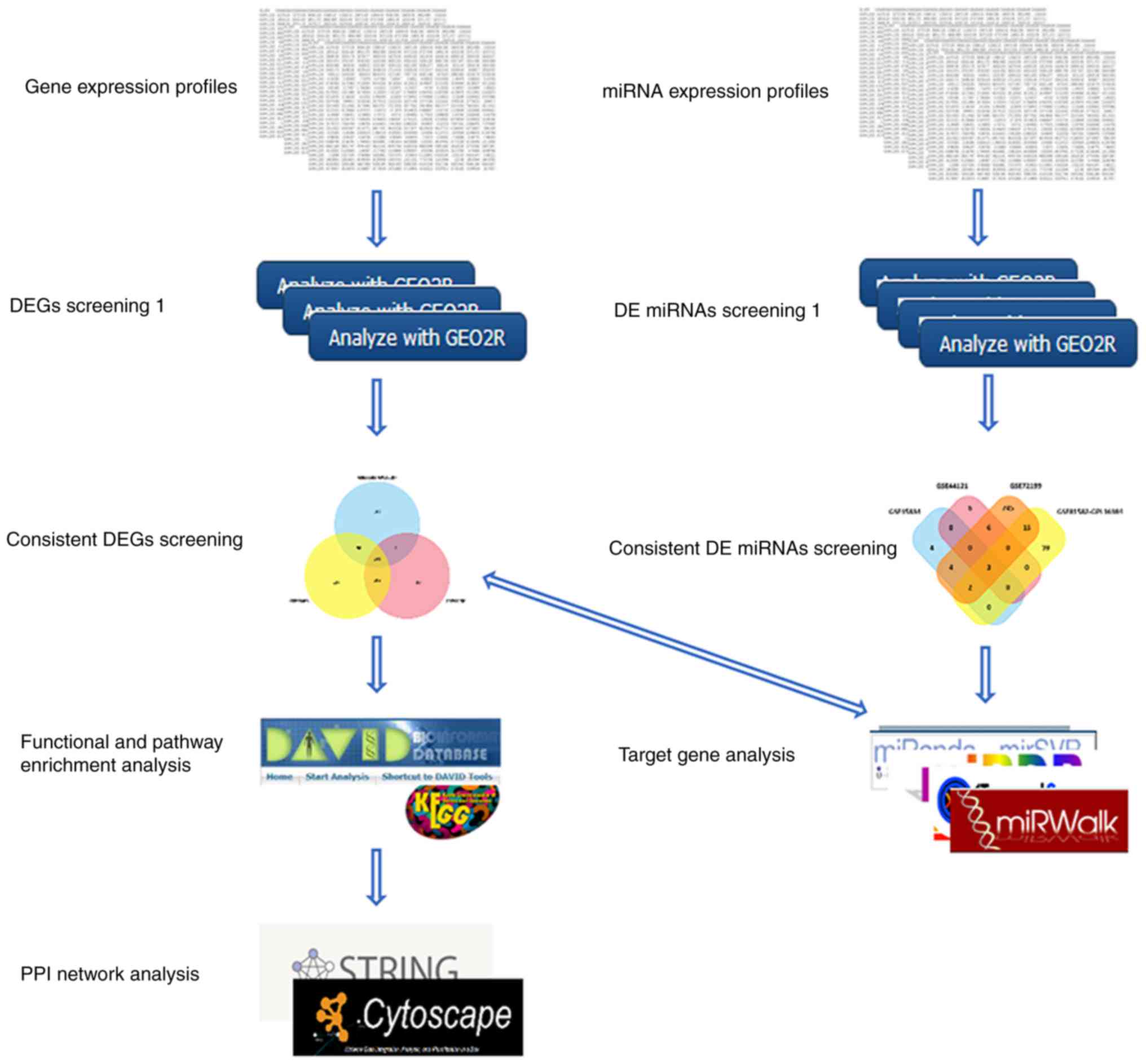
The bioinformatics analysis was conducted following
the procedure presented in Fig. 1.
The primary CRC tumor tissues and CRC liver metastatic tumor tissue
gene expression profiles of GSE41258, GSE68468 and
GSE81582-GPL15207, and the miRNA expression profiles of GSE35834,
GSE44121, GSE72199, and GSE81582-GPL16384, were obtained from
NCBI-GEO (http://www.ncbi.nlm.nih.gov/geo), a free database of
microarray profiles and next-generation sequencing. The microarray
data of GSE41258 and GSE68468 were based on the GPL96 platform
([HG-U133A] Affymetrix Human Genome U133A Array), including 186
primary CRC tumor tissues and 47 CRC liver metastatic tumor
tissues. GSE81582-GPL15207 was based on the GPL15207 platform
([PrimeView] Affymetrix Human Gene Expression Array), including 23
primary CRC tumor tissues and 19 CRC liver metastatic tumor
tissues. GSE35834 was based on the GPL8786 platform [(miRNA-1)
Affymetrix Multispecies miRNA-1 Array], including 31 primary CRC
tumor tissues and 24 CRC liver metastatic tumor tissues. GSE44121
was based on the GPL16231 platform (Nanostring nCounter Human
microRNA Expression Platform), including 9 primary CRC tumor
tissues and 9 CRC liver metastatic tumor tissues. GSE72199 was
based on the GPL15018 platform [Agilent-031181
Unrestricted_Human_miRNA_V16.0_Microarray 030840 (Feature Number
version)], including 28 primary CRC tumor tissues and 8 CRC liver
metastatic tumor tissues. GSE81582-GPL16384 was based on GPL16384
[(miRNA-3) Affymetrix Multispecies miRNA-3 Array], including 23
primary CRC tumor tissues and 19 CRC liver metastatic tumor
tissues.
Identification of DEGs and DE
miRNAs
The GEO2R online tool (https://www.ncbi.nlm.nih.gov/geo/geo2r/), which allows
users to compare two or more groups of samples in a GEO series, was
applied to detect DEGs and DE miRNAs of primary CRC tumors and CRC
liver metastatic tumors. P<0.05 and |log fold change (FC)| ≥1
were set as the cut-off criteria.
GO functional analysis and KEGG
pathway enrichment analysis of DEGs
GO functional analysis is a useful method for
annotating genes and identifying characteristic biological
attributes for high-throughput genome or transcriptome data
(18). KEGG incorporates a wide
range of databases, including those on genomes, biological
pathways, diseases, drugs and chemical substances (19). The GO functional analysis and KEGG
pathway enrichment analysis of DEGs were conducted using Database
for Annotation, Visualization and Integrated Discovery (DAVID;
http://david.ncifcrf.gov/), an online
bioinformatics database that aims to provide tools for the
functional interpretation of genes or proteins (20). P<0.05 was set as the cut-off
criterion.
PPI network and module analysis
The online database STRING (http://string-db.org) (21) was used to develop a PPI network.
Cytoscape software (22) was used
to construct a PPI network and analyze the interactions of the
DEGs. The cytoHubba plug-in was used to identify hub genes. The
Molecular Complex Detection (MCODE) plug-in was used to screen
modules of the PPI network in Cytoscape with a degree cut-off =2,
node score cut-off =0.2, k-core =2 and max depth =100. The KEGG
pathway enrichment analysis of genes in the top module was
performed using DAVID.
Predicting the target genes of DE
miRNAs
To search for the targets of DE miRNAs, the miRanda
(http://www.mocrorna.org) (23), PITA (http://genie.weizmann.ac.il/pubs/mir07/mir07_data.html)
(24), TargetScan (http://www.targetscan.org/) (25), miRDB (http://www.mirdb.org/) (26) and miRWalk (http://mirwalk.uni-hd.de/) (27) online databases were used. The genes
identified in ≥4 databases were considered as targets of DE miRNAs.
The regulatory network between DE miRNAs and DEGs, which screened
for predicted target genes, was further constructed to search for
key molecules and axes in CRC liver metastasis.
Comparison of the expression level of
target genes
The Gene Expression Profiling Interactive Analysis
(GEPIA; http://gepia.cancer-pku.cn/index.html) is an online
tool, which delivers fast and customizable analysis for RNA
sequencing expression data based on The Cancer Genome Atlas and
GTEx databases, which include data on 9,736 tumors and 8,587 normal
samples (28). GEPIA provides key
interactive and customizable functions, including differential
expression analysis between cancer and normal tissues. Through its
use, it could be determined whether the expression of target genes
of DE miRNAs differed between CRC tissues and normal mucosa. A
boxplot was constructed to visualize any altered expression.
Cell culture and transfection
The human CRC cell lines SW480 and LoVo, obtained
from the Cell Bank of Chinese Academy of Sciences (Shanghai,
China), were cultured in Dulbecco's modified Eagle's medium (DMEM;
Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) with 10%
fetal bovine serum (FBS) (ScienCell Research Laboratories, Inc.,
San Diego, CA, USA) and 1% penicillin/streptomycin (Gibco; Thermo
Fisher Scientific, Inc.). Cells were cultured at 37°C in an
incubator with 5% CO2.
SW480 and LoVo cells were cultured in 12-well plates
and transfected with miR-885 mimics, inhibitor and miRNA negative
control (NC) sequences that were purchased from Shanghai GenePharma
Co., Ltd. (Shanghai, China). Transfection was conducted using
Lipofectamine® 3000 (Thermo Fisher Scientific, Inc.) The
cells were harvested at 48 h after transfection for reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
analysis, and migration analysis.
