Introduction
Benign prostatic hyperplasia (BPH) is a condition
comprising a series of lower urinary tract symptoms caused by the
non-malignant enlargement of the prostate gland, which is a common
condition in older men (1). The
proportion of men older than 50 years with BPH increases with age,
with an incidence of >50% in men aged 60–69 years and 90% in
those aged 70–89 years (2).
However, the etiology of BPH remains unclear. Certain studies have
suggested a possible role of chronic inflammation in the
pathogenesis and progression of BPH (3). A number of studies have sought the
association between chronic inflammation and BPH, speculating that
BPH is an immune-mediated inflammatory disease (4–6).
Prostate inflammation involves a complex network of cytokines and
inflammatory mediators. This network leads to the sustained
mitogenic stimulation of epithelial and stromal cells, eventually
resulting in BPH (6).
Chemokines are chemotactic cytokines produced in
activated innate immune cells, and are potent inducers of
leukocytes, including neutrophils and monocytes, acting as
mediators in acute and chronic inflammation. Through interaction
with their receptors, chemokines regulate the directional movement
of various immune cells in tissues and organs so as to maintain the
homeostasis of the local microenvironment (7). C-C motif chemokine ligand (CCL2), also
known as monocyte chemoattractant protein 1, is a member of the CC
chemokine family and regulates the accumulation of monocytes,
macrophages and other inflammatory cells at sites of inflammation
by binding with CC chemokine receptor 2 (CCR2) (8). CCL5, also known as regulated upon
activation, normally T-expressed, and presumably secreted, is
another cytokine that belongs to the CC chemokine family. CCL5 is
chemotactic for T cells, eosinophils and basophils, and serves an
active role in the recruitment of leukocytes to inflammatory sites
(9). Prostate epithelial and
stromal cells are able to secrete high levels of CCL2 and CCL5 in
response to inflammatory stimulation. By binding with their
receptors, they can stimulate the growth of epithelial cells
through paracrine or autocrine pathways (10). Increased concentrations of CCL2 and
CCL5, and the increased production of proinflammatory cytokines,
such as interleukin (IL)6 and IL8, in the prostate gland, may
further lead to the accumulation of inflammatory cells, resulting
in a positive feedback loop that induces BPH (10).
A number of studies have investigated the
association between single nucleotide polymorphisms (SNPs) in
chemokines and their receptors, and prostate cancer (11–14).
However, few studies have assessed the association of chemokines
and their receptors with BPH risk. Therefore, in the present study,
three common functional polymorphisms (CCL2 rs1024611, CCR2
rs1799864 and CCL5 rs2107538) were selected and their associations
with the development and progression of BPH were investigated.
Materials and methods
Study subjects
A total of 269 subjects were enrolled in the present
study between January 2014 and December 2015. These subjects
comprised 109 patients with BPH and 160 healthy males as controls.
The patients were diagnosed with BPH by at least two attending
physicians in the Department of Urology, Lanzhou University Second
Hospital (Lanzhou, China) according to their clinical features. For
all patients, lower urinary tract symptoms were quantified using
the International Prostate Symptom Score (IPSS), IPSS-quality of
life (QOL) score and Overactive Bladder Symptom Score (OABSS)
(15,16). The serum prostate-specific antigen
(PSA) level was determined by ELISA in each subject. Patients with
serum PSA levels ≥4 ng/ml were further tested to exclude prostate
cancer by digital rectal examination and transrectal
ultrasound-guided prostate biopsy. The peak urinary flow rate
(Qmax) and the voided volumes were measured for all patients using
an uroflowmetry system (ZNC 961A; Yongxin Medical Equipment Co.,
Ltd., Jiangsu, China). The prostate volumes (PVs) of the patients
were calculated using transrectal ultrasonography and the post-void
residual volumes were measured by transabdominal ultrasonography.
Subjects with a history of urinary tract infection, prostate
cancer, neurogenic bladder dysfunction, acute/chronic prostatitis,
uncontrolled diabetes, lower tract surgery or cardiovascular
disease were excluded from the study. Based on previous studies
(17,18), all the BPH cases were separated into
two groups based on the measurements of each parameter: IPSS,
<20 vs. ≥20; PSA, <1.5 vs. ≥1.5 ng/ml; PV, <30 vs. ≥30 ml;
and Qmax, <10 vs. ≥10 ml/sec.
The healthy controls recruited in the present study
were older males visiting the physician for routine checkups. They
all underwent screening, had normal serum PSA levels (<4.0
ng/ml), and exhibited no evidence of any of the aforementioned
diseases.
All the participants provided written informed
consent prior to participation. The present study was approved by
the Ethics Committee at Lanzhou University Second Hospital.
SNP selection and genotyping
In the present study, three SNPs (CCL2 rs1024611,
CCR2 rs1799864, and CCL5 rs2107538) that have previously been
demonstrated to have an association with prostate cancer were
selected (11–14). Peripheral venous blood samples from
all participants were collected in EDTA blood collection tubes and
stored at −80°C prior to the extraction of genomic DNA. Genomic DNA
was extracted from the blood using the Tiangen DNA extraction kit
(Tiangen Biotech Co., Ltd., Beijing, China). The SNPs were
genotyped using the polymerase chain reaction (PCR) and multiple
ligase detection reaction (LDR) system at Shanghai Biowing Applied
Biotechnology Co., Ltd. (Shanghai, China). Based on the sequence
information of chemokine genes on the NCBI website (https://www.ncbi.nlm.nih.gov/), PCR primers and LDR
probes (Table I) were designed
using Primer Premier version 5.0 software (Premier Biosoft
International, Palo Alto, CA, USA) and synthesized by Shanghai
Biowing Applied Biotechnology Co., Ltd. (Shanghai, China). PCR
conditions consisted of 40 cycles at 94°C for 30 sec, 53°C for 90
sec and 65°C for 30 sec, followed by 1 cycle at 65°C for 10 min to
terminate the reaction. LDR conditions consisted of 40 cycles at
94°C for 15 sec and 50°C for 25 sec. The reagent used in these
methods, such as DNA polymerase and DNA ligase were supplied by
Shanghai Biowing Applied Biotechnology Co., Ltd. All PCR products
were analyzed for quality and concentration using 3.0% agarose gel
electrophoresis. Visualization was achieved using ethidium
bromide.
 | Table I.Primer and probe sequences used in
PCR-LDR. |
Table I.
