Introduction
Despite the increased understanding of its
pathogenic risk and the development of progressive therapeutic
strategies, colorectal cancer (CRC) is the third most commonly
diagnosed cancer in men and the second most commonly diagnosed
cancer in women worldwide (1).
Colon cancer is often referred to as a homogeneous
entity, which should be treated accordingly. However, recent
advances on CRC that have identified more subgroups of colon cancer
may challenge this concept. These studies revealed that colon
cancer can be divided into two subgroups (proximal and distal to
the splenic flexure), with specific molecular, clinical and
pathological characteristics (2–4).
Biological differences between the left and right colon may partly
explain the significant heterogeneity of the two sides. The
difference of embryonic derivation should take into account that
proximal and distal colon originate from the midgut and the
hindgut, respectively, which may be the initiating factor (5). Additionally, the differential
bacterial flora from the left and right colon may contribute to the
heterogeneity (6). Thus, the
location of the tumour may be an important factor that is worth
taking into consideration. However, little is known about the
molecular mechanism, especially with regard to non-coding RNA
molecules. To further detect the potentially location-related
mechanisms, cancer-specific RNAs from left and right colon cancer
were identified and a competing endogenous RNA (ceRNA) network was
established based on 3 types of RNAs, including long non-coding
RNAs (lncRNAs), microRNAs (miRNAs) and mRNAs, which were
differentially expressed in the two sides.
Non-coding RNAs (ncRNAs), which play crucial
biological roles (7), can be
divided into small ncRNAs (<200 bp) and long ncRNAs (>200 bp)
based on the number of base pairs. Numerous studies have revealed
that lncRNAs play important roles in the process of tumourigenesis
(8–10). The competing endogenous RNA (ceRNA)
hypothesis presented by Salmena et al in 2011 indicated a
regulatory RNA network (11). All
types of RNA transcripts, including mRNAs, lncRNAs and pseudogene
transcripts in the network, could communicate with each other and
compete for the binding of miRNA response elements (MREs). This
competition exerts a crucial role in tumourigenesis by affecting
the expression levels of various RNAs through MREs.
Several studies on the differences between the
proximal and distal colon in pathway activation and their clinical
implications have been reported (12,13).
However, there is still a lack of large sample size studies and
cancer-specific lncRNA biomarkers concerning the differences of the
colon sides, and almost none of the studies focused on the
potential ceRNA network. To detect the relationship between RNAs of
these two sides, data was downloaded from The Cancer Genome Atlas
(TCGA) (http://cancergenome.nih.gov), which
contains mRNA, miRNA and lncRNA data of 186 samples of tumour
tissues and 17 adjacent non-tumour colon tissues in the left colon
and 229 samples of tumour tissues and 21 adjacent non-tumour colon
tissue in the right colon. To the best of our knowledge, our study
is the first to use a large-scale sequencing database to explore
the side-specific lncRNA expression profiles and ceRNA
co-expression network in the proximal and distal colon. The present
study may further our insight into the potentially location-related
mechanisms and help clarify the functions of lncRNAs in colon
cancer.
Materials and methods
Patients and samples
RNA expression and clinical data were downloaded,
including sex, TNM stage, survival information, from TCGA database.
The criteria of exclusion were set as follows: i) Histologic
diagnosis was not colon cancer; ii) tissue samples without enough
data for analysis; iii) patients who suffered other malignancies;
and iv) patients who had received preoperative chemotherapy. A
total of 186 colon tumour tissues and 17 adjacent non-tumour colon
tissues in the left colon and 229 colon tumour tissues and 21
adjacent non-tumour colon tissues in the right colon were collected
in the present study. Our study fully abided by the publication
guidelines of TCGA, and thus the approval of an Ethics Committee
was not required.
A total of 116 paired colon cancer tissue samples
(58 pairs from both sides) were surgically obtained between June
2005 and June 2018 at The First Affiliated Hospital of Nanjing
Medical University (Jiangsu, China). Our study was approved by the
Research Ethics Committee of Nanjing Medical University, and
informed consent was obtained from all patients. All the tissues
were frozen in liquid nitrogen immediately after surgical excision
and stored at −80°C. The TNM stage was classified on the basis of
the National Comprehensive Cancer Network (NCCN) guidelines.
Patients who received any preoperative treatments were not included
in the present study.
RNA sequence data sets and
computational analysis
The colon cancer (COAD) RNA expression prolife data
(level 3) was downloaded from TCGA database (September 2017)
(http://cancergenome.nih.gov). We
obtained the normalized count data of RNA sequencing, including
lncRNA and mRNA expression profiles. Level 3 miRNAseq data was
obtained from TCGA by an Illumina HiSeq 2000 miRNA sequencing
(miRNAseq) platform (Illumina, Inc., Hayward, CA, USA). First, the
tumour samples were divided into 2 groups (left and right colon
cancer). Then, we compared the differentially expressed lncRNAs
(DElncRNA), mRNAs (DEmRNA) and miRNAs (DEmiRNA) between tumour
tissues and adjacent non-tumour tissues using the Empirical
Analysis of Digital Gene Expression Data package in R (edgeR, R
version 3.4.1, http://www.bioconductor.org/packages/) [absolute
log2(fold change)>2.0, FDR<0.01] in these 2
groups. In the next step, the intersecting lncRNAs were identified
in the aforementioned 2 groups. After removing the elements of the
intersection from DElncRNAs (RIDElncRNAs) of the left and right
side, the DElncRNAs included exclusively in left or right colon
cancer were obtained. Fig. 1
displays the bioinformatics analysis process.