Wound-healing assay
A scratch wound-healing assay was used to assess
SW480 and LoVo cell migration. Subsequent to transfection for 48 h,
the cells were digested and a total of 3×105 cells/well
were seeded into 12 well-plates and cultured overnight. Wounds were
subsequently created using 10-µl sterile tips. Cell migration
distances were measured using an inverted microscope (Olympus
Corp., Tokyo, Japan) at 0, 24 and 36 h.
Transwell migration assay
A Transwell migration assay was performed using
24-well Transwell plates with 8-µm pores (Corning Incorporated,
Corning, NY, USA) to assess the migration of SW480 and LoVo cells.
In total, 1×105 cells in 200 µl FBS-free DMEM were
placed in the upper Transwell chamber. Subsequently, 700 µl 30% FBS
DMEM was added to the lower chamber as a chemoattractant. Following
cell culture for 24 h, the non-invasive cells in the upper chamber
were washed with cotton swabs. Migrated cells at the bottom of the
membrane were fixed with 4% paraformaldehyde and stained with 0.1%
crystal violet for visualization. Cells were counted in five
respective fields at an ×200 magnification using a light microscope
(Olympus Corp.).
RNA extraction and RT-qPCR
Total RNA was extracted from cells with
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol. Subsequently, cDNA
was synthesized from 2 µg total RNA and quantification of miR-885
was performed using an miRNA 1st Strand cDNA Synthesis kit (by
stem-loop; Vazyme Biotech Co., Ltd., Nanjing, China). Real-time PCR
was performed using an Applied Biosystems StepOne Real-Time PCR
system, using ChamQ™ SYBR® qPCR Master Mix (High ROX
Premixed; Vazyme), containing 5 ng cDNA and 10 pM of each primer.
The cycling conditions consisted of 1 cycle at 95°C for 30 sec; 40
cycles at 95°C for 10 sec; and 60°C for 30 sec. Melting curve
analysis was conducted for each PCR reaction to confirm the
specificity of amplification. The concentration of miR-885 was
calculated based on the quantification cycle (Cq), and the relative
expression levels were calculated as 2−∆∆Cq [∆∆Cq =
(CqmiR-885 - CqU6) mimics or inhibitor -
(CqmiR-885 - CqU6) NC)] following
normalization with reference to the quantification of U6
expression.
As for the target genes, RT was conducted with
HiScript® II Q RT SuperMix for qPCR (+gDNA wiper;
Vazyme) according to the manufacturer's protocol. The expression
level of target genes was normalized to GAPDH and calculated as
2−∆∆Cq. The primers wereas follows: U6 forward (F):
TCGCTTCGGCAGCACATA and reverse (R): TTTGCGTGTCATCCTTGC]; miR-885
(F: GCGCGTCCATTACACTACCCT and R: AGTGCAGGGTCCGAGGTATT); IGFBP5 (F:
TGAAGGCTGAAGCAGTGAAGAAGG and R: TTGTCCACGCACCAGCAGATG); vWF (F:
GGCGGCAACAGGACCAACAC and R: GGAGGAGCCATCCAGGAGAAGG); and GAPDH (F:
ACAACTTTGGTATCGTGGAAGG and R: GCCATCACGCCACAGTTTC).
Statistical analysis
All data were analyzed using GraphPad Prism 6.0
statistical software (GraphPad Software, Inc., La Jolla, CA, USA)
and SPSS 22.0 (IBM Corp., Armonk, NY, USA). The two-tailed paired
Student's t-test was conducted for the analysis of two groups.
One-way analysis of variance (ANOVA) test and Bonferroni post hoc
test were used for evaluating differences among multiple groups.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Screening for DEGs
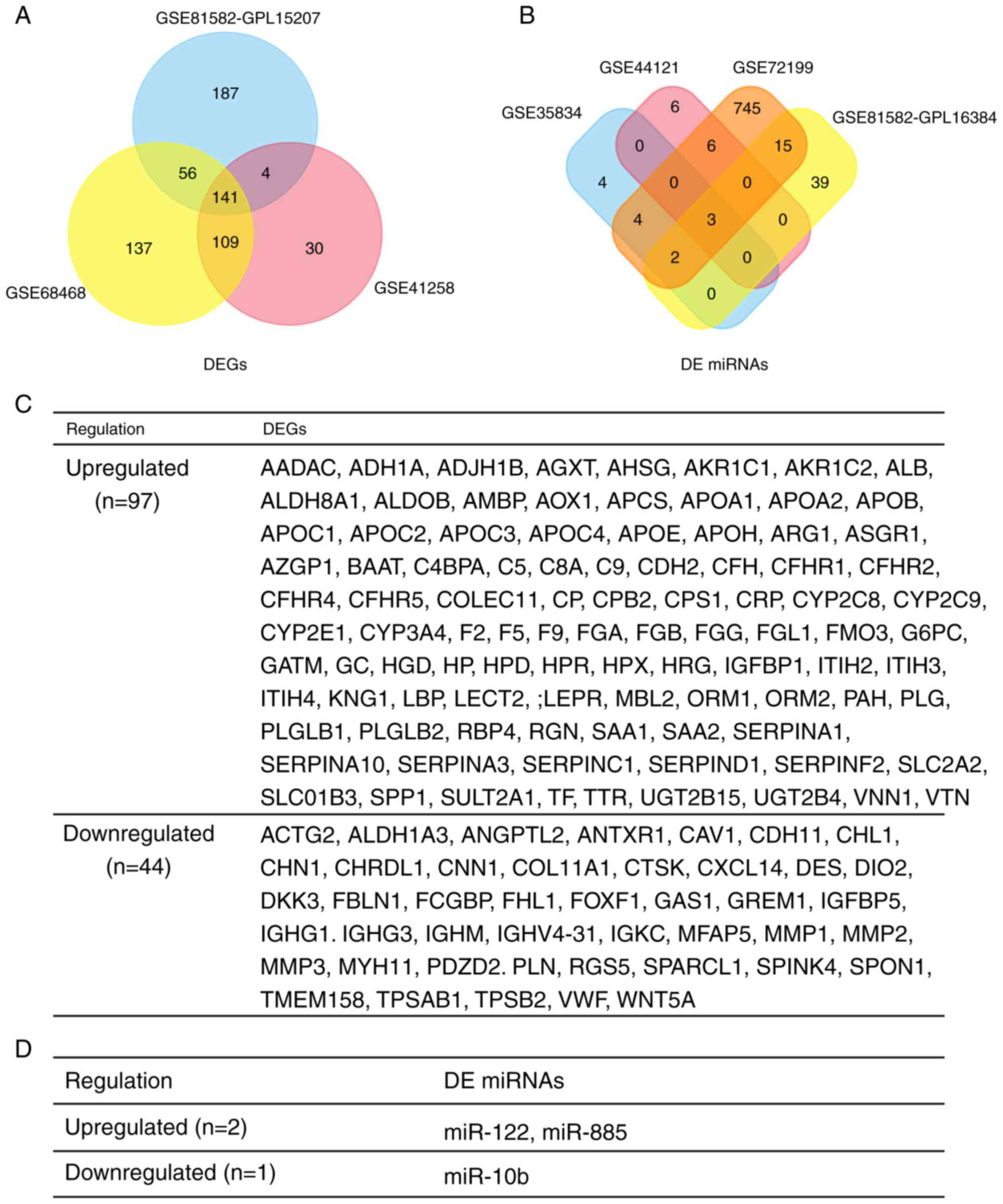
In total, 284, 444 and 397 DEGs were extracted from
the gene expression profile datasets GSE41258, GSE68468 and
GSE81582-GPL15207, respectively, using P<0.05 and |logFC| ≥1 as
cut-off criteria. Subsequently, 13, 15, 776 and 59 DE miRNAs were
extracted from the miRNA expression profile datasets GSE35834,
GSE44121, GSE72199 and GSE81582-GPL16384, respectively. Following
integrated bioinformatics analysis, a total of 141 consistent DEGs
(including 97 upregulated genes and 44 downregulated genes) and 3
consistent DE miRNAs (2 upregulated and 1 downregulated) were
identified from the 3 gene and 4 miRNA expression profile datasets
(Fig. 2A and B), respectively, in
the primary CRC tumor tissues, compared with liver metastatic CRC
tumor tissues (Fig. 2C and D).
GO functional analysis and KEGG
pathway enrichment analysis
DAVID was used to conduct GO functional analysis and
KEGG pathway enrichment analysis. GO describes genes from three
aspects, namely molecular function (MF), cellular component (CC)
and biological process (BP) (29).