Primer and probe sequences used in
PCR-LDR.
| A, Primer
sequences |
|---|
|
|---|
| SNP | Primer | Primer sequence
(5′-3′) | PCR length
(bp) |
|---|
| CCL2 rs1024611 | Forward |
cagtaaacacagggaaggtg | 95 |
|
| Reverse |
tcttgacagagcagaagtgg |
|
| CCR2 rs1799864 | Forward |
gctctactcgctggtgttc | 128 |
|
| Reverse |
agatcagagatggccaggt |
|
| CCL5 rs2107538 | Forward |
caaggagtggcagttagga | 170 |
|
| Reverse |
tatccagaggaccctcctc |
|
|
| B, Probe
sequences |
|
| SNP | Probe | Probe sequence
(5′-3′) | LDR length
(bp) |
|
| CCL2 rs1024611 | Modify |
P-tcactttccagaagactttcttttctttttttttttttttttttttttttttttttttttttttttttttttttttttt-FAM |
|
|
| C |
tttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttgagcagaagtgggaggcagacagctg | 170 |
|
| T |
tttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttgagcagaagtgggaggcagacagcta | 172 |
| CCR2 rs1799864 | Modify |
P-gaccagcatgttgcccacaaaaccatttttttttttttttttttttttttttttttttttttttttttttttttttttt-FAM |
|
|
| A |
tttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttgcagtttattaagatgaggat | 165 |
|
| G |
tttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttgcagtttattaagatgaggac | 167 |
| CCL5 rs2107538 | Modify |
P-taagatctgtaatgaataagcaggatttttttttttttttttttttttttttttttttttttttttttttttttttttt-FAM |
|
|
| C |
ttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttccttccatggatgagggaaaggagg | 180 |
|
| T |
ttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttttccttccatggatgagggaaaggaga | 182 |
Functional annotation of selected
SNP
To explore the potential function of the loci
identified from the study, the online tool RegulomeDB (http://www.regulomedb.org/) was used to predict the
putative function of the identified SNPs (19). Furthermore, assessment of whether
the loci of genes may influence their corresponding mRNA expression
levels was conducted using the online GTEx database (http://www.gtexportal.org/home/).
Statistical analysis
The genotype distributions of the SNPs in the
patient and control groups were analyzed with a Chi-square test to
investigate the Hardy-Weinberg equilibrium (HWE). Logistic
regression models were used to estimate the main effects of the
SNPs on BPH risk, assuming an additive and dominant model of
inheritance, and adjusting for age as a covariable. Logistic
regression analysis was also performed on the combined results of
the three polymorphisms. The risk of BPH was evaluated by
calculating the odds ratio (OR) and corresponding 95% confidence
interval (CI). Stratified analyses were executed according to IPSS,
PSA, PV and Qmax. Data obtained from the GTEx database were
calculated and reported as the mean ± standard deviation.
Comparisons between different genotype groups were evaluated by
one-way analysis of variance followed by Tukey's multiple
comparison tests. All analyses were conducted using IBM SPSS
version 20.0 software (IBM Corp., Armonk, NY, USA). P<0.05 was
considered to indicate a statistically significant difference.
Results
Clinical characteristics of the study
subjects
The clinical characteristics of the 109 cases and
160 controls are presented in Table
II. The mean age of the patients (65.62±8.20 years) was higher
compared with that of the controls (54.51±3.87 years), and the
association tests were adjusted accordingly.
 | Table II.Clinical characteristics of all
subjects. |
Table II.
Clinical characteristics of all
subjects.
|
Characteristics | Cases | Controls |
|---|
| No. of
subjects | 109 | 160 |
| Age (years) | 65.62±8.20 | 54.51±3.87 |
| IPSS | 16.09±8.15 | N/A |
|
0–19 | 70 (64.2) | N/A |
|
≥20 | 39 (35.8) | N/A |
| PSA (ng/ml) | 2.46±2.82 | N/A |
|
<1.5 | 52 (47.7) | N/A |
|
≥1.5 | 57 (52.3) | N/A |
| PV (ml) | 33.13±19.72 | N/A |
|
<30 | 56 (51.4) | N/A |
|
≥30 | 53 (48.6) | N/A |
| Qmax (ml/sec) | 10.32±3.41 | N/A |
|
<10 | 39 (35.8) | N/A |
|
≥10 | 70 (64.2) | N/A |
| OABSS | 4.96±3.19 | N/A |
| QoL | 4.44±0.87 | N/A |
| VV (ml) | 232.73±109.99 | N/A |
| PVR (ml) | 27.57±43.10 | N/A |
HWE test
The genotype distributions of the SNPs in the