Construction of the ceRNA network
According to the relationship among lncRNAs, miRNAs
and mRNAs and the theory that lncRNAs can regulate miRNAs by
binding them and further regulate mRNAs, a ceRNA network was
constructed. miRcode (http://www.mircode.org/) was used to predict the
differentially expressed miRNA targets to find the lncRNA-miRNA
interactions. TargetScan (http://www.TargetScan.org/), miRDB (http://www.mirdb.org/) and miRanda (http://www.microrna.org/microrna/home.do) were used to
predict miRNA-targeted mRNA. Then, the intersection with the
differentially expressed lnRNAs and mRNAs was retained. Finally,
the lncRNA/miRNA/mRNA ceRNA network was constructed using Cytoscape
v3.0 (http://www.cytoscape.org/). Fig. 2. reveals the flowchart of the ceRNA
network.
GO and pathway analysis
Differentially expressed mRNAs included in the ceRNA
network were entered into the Database for Annotation,
Visualization, and Integrated Discovery (DAVID) bioinformatics
resource (https://david.ncifcrf.gov/) for
functional enrichment analysis.
Clinical feature analysis of key
lncRNAs
Based on the bioinformatics analysis and the ceRNA
network, the relationship between the clinical features, including
sex, age, tumour staging, TNM staging and lymphatic metastasis, and
the expression of key lncRNAs was analysed. In addition, the
association between side-specific lncRNAs and colon cancer patient
survival time was analysed.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
validation
RNA from tissue samples was extracted by TRIzol
reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA,
USA). A PrimeScript RT kit (Takara Biotechnology Co., Ltd., Dalian,
China) was used to synthesize complementary DNA (cDNA). qPCR was
performed in a 20-µl volume system (2 µl cDNA; 1.2 µl primers; 6.8
µl dH2O; 10 µl SYBR-Green) using FastStart Universal
SYBR-Green Master Kit (Roche Diagnostics, Indianapolis, IN, USA)
and a StepOnePlus Real-time PCR System (Applied Biosystems; Thermo
Fisher Scientific, Inc.). Thermocycling conditions were as follows:
Hot-start DNA polymerase activation at 95°C for 10 min, 40 cycles
at 95°C for 15 sec and 60°C for 1 min, followed by one cycle of
melt curve analysis at 95°C for 15 sec, 60°C for 1 min, and 95°C
for 15 sec. The data were analysed using the 2−ΔΔCq
(14) method and the mRNA
expression levels were normalized to GAPDH. The rimer sequences
were as follows: LINC00402 forward, 5′-TAGGCAGGAAAGAGGTTG-3′ and
reverse, 5′-TGGTAGGTAGCAGGTGGT-3′; KCNQ1OT1 forward,
5′-AGGGTGACAGTGTTTCATAGGCT-3′ and reverse,
5′-GAGGCACATTCATTCGTTGGT-3′; GAPDH forward,
5′-ACAGTCAGCCGCATCTTCTT-3′ and reverse, 5′-GACAAGCTTCCCGTTCTCAG-3′.
All RT-qPCR reactions were performed in triplicate.
Statistical analysis
All the results were expressed as the mean ± SD. R
Studio (R version 3.4.1; http://www.rstudio.com), Statistical Programme for
Social Sciences 20.0 (IBM Corp., Armonk, NY, USA) and GraphPad
Prism 5.0 software (GraphPad Software, Inc., La Jolla, CA, USA)
were used to analyse the data. The prognostic characteristics of
lncRNAs based on the univariate Cox proportional hazards regression
model were detected. Then, multivariate Cox regression analysis was
applied for further study. The differences in the qRT-PCR results
were compared by paired Student's t-test. P<0.05 was considered
to indicate a statistically significant difference.
Results
DElncRNAs that are included
exclusively in left or right colon cancer
It was determined that 879 lncRNAs (DElncRNA) were
differentially expressed between tumour samples and adjacent
tissues in left colon cancer from TCGA database [absolute
log2(fold change)>2.0, FDR<0.01]. There were 916
in right colon cancer. After removing the elements of the
intersection from DElncRNAs (RIDElncRNAs) of left and right colon
cancer, the DElncRNAs included exclusively in left or right colon
cancer were obtained, including 277 DElncRNAs (RIDElncRNAs) in the
left colon cancer and 314 DElncRNAs (RIDElncRNAs) in the right
colon cancer. Finally, 18 RIDElncRNAs were used to construct the
ceRNA network of left colon cancer (Tables I and II), and 21 RIDElncRNAs were used to
construct the network of right colon cancer (Tables I and III).
 | Table I.Key lncRNAs involved in the ceRNA
network. |
Table I.