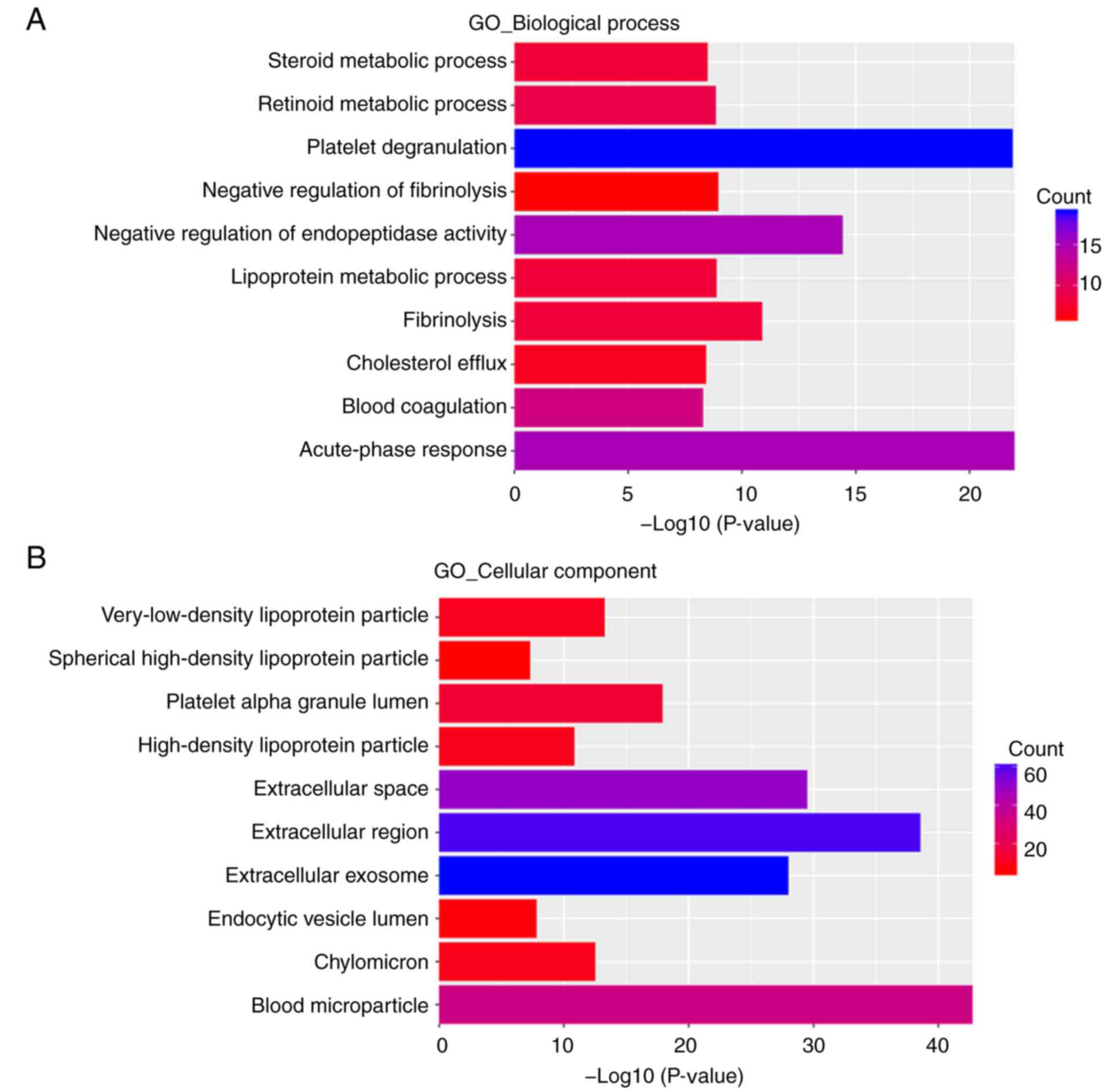
In the BP group, upregulated DEGs were primarily enriched in
‘acute-phase response’, ‘platelet degranulation’, ‘negative
regulation of endopeptidase activity’, ‘fibrinolysis’, ‘negative
regulation of fibrinolysis’, ‘lipoprotein metabolic process’,
‘retinoid metabolic process’, ‘steroid metabolic process’,
‘cholesterol efflux’ and ‘blood coagulation’ (Fig. 3A). Downregulated DEGs were primarily
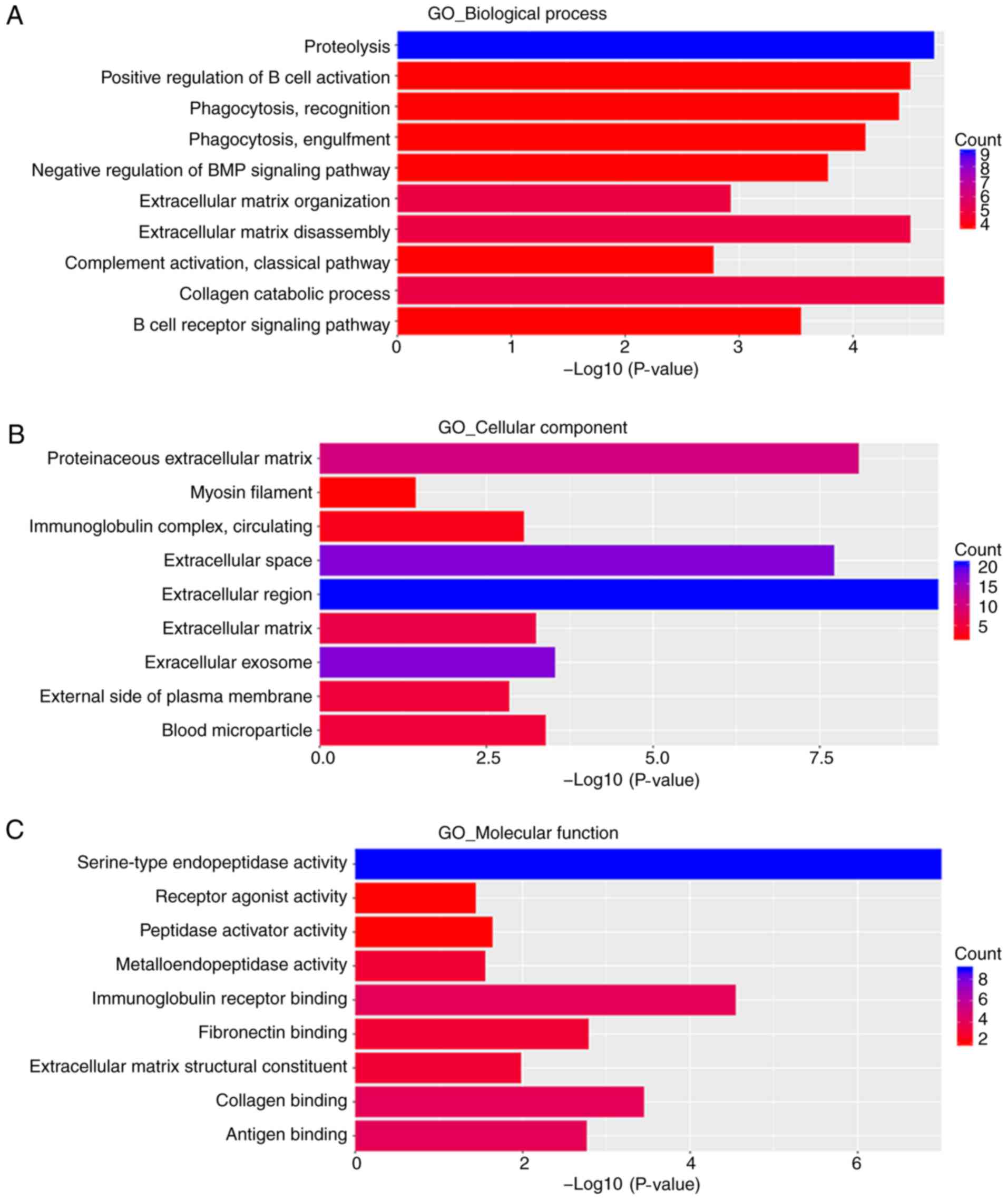
enriched in ‘collagen catabolic process’, ‘proteolysis’,
‘extracellular matrix disassembly’, ‘positive regulation of B cell
activation’, ‘phagocytosis, recognition’, ‘phagocytosis,
engulfment’, ‘negative regulation of BMP signaling pathway’, ‘B
cell receptor signaling pathway’, ‘extracellular matrix
organization’ and ‘complement activation, classical pathway’
(Fig. 4A).
In the CC group, upregulated DEGs were primarily
enriched in ‘blood microparticle’, ‘extracellular region’,
‘extracellular space’ and ‘extracellular exosome’ (Fig. 3B). Downregulated DEGs were primarily
enriched in the ‘extracellular region’, ‘proteinaceous
extracellular matrix’, ‘extracellular space’, ‘extracellular
exosome’ and ‘blood microparticle’ (Fig. 4B).
In the MF group, upregulated DEGs were primarily
enriched in ‘serine-type endopeptidase inhibitor activity’, ‘lipase
inhibitor activity’, ‘heparin binding’, ‘endopeptidase inhibitor
activity’, ‘receptor binding’, ‘phosphatidylcholine-sterol
O-acyltransferase activator activity’, ‘lipid transporter
activity’, ‘phospholipid binding’, ‘monooxygenase activity’ and
‘cholesterol transporter activity’ (Fig. 3C). Downregulated DEGs were primarily
enriched in ‘serine-type endopeptidase activity’, ‘immunoglobulin
receptor binding’, ‘collagen binding’, ‘fibronectin binding’,
‘antigen binding’, ‘extracellular matrix structural constituent’,
‘peptidase activator activity’, ‘metalloendopeptidase activity’ and
‘receptor agonist activity’ (Fig.
4C).
In the KEGG pathway enrichment analysis, upregulated
DEGs were primarily enriched in ‘complement and coagulation
cascades’, ‘drug metabolism-cytochrome P450’, ‘metabolism of
xenobiotics by cytochrome P450’, ‘chemical carcinogenesis’,
‘retinol metabolism’, ‘steroid hormone biosynthesis’, ‘tyrosine
metabolism’, ‘staphylococcus aureus infection’, ‘linoleic acid
metabolism’ and ‘metabolic pathways’ (Fig. 3D).