disease and control groups were evaluated, and the results revealed
good conformity to the HWE for all the SNPs (P>0.05). Therefore,
the study population included in the present study is a typical
Mendelian population (Table
III).
 | Table III.Exact Hardy-Weinberg equilibrium test
for SNPs (n=269). |
Table III.
Exact Hardy-Weinberg equilibrium test
for SNPs (n=269).
| SNP | Genotype or
allele | All subjects | Cases | Controls |
|---|
| CCL2 rs1024611 | A/A | 56 | 25 | 31 |
|
| A/G | 135 | 44 | 91 |
|
| G/G | 78 | 40 | 38 |
|
| A | 247 | 94 | 153 |
|
| G | 291 | 124 | 167 |
| P-value |
| 0.9 | 0.078 | 0.11 |
| CCR2 rs1799864 | G/G | 157 | 63 | 94 |
|
| G/A | 101 | 42 | 59 |
|
| A/A | 11 | 4 | 7 |
|
| G | 415 | 168 | 247 |
|
| A | 123 | 50 | 73 |
| P-value |
| 0.39 | 0.43 | 0.66 |
| CCL5 rs2107538 | G/G | 102 | 52 | 50 |
|
| G/A | 130 | 43 | 87 |
|
| A/A | 37 | 14 | 23 |
|
| G | 334 | 147 | 187 |
|
| A | 204 | 71 | 133 |
| P-value |
| 0.7 | 0.28 | 0.15 |
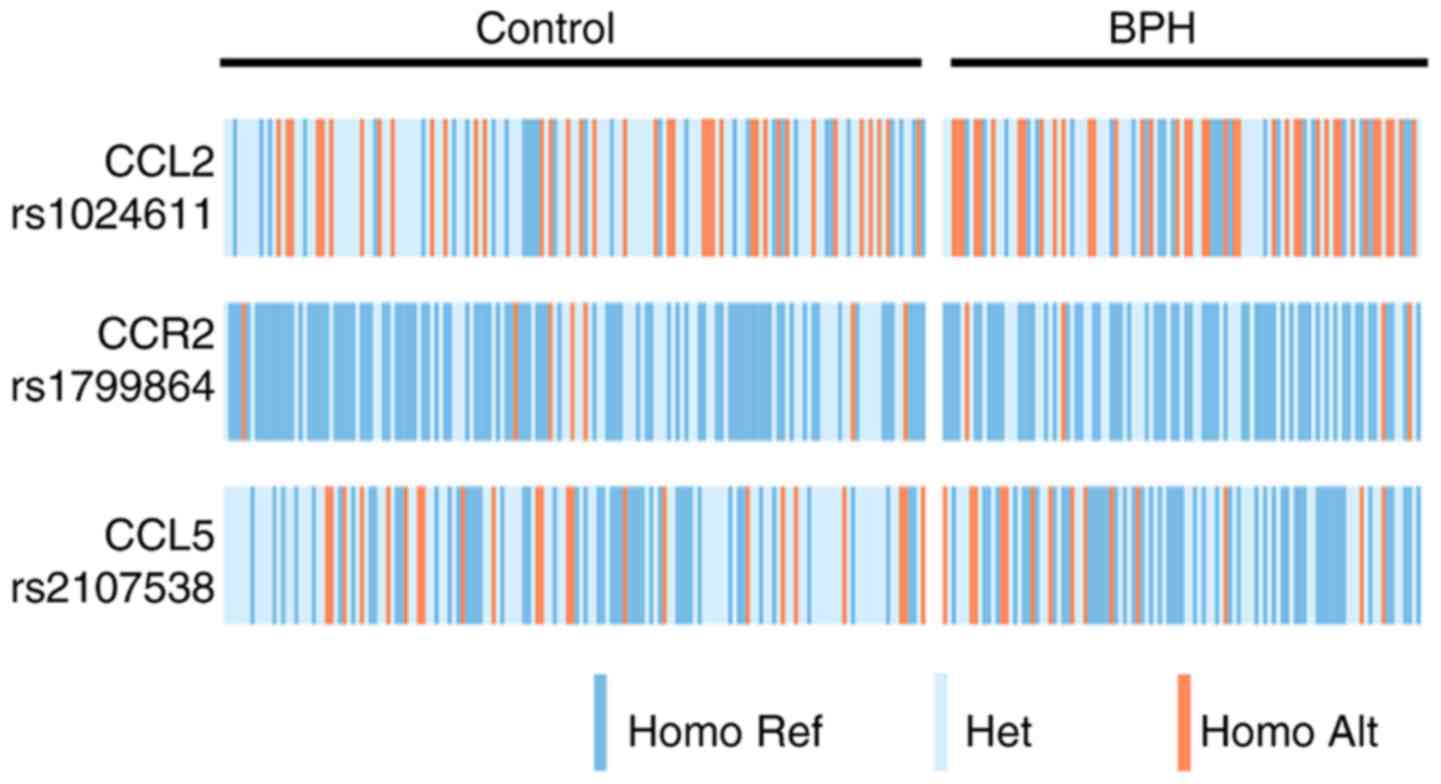
Genetic association with BPH risk
The genotype distributions of the three studied
genetic polymorphisms in the BPH and control groups are presented
in Fig. 1. The specific
associations between the SNPs and BPH risk were calculated
(Table IV). The G/G, G/A and A/A
genotype frequencies of the CCL5 rs2107538 polymorphism were 47.7,
39.5 and 12.8%, respectively, in the patients, and 31.2, 54.4 and
14.4%, respectively, in the controls. The allele frequencies of the
patient and control groups were 67.4 and 58.4% for the G allele,
and 32.6 and 41.6% for the A allele, respectively. The CCL5
rs2107538 polymorphism exhibited a significant association with a
decreased risk of BPH in the heterozygous (OR=0.37, 95%
CI=0.17–0.78), dominant (OR=0.39, 95% CI=0.19–0.79) and allelic
form (OR=0.58, 95% CI=0.35–0.96). However, no significant
differences in the genotype or allele distributions for the CCL2
rs1024611 and CCR2 rs1799864 polymorphisms were observed between
the patients and controls.
 | Table IV.Genotypes and allele frequencies of
SNPs and their association with BPH risk. |
Table IV.
Genotypes and allele frequencies of
SNPs and their association with BPH risk.
| SNP | Genotype or
allele | Controls, n=160
(%) | Cases, n=109
(%) | Models | OR (95%
CI)a |
P-valuea |
|---|
| CCL2 rs1024611 | A/A | 31 (19.4) | 25 (22.9) | Heterozygous | 0.46
(0.19–1.12) |
0.086 |
|
| A/G | 91 (56.9) | 44 (40.4) | Homozygous | 1.27
(0.49–3.28) | 0.62 |
|
| G/G | 38 (23.8) | 40 (36.7) | Dominant | 0.68
(0.30–1.56) | 0.37 |
|
| A | 153 (47.8) | 94 (43.1) |
| 1 |
|
|
| G | 167 (52.2) | 124 (56.9) |
| 1.22
(0.76–1.97) | 0.42 |
| CCR2 rs1799864 | G/G | 94 (58.8) | 63 (57.8) | Heterozygous | 1.11
(0.54–2.27) | 0.78 |
|
| G/A | 59 (36.9) | 42 (38.5) | Homozygous | 1.31
(0.28–6.11) | 0.73 |
|
| A/A | 7 (4.4) | 4 (3.7) | Dominant | 1.13
(0.57–2.25) | 0.72 |
|
| G | 247 (77.2) | 168 (77.1) |
| 1 |
|
|
| A | 73 (22.8) | 50 (22.9) |
| 1.12
(0.64–1.98) | 0.69 |
| CCL5 rs2107538 | G/G | 50 (31.2) | 52 (47.7) | Heterozygous | 0.37
(0.17–0.78) | 0.01 |
|
| G/A | 87 (54.4) | 43 (39.5) | Homozygous | 0.47
(0.16–1.35) | 0.16 |
|
| A/A | 23 (14.4) | 14 (12.8) | Dominant | 0.39
(0.19–0.79) |
0.008 |
|
| G | 187 (58.4) | 147 (67.4) |
| 1 |
|
|
| A | 133 (41.6) | 71 (32.6) |
| 0.58
(0.35–0.96) |
0.033 |
Furthermore, all possible interactions of CCL5
rs2107538 with CCL2 rs1024611 and CCR2 rs1799864 gene polymorphisms
were investigated. The combined genotype G/A-A/G of CCL5 rs2107538
and CCL2 rs1024611 exhibited a decreased risk of BPH. However, an
increased risk of BPH was implied for the G/G-G/G combination of
these polymorphisms (Table V). The
combined genotype G/A-G/G of CCL5 rs2107538 and CCR2 rs1799864 was
also associated with a lower risk for BPH (Table VI). These results suggest that the
interaction between the CCL5 rs2107538 and CCR2 rs1799864
polymorphisms may give rise to a synergistic effect.
 | Table V.Association of BPH risk with
different combinations of CCL5 rs2107538 and CCL2 rs1024611
genotypes. |
Table V.
Association of BPH risk with
different combinations of CCL5 rs2107538 and CCL2 rs1024611
genotypes.
| CCL5 rs2107538 and
CCL2 rs1024611 | Controls, n=160
(%) | Cases, n=109
(%) | OR (95%
CI)a |
P-valuea |
|---|
| G/G-A/A | 11 (6.9) | 14 (12.8) | 1 |
|
| G/G-A/G | 37 (23.1) | 16 (14.7) | 0.26
(0.07–1.01) | 0.05 |
| G/G-G/G | 2 (1.2) | 22 (20.2) | 10.83
(1.50–78.13) |
0.018 |
| G/A-A/A | 12 (7.5) | 7 (6.4) | 0.65
(0.12–3.53) | 0.62 |
| G/A-A/G | 44 (27.5) | 20 (18.3) | 0.20
(0.05–0.77) |
0.019 |
| G/A-G/G | 31 (19.4) | 16 (14.7) | 0.28
(0.07–1.11) | 0.07 |
| A/A-A/A | 8 (5.0) | 4 (3.7) | 0.13
(0.02–1.10) | 0.06 |
| A/A-A/G | 10 (6.2) | 8 (7.3) | 0.55
(0.11–2.83) | 0.47 |
| A/A-G/G | 5 (3.1) | 2 (1.8) | 0.46
(0.04–5.00) | 0.52 |
 | Table VI.Association of BPH risk with
different combinations of CCL5 rs2107538 and CCR2 rs1799864
genotypes. |
Table VI.
Association of BPH risk with
different combinations of CCL5 rs2107538 and CCR2 rs1799864
genotypes.
| CCL5 rs2107538 and
CCR2 rs1799864 | Controls, n=160
(%) | Cases, n=109
(%) | OR (95%
CI)a |
P-valuea |
|---|
| G/G-G/G | 30 (18.8) | 30 (27.5) | 1 |
|
| G/G-G/A | 18 (11.2) | 20 (18.3) | 0.77
(0.26–2.33) | 0.65 |
| G/G-A/A | 2 (1.2) | 2 (1.8) | 1.51
(0.11–20.36) | 0.76 |
| G/A-G/G | 53 (33.1) | 26 (23.9) | 0.29
(0.11–0.78) |
0.014 |
| G/A-G/A | 31 (19.4) | 16 (14.7) | 0.45
(0.15–1.37) | 0.16 |
| G/A-A/A | 3 (1.9) | 1 (0.9) | 0.26
(0.02–3.76) | 0.32 |
| A/A-G/G | 11 (6.9) | 7 (6.4) | 0.36
(0.08–1.56) | 0.17 |
| A/A-G/A | 10 (6.2) | 6 (5.5) | 0.42
(0.08–2.28) | 0.32 |
| A/A-A/A | 2 (1.2) | 1 (0.9) | 0.94
(0.07–12.16) | 0.96 |
Genetic association with
clinicopathological characteristics
The associations between the three SNPs and the
clinicopathological characteristics (IPSS, PSA, PV and Qmax) of the
patients with BPH were investigated. The results indicated that
CCR2 rs1799864 was significantly associated with IPSS, with a
decreasing risk of high IPSS progression in the dominant (OR=0.39,
95% CI=0.17–0.91) and the allelic forms (OR=0.43, 95% CI=0.20–0.90)
(Table VII).
 | Table VII.Genotype and allele frequencies of
SNPs and their association with IPSS. |
Table VII.
Genotype and allele frequencies of
SNPs and their association with IPSS.
| SNP | Genotype or
allele | IPSS<20, n=70
(%) | IPSS≥20, n=39
(%) | Models | OR (95%
CI)a |
P-valuea |
|---|
| CCL2 rs1024611 | A/A | 14 (20) | 11 (28.2) | Heterozygous | 0.57
(0.21–1.59) | 0.29 |
|
| A/G | 30 (42.9) | 14 (35.9) | Homozygous | 0.69
(0.25–1.92) | 0.47 |
|
| G/G | 26 (37.1) | 14 (35.9) | Dominant | 0.63
(0.25–1.56) | 0.32 |
|
| A | 58 (41.4) | 36 (46.2) |
| 1 |
|
|
| G | 82 (58.6) | 42 (53.8) |
| 0.83
(0.47–1.45) | 0.51 |
| CCR2 rs1799864 | G/G | 35 (50) | 28 (71.8) | Heterozygous | 0.44
(0.19–1.03) | 0.06 |
|
| G/A | 31 (44.3) | 11 (28.2) | Homozygous | 0.00 (0.00-NA) | 1.00 |
|
| A/A | 4 (5.7) | 0 (0) | Dominant | 0.39
(0.17–0.91) |
0.026 |
|
| G | 101 (72.1) | 67 (85.9) |
| 1 |
|
|
| A | 39 (27.9) | 11 (14.1) |
| 0.43
(0.20–0.90) |
0.024 |
| CCL5 rs2107538 | G/G | 37 (52.9) | 15 (38.5) | Heterozygous | 1.73
(0.73–4.08) | 0.21 |
|
| G/A | 25 (35.7) | 18 (46.1) | Homozygous | 1.79
(0.53–6.07) | 0.35 |
|
| A/A | 8 (11.4) | 6 (15.4) | Dominant | 1.74
(0.78–3.90) | 0.17 |
|
| G | 99 (70.7) | 48 (61.5) |
| 1 |
|
|
| A | 41 (29.3) | 30 (38.5) |
| 1.47
(0.82–2.65) | 0.20 |
A significant association was detected between CCL5
rs2107538 and PV, as demonstrated in Table VIII, reflecting an increasing
effect on the risk of large PV progression in the heterozygous
(OR=3.02, 95% CI=1.28–7.11) and dominant model (OR=2.83, 95%
CI=1.28–6.26). The allele frequency analysis confirmed that CCL5
rs2107538 was associated with PV (OR=1.94, 95% CI=1.08–3.49).
Furthermore, a significant association between CCR2 rs1799864 and
Qmax (≥10 vs. <10 ml/sec) was identified. Table IX demonstrates that CCR2 rs1799864
exhibited a decreasing risk of low Qmax progression in the
heterozygous (OR=0.38, 95% CI=0.16–0.92) and dominant model
(OR=0.39, 95% CI=0.17–0.91). However, no association was detected
between the SNPs and PSA (Table
X).
 | Table VIII.Genotype and allele frequencies of
SNPs and their association with PV. |
Table VIII.