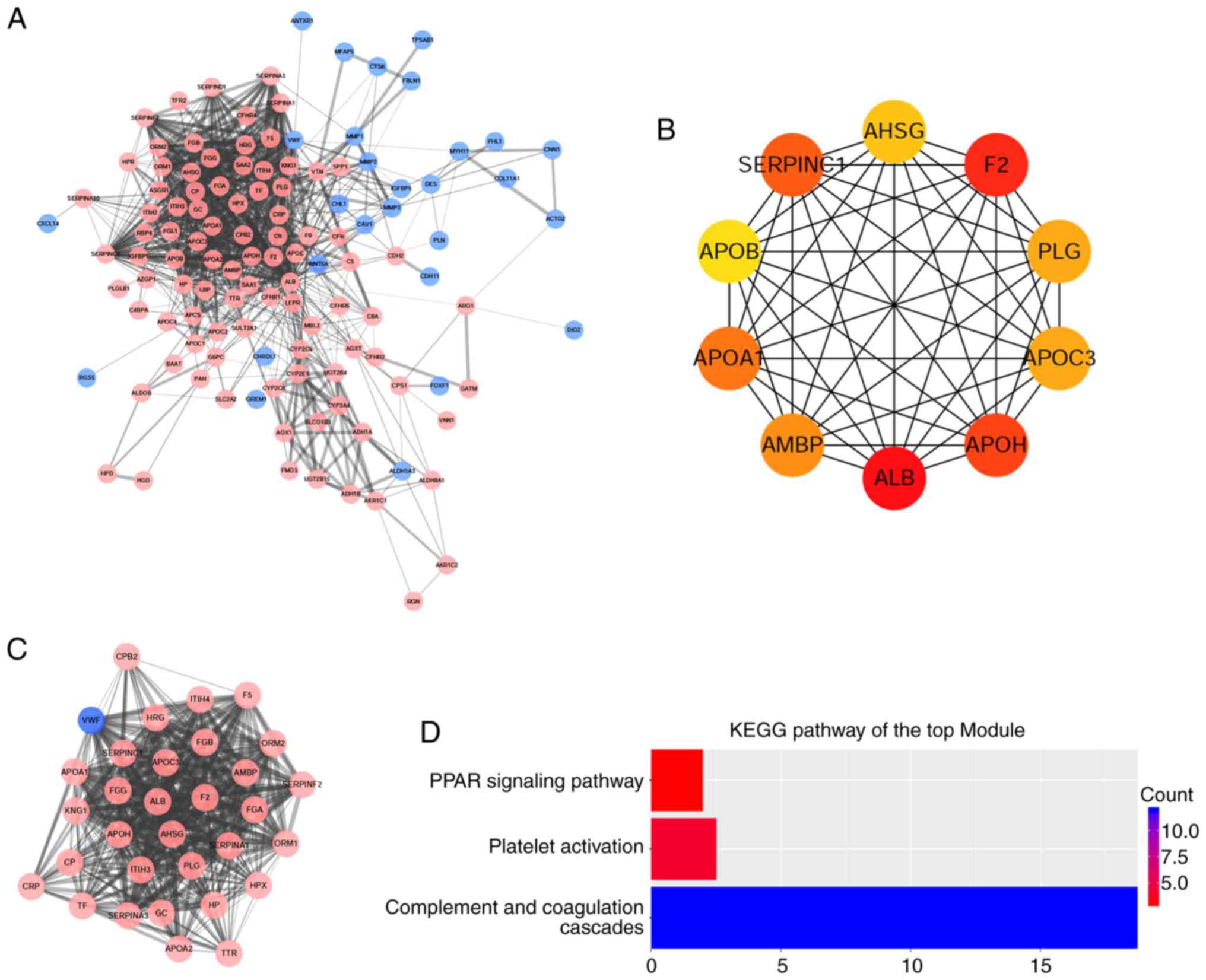
Hub gene identification with DEGs PPI
and modular analysis
Using the STRING online database and Cytoscape
software, a total of 122 DEGs (94 upregulated and 28 downregulated
genes) of the 141 commonly altered DEGs were filtered into the DEGs
PPI network complex, containing 93 nodes and 971 edges (Fig. 5A), and 19 of the 141 DEGs were not
included in the DEGs PPI network. The top 10 hub genes, including
albumin (ALB), coagulation factor II, thrombin (F2), alipoprotein H
(APOH), serpin family C member 1 (SERPINC1), apolipoprotein A1
(APOA1), α-1-microglobulin/bikunin precursor (AMBP), apolipoprotein
C3 (APOC3), plasminogen (PLG), α-2 HS glycoprotein (AHSG) and
apolipoprotein B-100 (APOB), were identified using the cytoHubba
plug-in, with a higher degree of connectivity (Fig. 5B). On the basis of the degree of
importance, the most significant module was detected from the PPI
network complex using the MCODE plug-in, which included 32 nodes
and 399 edges (Fig. 5C). The KEGG
pathway enrichment analysis demonstrated that the module primarily
associated with ‘complement and coagulation cascades’, ‘platelet
activation’ and ‘the PPAR signaling pathway’ (Fig. 5D).
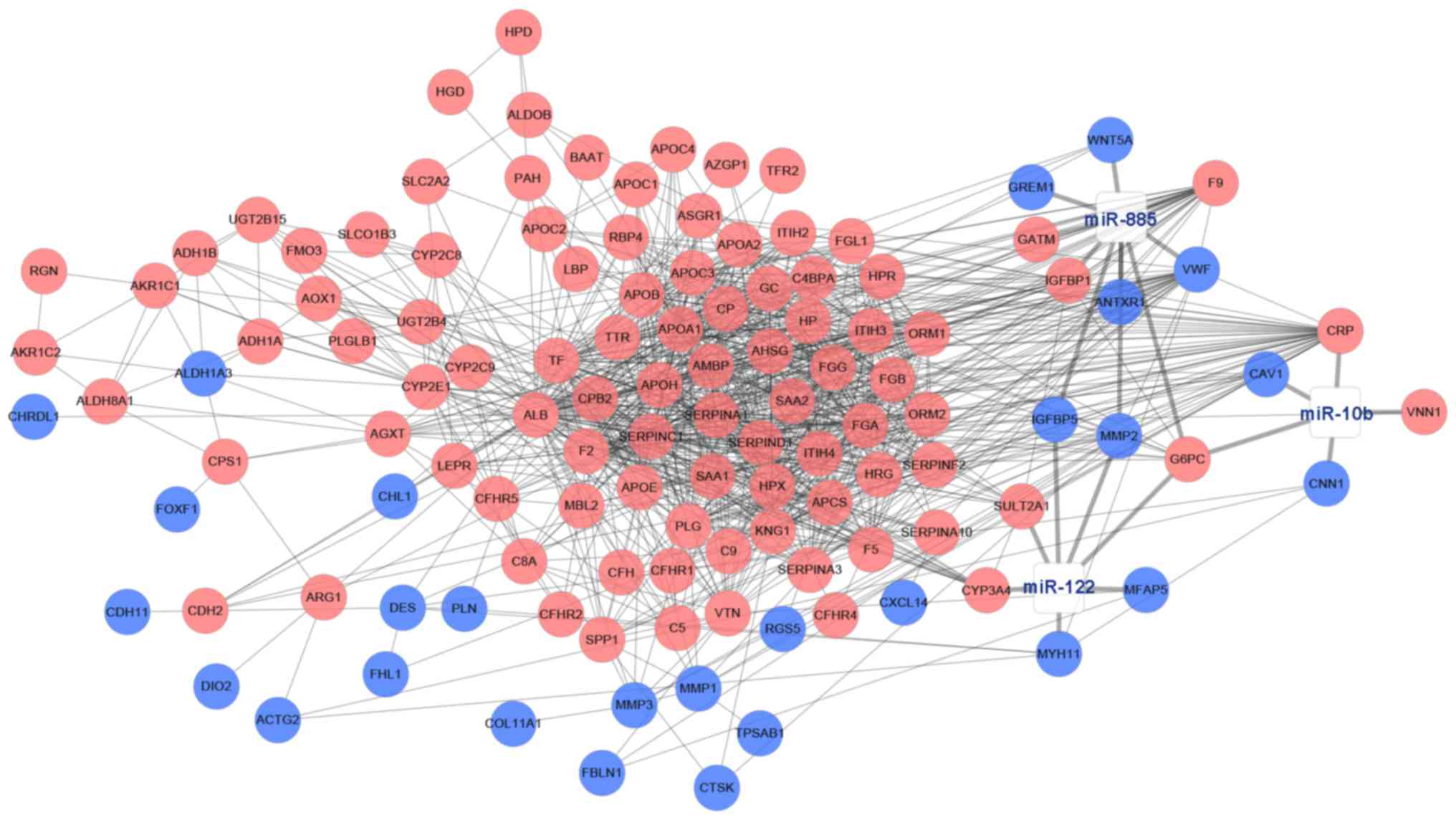
DE miRNA-DEGs regulatory network
construction
Subsequent to predicting the target genes of 3 DE
miRNAs (miR-10b, miR-122 and miR-885) from five databases,
integrated analysis between predicted target genes and DEGs were
conducted to select the target DEGs. In total, 20 target DEGs were
obtained, and a novel DE miRNA-DEGs regulatory network in CRC liver
metastasis was constructed (Fig.
6).