Genotype and allele frequencies of
SNPs and their association with PV.
| SNP | Genotype or
allele | PV<30 ml, n=56
(%) | PV≥30 ml, n=53
(%) | Models | OR (95%
CI)a |
P-valuea |
|---|
| CCL2 rs1024611 | A/A | 15 (26.8) | 10 (18.9) | Heterozygous | 1.41
(0.51–3.89) | 0.51 |
|
| A/G | 22 (39.3) | 22 (41.5) | Homozygous | 1.72
(0.61–4.84) | 0.31 |
|
| G/G | 19 (33.9) | 21 (39.6) | Dominant | 1.55
(0.61–3.91) | 0.36 |
|
| A | 52 (46.4) | 42 (39.6) |
| 1 |
|
|
| G | 60 (53.6) | 64 (60.4) |
| 1.36
(0.79–2.36) | 0.27 |
| CCR2 rs1799864 | G/G | 33 (58.9) | 30 (56.6) | Heterozygous | 1.10
(0.50–2.45) | 0.81 |
|
| G/A | 21 (37.5) | 21 (39.6) | Homozygous | 1.35
(0.17–10.69) | 0.78 |
|
| A/A | 2 (3.6) | 2 (3.8) | Dominant | 1.12
(0.52–2.44) | 0.77 |
|
| G | 87 (77.7) | 81 (76.4) |
| 1 |
|
|
| A | 25 (22.3) | 25 (23.6) |
| 1.11
(0.58–2.12) | 0.75 |
| CCL5 rs2107538 | G/G | 34 (60.7) | 18 (34) | Heterozygous | 3.02
(1.28–7.11) |
0.011 |
|
| G/A | 16 (28.6) | 27 (50.9) | Homozygous | 2.31
(0.68–7.85) | 0.18 |
|
| A/A | 6 (10.7) | 8 (15.1) | Dominant | 2.83
(1.28–6.26) |
0.0094 |
|
| G | 84 (75.0) | 63 (59.4) |
| 1 |
|
|
| A | 28 (25.0) | 43 (40.6) |
| 1.94
(1.08–3.49) |
0.027 |
 | Table IX.Genotype and allele frequencies of
SNPs and their association with Qmax. |
Table IX.
Genotype and allele frequencies of
SNPs and their association with Qmax.
| SNP | Genotype or
allele | Qmax≥10 ml/sec,
n=70 (%) | Qmax<10 ml/sec,
n=39 (%) | Models | OR (95%
CI)a |
P-valuea |
|---|
| CCL2 rs1024611 | A/A | 18 (25.7) | 7 (17.9) | Heterozygous | 1.87
(0.64–5.44) | 0.25 |
|
| A/G | 25 (35.7) | 19 (48.7) | Homozygous | 1.25
(0.42–3.79) | 0.69 |
|
| G/G | 27 (38.6) | 13 (33.3) | Dominant | 1.56
(0.58–4.17) | 0.38 |
|
| A | 61 (43.6) | 33 (42.4) |
| 1 |
|
|
| G | 79 (56.4) | 45 (57.6) |
| 1.07
(0.61–1.88) | 0.82 |
| CCR2 rs1799864 | G/G | 35 (50) | 28 (71.8) | Heterozygous | 0.38
(0.16–0.92) |
0.031 |
|
| G/A | 32 (45.7) | 10 (25.6) | Homozygous | 0.48
(0.05–4.96) | 0.54 |
|
| A/A | 3 (4.3) | 1 (2.6) | Dominant | 0.39
(0.17–0.91) |
0.025 |
|
| G | 102 (72.9) | 66 (84.6) |
| 1 |
|
|
| A | 38 (27.1) | 12 (15.4) |
| 0.49
(0.24–1.02) |
0.055 |
| CCL5 rs2107538 | G/G | 34 (48.6) | 18 (46.1) | Heterozygous | 0.85
(0.35–2.02) | 0.71 |
|
| G/A | 29 (41.4) | 14 (35.9) | Homozygous | 1.74
(0.52–5.83) | 0.37 |
|
| A/A | 7 (10) | 7 (17.9) | Dominant | 1.02
(0.46–2.27) | 0.96 |
|
| G | 97 (69.3) | 50 (64.1) |
| 1 |
|
|
| A | 43 (30.7) | 28 (35.9) |
| 1.20
(0.66–2.16) | 0.56 |
 | Table X.Genotype and allele frequencies of
SNPs and their association with PSA. |
Table X.
Genotype and allele frequencies of
SNPs and their association with PSA.
| SNP | Genotype or
allele | PSA<1.5 ng/ml,
n=52 (%) | PSA≥1.5 ng/ml, n=57
(%) | Models | OR (95%
CI)a |
P-valuea |
|---|
| CCL2 rs1024611 | A/A | 13 (25) | 12 (21.1) | Heterozygous | 1.01
(0.37–2.75) | 0.99 |
|
| A/G | 22 (42.3) | 22 (38.6) | Homozygous | 1.51
(0.54–4.20) | 0.43 |
|
| G/G | 17 (32.7) | 23 (40.4) | Dominant | 1.22
(0.49–3.03) | 0.67 |
|
| A | 48 (46.2) | 46 (40.4) |
| 1 |
|
|
| G | 56 (53.8) | 68 (59.6) |
| 1.30
(0.75–2.25) | 0.35 |
| CCR2 rs1799864 | G/G | 32 (61.5) | 31 (54.4) | Heterozygous | 1.39
(0.62–3.10) | 0.42 |
|
| G/A | 18 (34.6) | 24 (42.1) | Homozygous | 1.24
(0.16–9.78) | 0.84 |
|
| A/A | 2 (3.8) | 2 (3.5) | Dominant | 1.38
(0.63–3.00) | 0.42 |
|
| G | 82 (78.8) | 86 (75.4) |
| 1 |
|
|
| A | 22 (21.2) | 28 (24.6) |
| 1.26
(0.66–2.41) | 0.49 |
| CCL5 rs2107538 | G/G | 28 (53.9) | 24 (42.1) | Heterozygous | 1.50
(0.65–3.45) | 0.34 |
|
| G/A | 18 (34.6) | 25 (43.9) | Homozygous | 1.40
(0.42–4.72) | 0.58 |
|
| A/A | 6 (11.5) | 8 (14) | Dominant | 1.48
(0.68–3.20) | 0.32 |
|
| G | 74 (71.2) | 73 (64.0) |
| 1 |
|
|
| A | 30 (28.8) | 41 (36.0) |
| 1.30
(0.72–2.32) | 0.38 |
Functional prediction of genetic
variants of CCL2, CCR2 and CCL5
To investigate the role of the CCL2 rs1024611, CCR2
rs1799864 and CCL5 rs2107538 polymorphisms in the regulation of
gene expression, a search of the GTEx database for data on mRNA
expression was conducted (Fig. 2).
For CCL5 rs2107538, the mRNA expression levels were downregulated
following the G>A conversion in whole blood cells (P<0.001,
Fig. 2C), but were not changed in
prostate tissue (P=0.20; data not shown). The CCL2 rs1024611 A>G
and CCR2 rs1799864 G>A changes had no impact on gene expression
in whole blood cells (P=0.16, Fig.
2A; and P=0.59, Fig. 2B,
respectively) or prostate tissues (P=0.42 and P=0.12, respectively;
data not shown). These results suggest that the downregulation of
mRNA expression by CCL5 rs2107538 is possibly part of the molecular
mechanism underlying its protective effects against BPH.
Subsequently, using the RegulomeDB database, CCL5 rs2107538 was
predicted to have the ability to influence the activity of
transcription factor binding sites in its promoter region. This SNP
was revealed to be on a binding site for GATA binding protein 2
(GATA2), and therefore may regulate the transcriptional activity of
CCL5. This further explains the underlying mechanism for the
downregulation of CCL5.
Discussion
In the present study, three common functional
polymorphisms (CCL2 rs1024611, CCR2 rs1799864 and CCL5 rs2107538)
were selected and their association with the risk of BPH
development and progression was explored.
The CCL2 rs1024611 polymorphic site is located in
the promoter region of the CCL2 gene, where an A>G substitution
can lead to the upregulation of its expression in vivo
(20). This polymorphism has been
reported to participate in various diseases, such as breast cancer
(7), IgA nephropathy (8) and psoriasis vulgaris (21). The association between the CCL2
rs1024611 polymorphism and prostate cancer risk has been examined
in several studies, but none have been able to demonstrate a
significant link (11,13,14).
The data analysis in the present study also demonstrated no
association between CCL2 rs1024611 polymorphism and BPH
susceptibility and progression, either in the genotype or the
allelic form.
The CCR2 rs1799864 polymorphism is a non-synonymous
mutation in exon 2, resulting in substitution of the amino acid
valine with isoleucine at position 64 (22). Previous studies have reported that
this genetic variation is a risk factor for psoriasis vulgaris
(21), ischemic stroke (23) and carotid atherosclerosis (24). However, in the present analysis, the
polymorphism rs1799864 in CCR2 was not associated with BPH
susceptibility in either the genotype or the allelic form, in
agreement with the study by Singh et al (25). However, it was significantly
associated with IPSS and Qmax. These results suggest that the
polymorphism may be associated with the severity of symptoms,
rather than the development of BPH per se. In particular,
the frequency of the minor allele of CCR2 rs1799864 was
significantly lower in patients with high IPSS than in those with
low IPSS. In addition, the G/A genotype frequency was significantly
lower in BPH patients with low Qmax. This result suggests that
patients with the A allele or G/A genotype of CCR2 rs1799864 may
not manifest severe symptoms, either because the BPH is less severe
or the sensitivity is less pronounced. A previous study reported
that CCR2 was abundantly expressed in normal prostate tissue, and
that the expression of CCR2 was elevated in the prostates of
patients with BPH (26). A possible
explanation of the present findings is that the CCR2 rs1799864
polymorphism may interfere with certain factors related to BPH
progression. It may be hypothesized that CCR2 is overexpressed in
BPH, but the expression level may be lower in patients with the A
allele or G/A genotype of CCR2 rs1799864. Similarly, a previous
study has demonstrated that individuals carrying the heterozygous
(G/A) or homozygous (A/A) genotypes and the A allele of the CCR2
rs1799864 polymorphism appear to have a lower risk of cervical
lesions, reflecting a protective effect against the development of
cervical lesions and the susceptibility to HPV 16 infection
(27).
The CCL5 rs2107538 polymorphism has been confirmed
in the promoter region of the gene and is able to alter
transcriptional activity and subsequent gene expression in human
cell lines (28). Numerous studies
have investigated the role of the CCL5 rs2107538 polymorphism in
various diseases. Areeshi et al (29) performed a meta-analysis to evaluate
the association between the polymorphism and tuberculosis
susceptibility, and identified a significant link with increased
risk of tuberculosis. In the study by Sáenz-López et al
(11), the G/A + A/A genotype of
CCL5 rs2107538 was reported to be significantly associated with an
increased risk of prostate cancer. By contrast, a case-control
study by Kidd et al (30)
suggested that the inheritance of CCL5 rs2107538 (AA, GA + AA) was
linked with a lower risk of prostate cancer. However, to the best
of our knowledge, no study has been performed on the association
between the CCL5 rs2107538 SNP and BPH risk. In the present study,
this polymorphism was revealed to be significantly associated with
BPH risk in the heterozygous, dominant and allele models. Subjects
with the A allele and heterozygous G/A genotype exhibited 0.58- and
0.37-fold lower risks of developing BPH in comparison with subjects
with the wild-type G allele and homozygous G/G genotype,
respectively. In the stratification analysis, it was further
observed that the frequency of the minor allele A and heterozygous
G/A genotype of CCL5 rs2107538 was significantly higher in the BPH
patients with a large PV than in those with a small PV. These
results suggest that the CCL5 rs2107538 SNP may be a marker for
protection against the occurrence of BPH and a risk factor for
severe progression. The observed protective effects of CCL5
rs2107538 may be due to decreased transcriptional levels, decreased
protein levels and, ultimately, decreased pro-inflammatory
function. An analysis of the GTEx database further revealed that
the mRNA expression levels were downregulated for CCL5 rs2107538
following the G>A change in whole blood cells. Results from
RegulomeDB database predicted that CCL5 rs2107538 may have the
ability to influence the activity of transcription factor binding
sites in its promoter region. Subsequently, we identified a binding
site for GATA2, which belongs to the GATA family of nuclear
regulatory proteins, and is a lineage marker serving an essential
regulatory role in the development of hematopoietic (31) and genitourinary systems (32). GATA2 is overexpressed in prostate
cancer (33), and is particularly
upregulated in metastatic prostate cancer tissues (34). The underlying mechanism of the CCL5
rs2107538 on mRNA expression levels is possibly due to a change in
the binding affinity for GATA2.
In the study by Zhernakova et al (35), this SNP was observed to correlate
with a reduction in serum protein levels in patients with type 1
diabetes and controls. The findings of the present study on the
protective effect of this SNP against BPH are consistent with other
published reports that reveal a reduced risk of prostate cancer
(30), type 1 diabetes (35) and lymphoma (36). In addition, we hypothesize that,
although CCL5 may be downregulated in BPH, the level may be higher
in patients with CCL5 rs2107538 genotypes A or G/A.