Comparison of DE miRNAs and the
expression level of target genes
In order to further examine the role of DE miRNAs
and these target DEGs in CRC, further analysis of the expression
level of DE miRNAs in CRC tumor tissues and normal mucosa in
GSE35834 and GSE81582-GPL16384 was conducted. Similar significant
expression differences of these DE miRNAs were identified between
CRC tumor and normal mucosa (Fig. 7A
and B). The GEPIA database was used to compare the expression
level of these genes between normal mucosa and CRC tissues. Among
the 20 target DEGs, consistent with the downregulation in liver
metastases, the expression of IGFBP5, microfibril associated
protein 5 (MFAP5), myosin heavy chain 11 (MYH11) and vWF were
significantly downregulated (P<0.05; |logFC| ≥1) in CRC tissues
compared with normal mucosa (Fig.
7C).
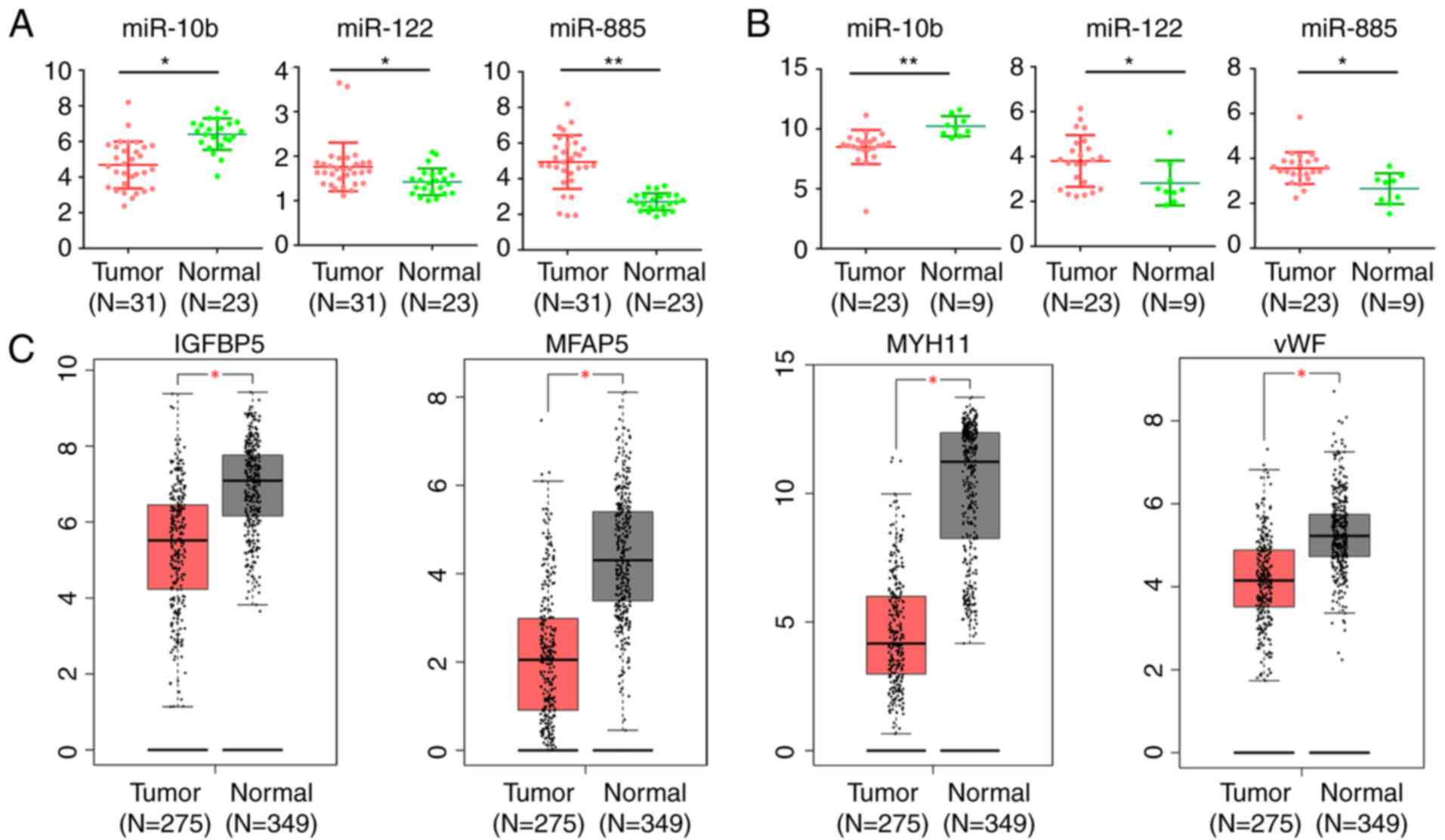 | Figure 7.Expression level of DE miRs and
target DEGs in CRC and normal mucosa. Expression level of miR-10b,
miR-122 and miR-885 in CRC and normal mucosa in (A) GSE35834 and
(B) GSE81582-GPL16384. *P<0.05, **P<0.01. (C) The expression
level of IGFBP5, MFAP5, MYH11 and vWF in CRC and normal mucosa
based on TCGA database analyzed by GEPIA. The red plot represents
tumor tissues and the grey plot represents normal mucosa.
*P<0.05 and |logFC|≥1 were set as the criteria. DE,
differentially expressed; miR, microRNA; DEGs, differentially
expressed genes; CRC, colorectal cancer; IGFBP5, insulin-like
growth factor binding protein 5; MFAP5, microfibril associated
protein 5; MYH11, myosin heavy chain 11; TCGA, The Cancer Genome
Atlas; FC, fold change; vWF, von Willebrand factor; GEIPA, Gene
Expression Profiling Interactive Analysis. |
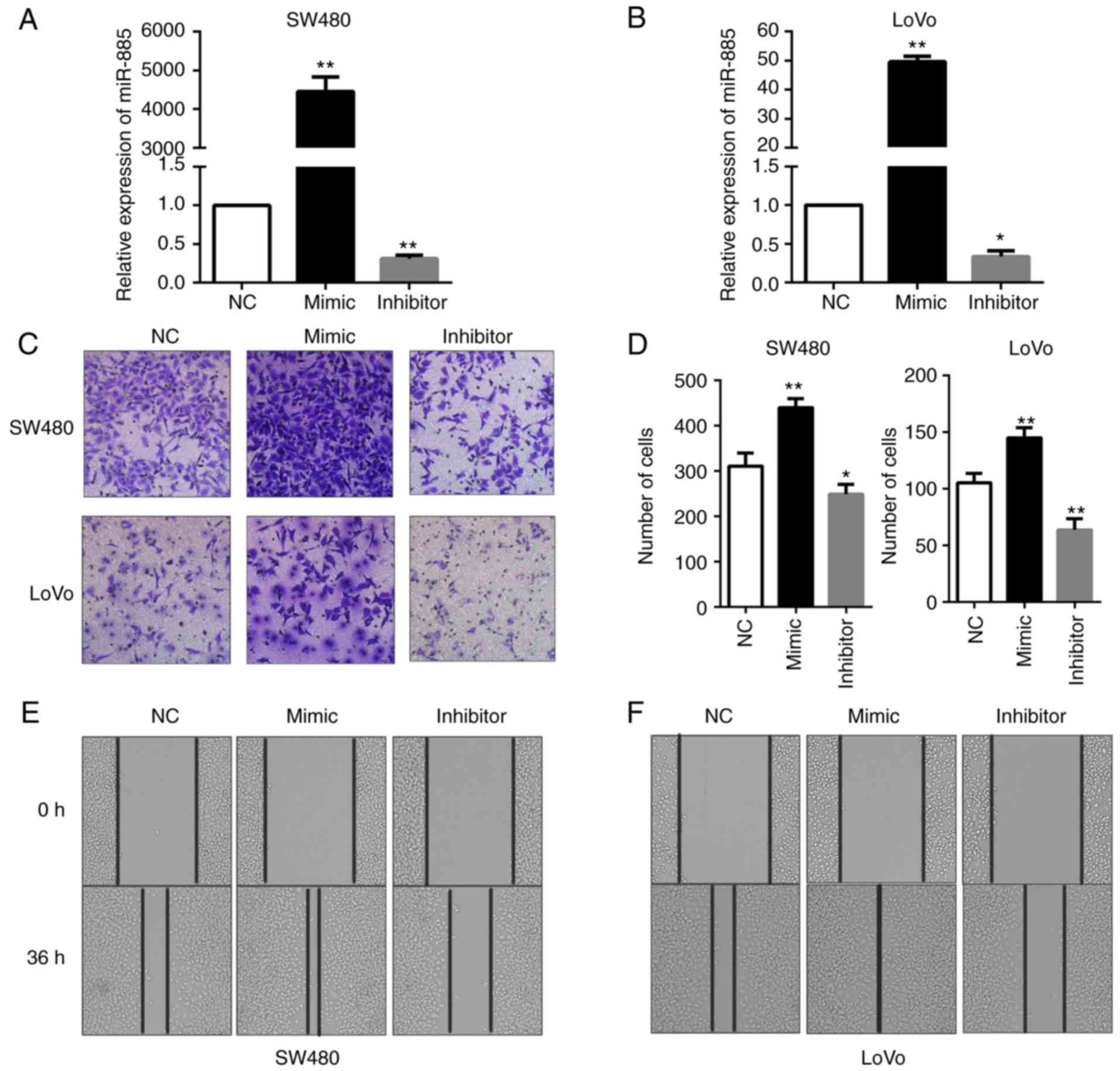
miR-885 promotes CRC cell migration in
vitro
The role of miR-122 and miR-10b in CRC has been
widely investigated; however, few previous studies have focused on
the role of miR-885 in CRC. To evaluate the possible functions of
miR-885 in CRC progression, miR-885 mimics and inhibitor were
transfected into SW480 and LoVo cells to obtain cells with
overexpression and low expression of miR-885 (Fig. 8A and B). The effects of miR-885 on
the migration of CRC cells were observed by Transwell migration
assays and wound healing assays. It was revealed that
overexpression of miR-885 promoted the migration abilities of the
CRC cells in vitro. Additionally, the inhibition of miR-885
suppressed the migration abilities of CRC (Fig. 8C-F).