Although no association of the CCL2 rs1024611 and
CCR2 rs1799864 polymorphisms with BPH was observed, the joint
analysis indicated that the combined occurrence of genotypes
G/A-A/G of CCL5 rs2107538 and CCL2 rs1024611, and G/A-G/G of CCL5
rs2107538 and CCR2 rs1799864 were ‘protective’ against the
occurrence of BPH. However, the G/G genotype of CCL2 rs1024611 may
be a risk factor of BPH when combined with the G/G genotype of CCL5
rs2107538. These joint results revealed possible associations
between these three genes in the risk of BPH.
The major limitation of the present study is the
small sample size used for comparison. Furthermore, the functional
role of CCL2 rs1024611, CCR2 rs1799864 and CCL5 rs2107538 in BPH
development and progression remains poorly understood. However, to
the best of our knowledge, this is the first report on the
association of the CCL2 rs1024611 and CCL5 rs2107538 polymorphisms
with BPH. The results revealed significant associations between the
CCR2 rs1988864 and CCL5 rs2107538 polymorphisms and the development
and progression of BPH. Further studies with larger sample sizes
from various populations are required to elucidate the role of
these polymorphisms. Knowledge of these genetic variants may be
important for the development of novel treatments for BPH and
improvement of the quality of life of patients with this
condition.
In conclusion, the present study suggests that the
CCL5 rs2107538 polymorphism may be a protective factor against the
occurrence of BPH. However, subgroup analysis revealed that the
CCL5 rs2107538 polymorphism may increase the risk of developing a
larger PV in patients with BPH. The CCR2 rs1799864 polymorphism was
revealed as a protective factor against high IPSS and low Qmax. No
association was observed between the CCL2 rs1024611 polymorphism
and BPH. These results suggest that CCR2 and CCL5 serve a role in
BPH development and progression.
Acknowledgments
The authors would like to express their deepest
gratitude to all the people who contributed to this paper.
Funding
The present study was funded by the National Natural
Science Foundation of China (81672519), the Doctoral Supervisor
Research Foundation of Lanzhou University Second Hospital
(bdkyjj-01) and the Fundamental Research Funds for the Central
Universities (lzujbky-2018-kb14).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
YP, YG and HL were involved in conceptualization,
methodology, formal analysis and investigations, writing the
original draft of the manuscript and project administration. SJ, CP
and WL conducted the formal analysis and investigations, data
curation and project administration. YZ and HW conducted
investigations and data curation. DK and RR contributed to the
conceptualization, writing, reviewing and editing of the
manuscript. ZW was involved in conceptualization, methodology and
validation, as well as writing, reviewing and editing the
manuscript, supervision and project administration. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
All participants provided signed informed consent
for the donation of their data for clinical research. The study was
conducted in accordance with the Declaration of Helsinki, and was
approved by the Ethics Committee at Lanzhou University Second
Hospital.
Patient consent for publication
Consent for publication was obtained from all
patients.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
BPH
|
benign prostatic hyperplasia
|
|
CCL2
|
C-C motif chemokine ligand 2
|
|
CCR2
|
C-C motif chemokine receptor 2
|
|
CCL5
|
C-C motif chemokine ligand 5
|
|
SNP
|
single nucleotide polymorphism
|
|
OABSS
|
Overactive Bladder Symptom Score
|
|
IPSS
|
International Prostate Symptom
Score
|
|
PSA
|
prostate specific antigen
|
|
QOL
|
quality of life
|
|
Qmax
|
peak urinary flow rate
|
|
VV
|
voided volume
|
|
PV
|
prostate volume
|
|
PVR
|
post-void residual volume
|
References
|
1
|
Thorpe A and Neal D: Benign prostatic
hyperplasia. Lancet. 361:1359–1367. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Roehrborn CG: Male lower urinary tract
symptoms (LUTS) and benign prostatic hyperplasia (BPH). Med Clin
North Am. 95:87–100. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
He Q, Wang Z, Liu G, Daneshgari F,
MacLennan GT and Gupta S: Metabolic syndrome, inflammation and
lower urinary tract symptoms: Possible translational links.
Prostate Cancer Prostatic Dis. 19:7–13. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fibbi B, Penna G, Morelli A, Adorini L and
Maggi M: Chronic inflammation in the pathogenesis of benign
prostatic hyperplasia. Int J Androl. 33:475–488. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bostanci Y, Kazzazi A, Momtahen S, Laze J
and Djavan B: Correlation between benign prostatic hyperplasia and
inflammation. Curr Opin Urol. 23:5–10. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kramer G, Mitteregger D and Marberger M:
Is benign prostatic hyperplasia (BPH) an immune inflammatory
disease? Eur Urol. 51:1202–1216. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sambyal V, Guleria K, Kapahi R, Manjari M,
Sudan M, Uppal MS and Singh NR: Association of the-2518 A/G
polymorphism of MCP-1 with breast cancer in Punjab, North-West
India. Asian Pac J Cancer Prev. 16:7243–7248. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gao J, Liu X, Wei L, Niu D, Wei J, Wang L,
Ge H, Wang M, Yu Q, Jin T, et al: Genetic variants of MCP-1 and
CCR2 genes and IgA nephropathy risk. Oncotarget. 7:77950–77957.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kouhpayeh HR, Taheri M, Baziboroon M,
Naderi M, Bahari G and Hashemi M: CCL5 rs2107538 polymorphism
increased the risk of tuberculosis in a sample of Iranian
population. Prague Med Rep. 117:90–97. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
De Nunzio C, Presicce F and Tubaro A:
Inflammatory mediators in the development and progression of benign
prostatic hyperplasia. Nat Rev Urol. 13:613–626. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sáenz-López P, Carretero R, Cózar JM,
Romero JM, Canton J, Vilchez JR, Tallada M, Garrido F and
Ruiz-Cabello F: Genetic polymorphisms of RANTES, IL1-A,
MCP-1 and TNF-A genes in patients with prostate cancer.
BMC Cancer. 8:3822008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sun T, Mary LG, Oh WK, Freedman ML,
Pomerantz M, Pienta KJ and Kantoff PW: Inherited variants in the
chemokine CCL2 gene and prostate cancer aggressiveness in a
Caucasian cohort. Clin Cancer Res. 17:1546–1552. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kucukgergin C, Isman FK, Cakmakoglu B,
Sanli O and Seckin S: Association of polymorphisms in MCP-1,
CCR2, and CCR5 genes with the risk and
clinicopathological characteristics of prostate cancer. DNA Cell
Biol. 31:1418–1424. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mandal RK, Agrawal T and Mittal RD:
Genetic variants of chemokine CCL2 and chemokine receptor
CCR2 genes and risk of prostate cancer. Tumour Biol.