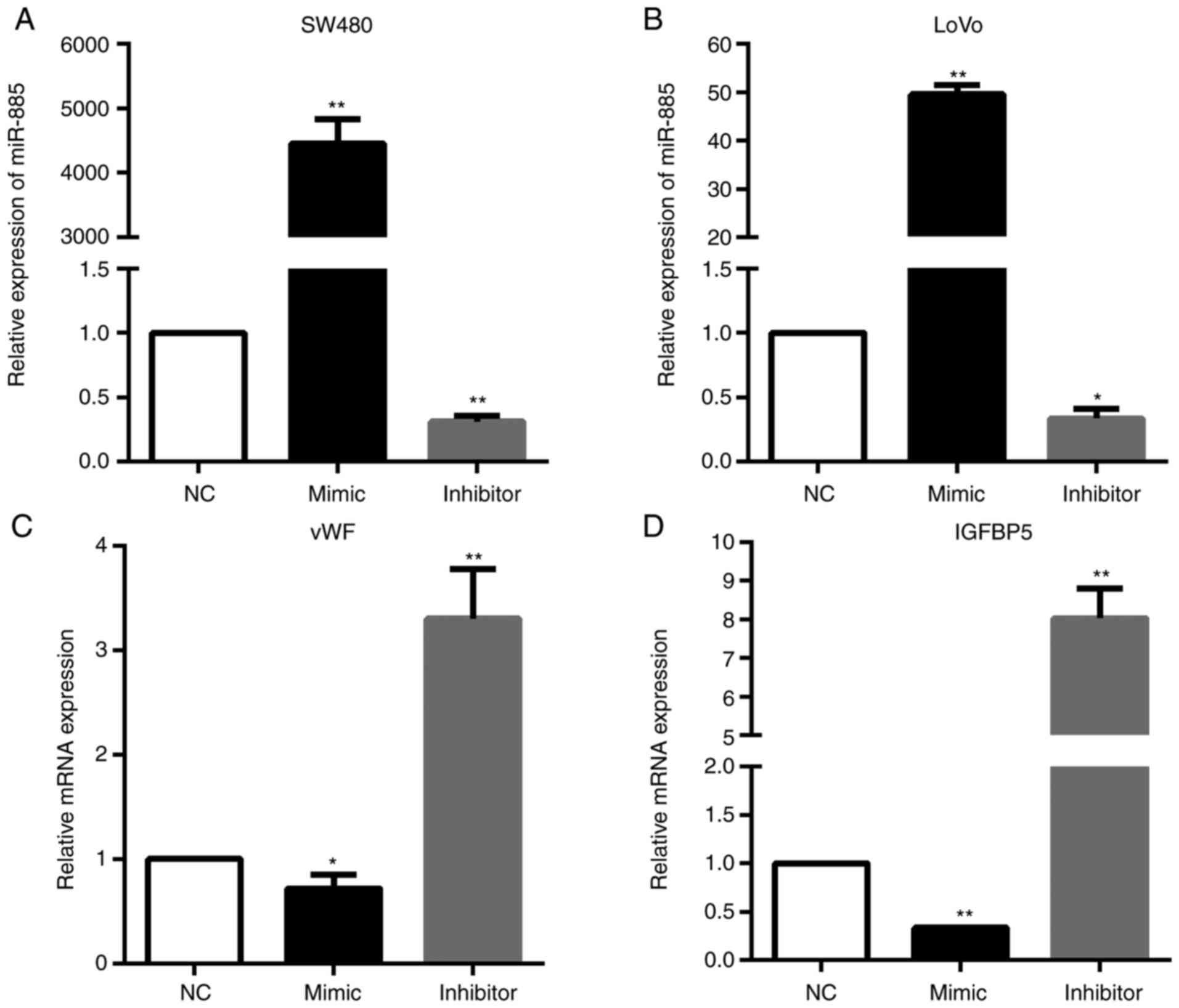
miR-885 decreases vWF and IGFBP5
expression in CRC
In order to verify the regulatory networks of
miR-885/vWF and miR-885/IGFBP5, PCR analysis was performed. The
results demonstrated that the vWF and IGFBP5 mRNA expression levels
were significantly downregulated in miR-885-overexpressing cells;
whereas, these expression levels were upregulated in the cells with
low expression of miR-885 (Fig.
9).
Discussion
A number of pre-clinical and clinical studies have
been conducted to reveal the underlying mechanisms of CRC liver
metastasis in the past decades; however, the incidence and
mortality of CRC liver metastasis remain high. This is primarily
due to the majority of the studies focusing on a single genetic
event, or the results were generated from a single cohort study
(30). In the present study, 3 gene
expression profile datasets were integrated and 141 commonly
altered DEGs (97 upregulated and 44 downregulated) were identified.
The 141 DEGs were classified into three groups (BP, CC and MF
groups) by GO terms, and the KEGG pathway enrichment analysis of
the DEGs was conducted using the DAVID database. Finally, the DEGs
PPI network was constructed, and the top 10 hub genes and the most
significant module was filtered from the PPI network.
Through integrated bioinformatics analysis, 10 hub
genes were identified, including ALB, F2, APOH, SERPINC1, APOA1,
AMBP, APOC3, PLG, AHSG and APOB with a high degree of connectivity.
Among them, APOH, APOA1, APOC3 and APOB belong to the
apolipoprotein family, and their biological functions are primarily
involved in the lipoprotein metabolism process and cholesterol
transport (31). Multiple previous
studies have demonstrated that apolipoproteins additionally serve a
role in the occurrence and development of varies malignancies
(32). APOA1 is the principal
protein constituent of high-density lipoprotein that interact with
ATP-binding cassette subfamily A member 1, ATP-binding cassette
subfamily G member 1 and lecithin cholesterol acyltransferase to
promote cholesterol efflux from cells to the liver for excretion
(33). By eliminating excessive
cholesterol, APOA1 exerts anti-inflammatory, anti-apoptotic and
antioxidant activities, thus serving a protective role against
cancer; chronic inflammation, oxidative stress, lipids and
cholesterol have all been associated with tumorigenesis (33). In addition, APOA1 suppresses tumor
growth by inhibiting myeloid-derived suppressor cells and their
immuno-suppression for tumor growth, invasion and metastasis, by
decreasing MMP-9 expression (34).
A recent study demonstrated that serum APOA1 was significantly
decreased in patients with CRC (35), and that a low serum APOA1 expression
level was associated with poor survival and advanced stage in CRC
(36). However, the present
bioinformatics analysis demonstrated a significant increase in
APOA1 expression level in liver metastatic CRC tissues, indicating
a stimulatory effect of APOA1 in CRC metastasis. The role of APOB,
APOC3 and APOH in the tumorigenesis and metastasis of CRC remains
unclear. However, a high APOB expression level was associated with
increased lung cancer incidence (32), and the APOB-100 gene polymorphisms
may be associated with increased risk of gallstones and gallbladder
cancer (37). APOC3 was
overexpressed in multiple hepatocellular carcinoma (HCC) family
case studies and may be involved in HCC familial aggregation
(38). APOH, additionally termed β2
glycoprotein I, was observed to affect endothelial cell growth and
angiogenesis (39), and serve roles
in lipopolysaccharide-induced signal transduction involved in
promoting the development of HCC (40). Therefore, further pre-clinical and
clinical studies are required to determine the exact role and
mechanisms of these apolipoproteins in CRC liver metastasis.
Furthermore, other hub genes may additionally be
involved in the metastasis of CRC. AMBP is a glycoprotein that is
normally highly expressed in the liver and may be detected in
plasma and urine (41). AMBP is
able to hydrolyze into two distinct functional proteins,
α-1-microglobulin and bikunin, which are involved in multiple
biological processes, including cell growth, cellular calcium
uptake and inflammation (41). At
present, there is no study, to the best of our knowledge, examining
the role of AMBP in CRC. However, it was documented that AMBP was
overexpressed in patients with gastric cancer, and a high
expression level of serum AMBP was associated with poor response to
paclitaxel-capecitabine chemotherapy in patients with advanced
gastric cancer (41). AHSG is a
negative acute phase protein in humans, primarily produced by the
liver. Accumulating evidence suggests that it is a multifunctional
protein capable of modulating the etiology of diabetes and other
metabolic diseases (42), and
promoting the invasion of tumor cells (43). Furthermore, upregulated AHSG was
determined in CRC, which may be used as a diagnostic marker for CRC
(44). The PLG system serves a
crucial role in the process of immune response, angiogenesis,
invasion and metastasis, which are crucial for cancer progression
(45). A previous study
demonstrated that PLG deficiency markedly reduced the number of
pulmonary metastases in a mouse breast cancer model, indicating the
stimulatory role of PLG in tumor metastasis (46).
Additionally, the most significant module was
filtered from the PPI network, among which the majority of the
corresponding genes were mostly associated with complement and
coagulation cascades. It was demonstrated that cancer increases the
risk of thrombosis by a 4.1-fold (47), and results in the hyperactivation of
coagulation and clotting abnormalities in cancer, referred to as
Trousseau's syndrome. Additional symptoms include venous
thromboembolism, deep vein thrombosis, disseminated intravascular
coagulation and pulmonary embolism (48). Recent evidence has demonstrated that
hypercoagulation and activation of complement cascades promote the
pro-tumorigenic phenotype of immune cells and protect tumor cells
from immune attack, ultimately favoring tumor development,
progression and metastases formation (49).
Tumor-promoting inflammation serves an important
role in carcinogenesis and cancer progression. As an important
component of tumor-promoting inflammation, activation of the
complement system promotes cancer cell proliferation,
dedifferentiation and migration (50). Complement activation regulates the
adaptive immune response and may play a role in regulating the
T-cell response to tumors (50). In
addition, specific experimental and clinical evidence suggests a
reciprocal interaction between complement and coagulation.
Complement may induce hyperactivation of the coagulation cascade by
modifying cellular membranes, inducing platelet activation and
aggregation, and stimulating the production of tissue factor in
human neutrophils (49).
The formation of CRC metastases in liver requires
primary CRC cells to adapt to growing in a different and harsh
environment. This adaptation requires multiple alterations in gene
expression, which may be translationally and post-transcriptionally
regulated through the up- or downregulation of multiple miRNAs
(51). Therefore, 3 gene expression
profile datasets were integrated, common DEGs were identified and
bioinformatics methods were used to analyze these datasets
thoroughly. Furthermore, 3 miRNA expression profile datasets were
integrated and 3 common DE miRNAs (two upregulated miRNAs, miR-122
and miR-885, and one downregulated miRNA, miR-10b) were identified.
Additionally, similarly significant expression differences of these
DE miRNAs were additionally observed between CRC tumor and normal
mucosa, suggesting an oncogenetic or tumor suppressive role of
these DE miRNAs. In agreement with the present study, recent
studies observed an upregulation of miR-885 and miR-122, and a
downregulation of miR-10b in liver metastatic CRC tissues,
suggesting that they serve as metastasis-specific miRNAs (52,53).
In addition, high miR-885 expression was statistically
significantly associated with poor prognosis in patients with CRC,
and serum miR-885 expression was statistically significantly
associated with lymph node metastasis, distant metastasis, tumor,
node, metastasis (TNM) stage, liver metastasis and lymphatic
invasion (52). A recent study
demonstrated that miR-885 serves a crucial role in liver metastasis
by enhancing cell invasiveness, and regulating the
epithelial-mesenchymal transition pathway by silencing CPEB2
(17). However, in contrast to the
consistently decreased expression level in liver metastatic CRC
tissues in different previous studies, the expression of miR-10b is
paradoxical in CRC tissues compared with normal adjacent mucosa
(12,54), and high miR-10b expression was
statistically significantly associated with poor prognosis in
patients with CRC (12,54). Additionally, increased miR-10b
expression was statistically significantly different among T stage,
distant metastasis and advanced TNM stage (52,55).
Therefore, whether miR-10b is an oncogenic miRNA or a tumor
suppressor in CRC requires further investigation.
Additionally, to unveil a comprehensive regulatory
network and screen out key molecules and axes in CRC liver
metastasis, a DE miRNA-DEGs regulatory network was constructed, in
which, the miR-122/IGFBP5, miR-122/MFAP5, miR-122/MYH11,
miR-885/IGFBP5 and miR-885/vWF axes may serve critical roles in the
occurrence and metastatic process of CRC. IGFBP5 is a member of the
family of insulin-like growth factor-binding proteins, which has
been demonstrated to regulate cell growth, differentiation,
apoptosis and modulate the metastatic process in the development of
multiple cancer types (56).
However, the expression of IGFBP5 varies from different types of
cancer and it serves as either an oncogene or tumor suppressor in
different tumors (56). Unlike the
present predictions, a previous study demonstrated that IGFBP5 was
upregulated in CRC (57) and
promoted CRC metastasis (58). vWF
is a protein that modulates adherence of platelets to the
subendothelium during primary hemostasis, which is crucial in the
initiation of the hemostatic or thrombotic process (59). vWF serves pro- and antitumor roles
in different cancer types (60).
Clinical studies demonstrated that high vWF plasma expression
levels were associated with advanced tumor stage and the presence
of distant metastasis in CRC (61).
However, the cleavage of vWF by a disintegrin and metalloproteinase
28 enhanced tumor metastasis, indicating an anti-metastatic role of
vWF (62). The present experiments
demonstrated that miR-885 promotes CRC migration, which may
decrease vWF and IGFBP5 expression, at least partially, indicating
that the miR-885/vWF and miR-885/IGFBP5 axes may serve roles in the
metastasis of CRC. However, the present results require in
vivo and further experiments, including luciferase reporter
assays, for confirmation.
In summary, the present bioinformatics analysis
identified 10 hub genes, including ALB, F2, APOH, SERPINC1, APOA1,
APOC3, AMBP, PLG, AHSG and APOB, and 3 DE miRNAs, namely miR-10b,
miR-122 and miR-885. The hub genes and DE miRNAs may be used as
novel biomarkers for predicting the liver metastasis of CRC.
Additionally, a DE miRNA-DEGs regulatory network was constructed,
which may help to elucidate the underlying molecular mechanisms of
liver metastasis of CRC. Furthermore, the present experiments
demonstrated that miR-885 promoted CRC cell migration by, at least
partially, decreasing vWF and IGFBP5 expression. In order to obtain
more accurate correlation results, a large number of clinical
samples and further experiments are required to validate the
present results and elucidate the underlying mechanisms of how
these key genes and miRNAs impact liver metastasis of CRC. The
present study may provide insight for future diagnosis and genomic
therapy for liver metastatic CRC.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Natural
Science Foundation of China (grant no. 81570568).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
TZ, JGuo, JW and HL conceived and designed the
study. TZ, JGuo, GW and JGu performed the data acquisition and
analysis. JGu and ZW performed the experiments. TZ and JGuo wrote
the paper. HL, JW, ZW and GW reviewed and edited the manuscript.
All authors read and approved the manuscript and agree to be
accountable for all aspects of the research in ensuring that the
accuracy or integrity of any part of the work is appropriately
investigated and resolved.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Manfredi S, Lepage C, Hatem C, Coatmeur O,
Faivre J and Bouvier AM: Epidemiology and management of liver
metastases from colorectal cancer. Ann Surg. 244:254–259. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bakalakos EA, Kim JA, Young DC and Martin
EW Jr: Determinants of survival following hepatic resection for
metastatic colorectal cancer. World J Surg. 22:399–404; discussion.
404–395. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hur K: MicroRNAs: Promising biomarkers for
diagnosis and therapeutic targets in human colorectal cancer
metastasis. BMB Rep. 48:217–222. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kulasingam V and Diamandis EP: Strategies
for discovering novel cancer biomarkers through utilization of
emerging technologies. Nat Clin Pract Oncol. 5:588–599. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sheffer M, Bacolod MD, Zuk O, Giardina SF,
Pincas H, Barany F, Paty PB, Gerald WL, Notterman DA and Domany E:
Association of survival and disease progression with chromosomal
instability: A genomic exploration of colorectal cancer. Proc Natl
Acad Sci USA. 106:7131–7136. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tsafrir D, Tsafrir I, Ein-Dor L, Zuk O,
Notterman DA and Domany E: Sorting points into neighborhoods
(SPIN): Data analysis and visualization by ordering distance
matrices. Bioinformatics. 21:2301–2308. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sayagues JM, Corchete LA, Gutierrez ML,
Sarasquete ME, DelMarAbad M, Bengoechea O, Fermiñán E, Anduaga MF,
Del Carmen S, Iglesias M, et al: Genomic characterization of liver
metastases from colorectal cancer patients. Oncotarget.
7:72908–72922. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Pizzini S, Bisognin A, Mandruzzato S,
Biasiolo M, Facciolli A, Perilli L, Rossi E, Esposito G, Rugge M,
Pilati P, et al: Impact of microRNAs on regulatory networks and
pathways in human colorectal carcinogenesis and development of
metastasis. BMC genomics. 14:5892013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mokutani Y, Uemura M, Munakata K, Okuzaki
D, Haraguchi N, Takahashi H, Nishimura J, Hata T, Murata K,
Takemasa I, et al: Down-regulation of microRNA-132 is associated
with poor prognosis of colorectal cancer. Ann Surg Oncol.