36:375–381. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kaplan SA, Olsson CA and Te AE: The
American Urological Association symptom score in the evaluation of
men with lower urinary tract symptoms: At 2 years of followup, does
it work? J Urol. 155:1971–1974. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Homma Y, Yoshida M, Seki N, Yokoyama O,
Kakizaki H, Gotoh M, Yamanishi T, Yamaguchi O, Takeda M and
Nishizawa O: Symptom assessment tool for overactive bladder
syndrome-overactive bladder symptom score. Urology. 68:318–323.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kaplan SA, McConnell JD, Roehrborn CG,
Meehan AG, Lee MW, Noble WR, Kusek JW and Nyberg LM Jr; Medical
Therapy of Prostatic Symptoms (MTOPS) Research Group, : Combination
therapy with doxazosin and finasteride for benign prostatic
hyperplasia in patients with lower urinary tract symptoms and a
baseline total prostate volume of 25 ml or greater. J Urol.
175:217–221. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Siami P, Roehrborn CG, Barkin J, Damiao R,
Wyczolkowski M, Duggan A, Major-Walker K and Morrill BB; CombAT
study group, : Combination therapy with dutasteride and tamsulosin
in men with moderate-to-severe benign prostatic hyperplasia and
prostate enlargement: The CombAT (Combination of Avodart and
Tamsulosin) trial rationale and study design. Contemp Clin Trials.
28:770–779. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Boyle AP, Hong EL, Hariharan M, Cheng Y,
Schaub MA, Kasowski M, Karczewski KJ, Park J, Hitz BC, Weng S, et
al: Annotation of functional variation in personal genomes using
RegulomeDB. Genome Res. 22:1790–1797. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Grzegorzewska AE, Pajzderski D, Sowinska A
and Jagodzinski PP: Monocyte chemoattractant protein-1 gene
(MCP-1-2518 A/G) polymorphism and serological markers of hepatitis
B virus infection in hemodialysis patients. Med Sci Monit.
20:1101–1116. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang J, Song Q, Zhu K, Lu J, Xiong X and
Hao F: The association of genetic variants in chemokine genes with
the risk of psoriasis vulgaris in Chinese population: A
case-control study. Medicine. 96:e82832017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Nakayama EE, Tanaka Y, Nagai Y, Iwamoto A
and Shioda T: A CCR2-V64I polymorphism affects stability of
CCR2A isoform. AIDS. 18:729–738. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhao N, Liu X, Wang Y, Liu X, Li J, Yu L,
Ma L, Wang S, Zhang H, Liu L, et al: Association of inflammatory
gene polymorphisms with ischemic stroke in a Chinese Han
population. J Neuroinflammation. 9:1622012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Nyquist PA, Winkler CA, McKenzie LM, Yanek
LR, Becker LC and Becker DM: Single nucleotide polymorphisms in
monocyte chemoattractant protein-1 and its receptor act
synergistically to increase the risk of carotid atherosclerosis.
Cerebrovasc Dis. 28:124–130. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Singh V, Srivastava P, Srivastava N,
Kapoor R and Mittal RD: Association of inflammatory chemokine gene
CCL2I/D with bladder cancer risk in North Indian population.
Mol Biol Rep. 39:9827–9834. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Fujita K, Ewing CM, Getzenberg RH, Parsons
JK, Isaacs WB and Pavlovich CP: Monocyte chemotactic protein-1
(MCP-1/CCL2) is associated with prostatic growth dysregulation and
benign prostatic hyperplasia. Prostate. 70:473–481. 2010.PubMed/NCBI
|
|
27
|
Santos EU, Lima GD, Oliveira Mde L,
Heráclio Sde A, Silva HD, Crovella S, Maia Mde M and Souza PR: CCR2
and CCR5 genes polymorphisms in women with cervical lesions from
Pernambuco, Northeast Region of Brazil: A case-control study. Mem
Inst Oswaldo Cruz. 111:174–180. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Nickel RG, Casolaro V, Wahn U, Beyer K,
Barnes KC, Plunkett BS, Freidhoff LR, Sengler C, Plitt JR,
Schleimer RP, et al: Atopic dermatitis is associated with a
functional mutation in the promoter of the C-C chemokine RANTES. J
Immunol. 164:1612–1616. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Areeshi MY, Mandal RK, Panda AK and Haque
S: A meta-analysis of the association between the CC chemokine
ligand 5 (CCL5)-403 G>A gene polymorphism and tuberculosis
susceptibility. PLoS One. 8:e721392013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kidd LR, Jones DZ, Rogers EN, Kidd NC,
Beache S, Rudd JE, Ragin C, Jackson M, McFarlane-Anderson N,
Tulloch-Reid M, et al: Chemokine Ligand 5 (CCL5), chemokine
receptor (CCR5) genetic variants, prostate cancer risk among men of
African Descent: A case-control study. Hered Cancer Clin Pract.
10:162012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhou Y, Lim KC, Onodera K, Takahashi S,
Ohta J, Minegishi N, Tsai FY, Orkin SH, Yamamoto M and Engel JD:
Rescue of the embryonic lethal hematopoietic defect reveals a
critical role for GATA-2 in urogenital development. EMBO J.
17:6689–6700. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Robinson JL, Tzou KS, Parker AS, Heckman
MG, Wu KJ, Hilton TW, Pisansky TM, Schild SE, Peterson JL, Vallow
LA, et al: GATA2 expression and biochemical recurrence following
salvage radiation therapy for relapsing prostate cancer. Br J
Radiol. 90:201701742017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Rodriguez-Bravo V, Carceles-Cordon M,
Hoshida Y, Cordon-Cardo C, Galsky MD and Domingo-Domenech J: The
role of GATA2 in lethal prostate cancer aggressiveness. Nat Rev
Urol. 14:38–48. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Vidal SJ, Rodriguez-Bravo V, Quinn SA,
Rodriguez-Barrueco R, Lujambio A, Williams E, Sun X, de la
Iglesia-Vicente J, Lee A, Readhead B, et al: A targetable
GATA2-IGF2 axis confers aggressiveness in lethal prostate cancer.
Cancer Cell. 27:223–239. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhernakova A, Alizadeh BZ, Eerligh P,
Hanifi-Moghaddam P, Schloot NC, Diosdado B, Wijmenga C, Roep BO and
Koeleman BP: Genetic variants of RANTES are associated with serum
RANTES level and protection for type 1 diabetes. Genes Immun.
7:544–549. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Bracci PM, Skibola CF, Conde L, Halperin
E, Lightfoot T, Smith A, Paynter RA, Skibola DR, Agana L, Roman E,
et al: Chemokine polymorphisms and lymphoma: A pooled analysis.
Leuk Lymphoma. 51:497–506. 2010. View Article : Google Scholar : PubMed/NCBI
|