23:599–608. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yan W, Zhang W, Sun L, Liu Y, You G, Wang
Y, Kang C, You Y and Jiang T: Identification of MMP-9 specific
microRNA expression profile as potential targets of anti-invasion
therapy in glioblastoma multiforme. Brain Res. 1411:108–115. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Afanasyeva EA, Mestdagh P, Kumps C,
Vandesompele J, Ehemann V, Theissen J, Fischer M, Zapatka M, Brors
B, Savelyeva L, et al: MicroRNA miR-885-5p targets CDK2 and MCM5,
activates p53 and inhibits proliferation and survival. Cell Death
Differ. 18:974–984. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang Z, Yin J, Yang J, Shen W, Zhang C,
Mou W, Luo J, Yan H, Sun P, Luo Y, et al: miR-885-5p suppresses
hepatocellular carcinoma metastasis and inhibits Wnt/beta-catenin
signaling pathway. Oncotarget. 7:75038–75051. 2016.PubMed/NCBI
|
|
17
|
Lam CS, Ng L, Chow AK, Wan TM, Yau S,
Cheng NS, Wong SK, Man JH, Lo OS, Foo DC, et al: Identification of
microRNA 885-5p as a novel regulator of tumor metastasis by
targeting CPEB2 in colorectal cancer. Oncotarget. 8:26858–26870.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. Gene
Ontology Consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ogata H, Goto S, Sato K, Fujibuchi W, Bono
H and Kanehisa M: KE GG Kyoto encyclopedia of genes and genomes.
Nucleic Acids Res. 27:29–34. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43:D447–D452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
John B, Enright AJ, Aravin A, Tuschl T,
Sander C and Marks DS: Human MicroRNA targets. PLoS Biol.
2:e3632004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kertesz M, Iovino N, Unnerstall U, Gaul U
and Segal E: The role of site accessibility in microRNA target
recognition. Nat Genet. 39:1278–1284. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wong N and Wang X: miRDB: An online
resource for microRNA target prediction and functional annotations.
Nucleic Acids Res. 43:D146–D152. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Dweep H, Sticht C, Pandey P and Gretz N:
miRWalk-database: Prediction of possible miRNA binding sites by
‘walking’ the genes of three genomes. J Biomed Inform. 44:839–847.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Carbon S, Ireland A, Mungall CJ, Shu S,
Marshall B and Lewis S: AmiGO Hub and Web Presence Working Group:
AmiGO: Online access to ontology and annotation data.
Bioinformatics. 25:288–289. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Guo Y, Bao Y, Ma M and Yang W:
Identification of key candidate genes and pathways in colorectal
cancer by integrated bioinformatical analysis. Int J Mol Sci.
18:E7222017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yu JT, Tan L and Hardy J: Apolipoprotein E
in Alzheimer's disease: An update. Annu Rev Neurosci. 37:79–100.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Borgquist S, Butt T, Almgren P, Shiffman
D, Stocks T, Orho-Melander M, Manjer J and Melander O:
Apolipoproteins, lipids and risk of cancer. Int J Cancer.
138:2648–2656. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zamanian-Daryoush M and DiDonato JA:
Apolipoprotein A-I and cancer. Front Pharmacol. 6:2652015.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Mangaraj M, Nanda R and Panda S:
Apolipoprotein A-I A molecule of diverse function. Indian J Clin
Biochem. 31:253–259. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Peltier J, Roperch JP, Audebert S, Borg JP
and Camoin L: Quantitative proteomic analysis exploring progression
of colorectal cancer: Modulation of the serpin family. J
Proteomics. 148:139–148. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Sirnio P, Vayrynen JP, Klintrup K, Makela
J, Makinen MJ, Karttunen TJ and Tuomisto A: Decreased serum
apolipoprotein A1 levels are associated with poor survival and
systemic inflammatory response in colorectal cancer. Sci Rep.
7:53742017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Gong Y, Zhang L, Bie P and Wang H: Roles
of ApoB-100 gene polymorphisms and the risks of gallstones and
gallbladder cancer: A meta-analysis. PLoS One. 8:e614562013.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhong DN, Ning QY, Wu JZ, Zang N, Wu JL,
Hu DF, Luo SY, Huang AC, Li LL and Li GJ: Comparative proteomic
profiles indicating genetic factors may involve in hepatocellular
carcinoma familial aggregation. Cancer Sci. 103:1833–1838. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Beecken WD, Ringel EM, Babica J, Oppermann
E, Jonas D and Blaheta RA: Plasmin-clipped
β2-glycoprotein-I inhibits endothelial cell growth by
down-regulating cyclin A, B and D1 and up-regulating p21 and p27.
Cancer Lett. 296:160–167. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Jing X, Tian ZB, Gao PJ, Han NJ, Xu YH,
Zhang H, Ding XL, Wang XW, Man X and Zhang CP: Lipopolysaccharide
enhances beta2-glycoprotein I activation of nuclear factor κB in
liver cancer cells. Clin Lab. 61:1239–1245. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Huang H, Han Y, Gao J, Feng J, Zhu L, Qu
L, Shen L and Shou C: High level of serum AMBP is associated with
poor response to paclitaxel-capecitabine chemotherapy in advanced
gastric cancer patients. Med Oncol. 30:7482013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Rasul S, Wagner L and Kautzky-Willer A:
Fetuin-A and angiopoietins in obesity and type 2 diabetes mellitus.
Endocrine. 42:496–505. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Nangami GN, Watson K, Parker-Johnson K,
Okereke KO, Sakwe A, Thompson P, Frimpong N and Ochieng J: Fetuin-A
(alpha2HS-glycoprotein) is a serum chemo-attractant that also
promotes invasion of tumor cells through Matrigel. Biochem Biophys
Res Commun. 438:660–665. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Fan NJ, Kang R, Ge XY, Li M, Liu Y, Chen
HM and Gao CF: Identification alpha-2-HS-glycoprotein precursor and
tubulin beta chain as serology diagnosis biomarker of colorectal
cancer. Diagn Pathol. 9:532014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kumari S and Malla R: New insight on the
role of plasminogen receptor in cancer progression. Cancer Growth
Metastasis. 8:35–42. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Didiasova M, Wujak L, Wygrecka M and
Zakrzewicz D: From plasminogen to plasmin: Role of plasminogen
receptors in human cancer. Int J Mol Sci. 15:21229–21252. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Heit JA, Silverstein MD, Mohr DN,
Petterson TM, O'Fallon WM and Melton LJ III: Risk factors for deep
vein thrombosis and pulmonary embolism: A population-based
case-control study. Arch Intern Med. 160:809–815. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Dicke C and Langer F: Pathophysiology of
Trousseau's syndrome. Hamostaseologie. 35:52–59. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Guglietta S and Rescigno M:
Hypercoagulation and complement: Connected players in tumor
development and metastases. Semin Immunol. 28:578–586. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Afshar-Kharghan V: The role of the
complement system in cancer. J Clin Invest. 127:780–789. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Torres S, Garcia-Palmero I, Bartolome RA,
Fernandez-Acenero MJ, Molina E, Calvino E, Segura MF and Casal JI:
Combined miRNA profiling and proteomics demonstrates that different
miRNAs target a common set of proteins to promote colorectal cancer
metastasis. J Pathol. 242:39–51. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Hur K, Toiyama Y, Schetter AJ, Okugawa Y,
Harris CC, Boland CR and Goel A: Identification of a
metastasis-specific MicroRNA signature in human colorectal cancer.
J National Cancer Ins. 107:2015.
|
|
53
|
Vychytilova-Faltejskova P, Pesta M, Radova
L, Liska V, Daum O, Kala Z, Svoboda M, Kiss I and Slaby O:
Genome-wide microRNA expression profiling in primary tumors and
matched liver metastasis of patients with colorectal cancer. Cancer
Genomics Proteomics. 13:311–316. 2016.PubMed/NCBI
|
|
54
|
Abdelmaksoud-Dammak R, Chamtouri N, Triki
M, Saadallah-Kallel A, Ayadi W, Charfi S, Khabir A, Ayadi L,
Sallemi-Boudawara T and Mokdad-Gargouri R: Overexpression of
miR-10b in colorectal cancer patients: Correlation with TWIST-1 and
E-cadherin expression. Tumour Biol. 39:10104283176959162017.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Jiang H, Liu J, Chen Y, Ma C, Li B and Hao
T: Up-regulation of mir-10b predicate advanced clinicopathological
features and liver metastasis in colorectal cancer. Cancer Med.
5:2932–2941. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Gullu G, Karabulut S and Akkiprik M:
Functional roles and clinical values of insulin-like growth
factor-binding protein-5 in different types of cancers. Chin J
Cancer. 31:266–280. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Femia AP, Luceri C, Toti S, Giannini A,
Dolara P and Caderni G: Gene expression profile and genomic
alterations in colonic tumours induced by 1,2-dimethylhydrazine
(DMH) in rats. BMC Cancer. 10:1942010. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Yu L, Lu Y, Han X, Zhao W, Li J, Mao J,
Wang B, Shen J, Fan S, Wang L, et al: microRNA-140-5p inhibits
colorectal cancer invasion and metastasis by targeting ADAMTS5 and
IGFBP5. Stem Cell Res Ther. 7:1802016. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Schmugge M, Rand ML and Freedman J:
Platelets and von Willebrand factor. Transfus Apher Sci.
28:269–277. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
O'Sullivan JM, Preston RJS, Robson T and
O'Donnell JS: Emerging roles for von willebrand factor in cancer
cell biology. Semin Thromb Hemost. 44:159–166. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Wang WS, Lin JK, Lin TC, Chiou TJ, Liu JH,
Yen CC and Chen PM: Plasma von Willebrand factor level as a
prognostic indicator of patients with metastatic colorectal
carcinoma. World J Gastroenterol. 11:2166–2170. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Mochizuki S, Soejima K, Shimoda M, Abe H,
Sasaki A, Okano HJ, Okano H and Okada Y: Effect of ADAM28 on
carcinoma cell metastasis by cleavage of von Willebrand factor. J
NatI Cancer Inst. 104:906–922. 2012. View Article : Google Scholar
|