Key lncRNAs involved in the ceRNA
network.
| lncRNAs from left
colon cancer | Log2
(fold change) | -Log (FDR) | lncRNAs from right
colon cancer | Log2
(fold change) | -Log (FDR) |
|---|
| CMAHP | −2.31 | 20.48 | LINC00483 | −2.15 | 17.50 |
| PRSS30P |
2.05 |
4.64 | LINC00488 | −3.20 | 15.36 |
| GRIK1-AS1 | −2.66 | 16.87 | LPP-AS1 |
3.94 |
2.06 |
| MIR7-3HG | −3.05 | 15.69 | COL4A2-AS2 |
2.53 |
2.92 |
| WT1-AS |
3.21 |
2.76 | ST7-AS2 |
3.47 |
2.38 |
| MIR22HG | −2.17 | 38.57 | MIR205HG |
5.31 |
2.26 |
| MUC19 |
3.33 |
2.53 | WASIR2 |
3.06 |
6.24 |
| LY86-AS1 | −2.12 |
5.68 | OSBPL10-AS1 |
2.75 |
2.59 |
| LINC00473 | −2.79 | 16.34 | ERVH48-1 |
3.12 |
2.77 |
| LINC00393 |
3.54 |
2.23 | DSCAM-AS1 |
4.55 |
2.11 |
| STEAP2-AS1 |
2.88 |
4.84 | EGOT |
2.28 |
4.27 |
| ATP11A-AS1 |
3.72 |
2.40 | THOC7-AS1 |
2.27 |
2.14 |
| CYP1B1-AS1 | −2.28 | 25.21 | ZBTB20-AS1 |
2.37 |
2.07 |
| BOK-AS1 |
5.57 |
3.63 | AC007731.1 |
3.47 |
3.56 |
| LINC00402 | −3.18 | 19.67 | ITGB5-AS1 |
2.50 |
2.40 |
| AC112721.1 |
2.84 |
3.18 | ARHGEF38-IT1 |
2.21 |
4.51 |
| GDNF-AS1 | −2.59 | 17.19 | ANO1-AS2 |
2.02 |
2.28 |
| ITPK1-AS1 |
2.38 |
2.01 | C8orf49 |
3.46 |
2.29 |
|
|
|
| RMST | −2.01 |
3.57 |
|
|
|
| NOVA1-AS1 | −2.01 |
3.48 |
|
|
|
| KCNQ1OT1 |
2.33 |
6.52 |
 | Table II.lncRNAs that may target specific
miRNAs in left colon cancer. |
Table II.
lncRNAs that may target specific
miRNAs in left colon cancer.
| lncRNAs | miRNAs |
|---|
| CMAHP | hsa-mir-96,
hsa-mir-141, hsa-mir-144, hsa-mir-145, hsa-mir-150, hsa-mir-424,
hsa-mir-182, hsa-mir-183, hsa-mir-187, hsa-mir-192, hsa-mir-215,
hsa-mir-375 |
| PRSS30P | hsa-mir-143,
hsa-mir-150, hsa-mir-424, hsa-mir-21 |
| GRIK1-AS1 | hsa-mir-145,
hsa-mir-375 |
| MIR7-3HG | hsa-mir-145,
hsa-mir-150 |
| WT1-AS | hsa-mir-96,
hsa-mir-141, hsa-mir-145, hsa-mir-424, hsa-mir-17, hsa-mir-182,
hsa-mir-98, hsa-mir-193b, hsa-mir-429, hsa-mir-22, hsa-mir-32,
hsa-mir-375 |
| MIR22HG | hsa-mir-375,
hsa-mir-32 |
| MUC19 | hsa-mir-375,
hsa-mir-22, hsa-mir-32, hsa-mir-429, hsa-mir-193b, hsa-mir-215,
hsa-mir-192, hsa-mir-187, hsa-mir-98, hsa-mir-182, hsa-mir-17,
hsa-mir-424, hsa-mir-150, hsa-mir-152, hsa-mir-14, hsa-mir-144,
hsa-mir-143, hsa-mir-96, hsa-mir-454 |
| LY86-AS1 | hsa-mir-375,
hsa-mir-187, hsa-mir-183, hsa-mir-182, hsa-mir-424, hsa-mir-150,
hsa-mir-145, hsa-mir-141, hsa-mir-96, hsa-mir-454 |
| LINC00473 | hsa-mir-424,
hsa-mir-150, hsa-mir-145 |
| LINC00393 | hsa-mir-215,
hsa-mir-192 |
| STEAP2-AS1 | hsa-mir-375,
hsa-mir-424, hsa-mir-152, hsa-mir-143 |
| ATP11A-AS1 | hsa-mir-22,
hsa-mir-215, hsa-mir-192, hsa-mir-187, hsa-mir-424, hsa-mir-152,
hsa-mir-143, hsa-mir-96 |
| CYP1B1-AS1 | hsa-mir-21,
hsa-mir-429, hsa-mir-193b, hsa-mir-150, hsa-mir-152, hsa-mir-145,
hsa-mir-454 |
| BOK-AS1 | hsa-mir-150 |
| LINC00402 | hsa-mir-22,
hsa-mir-429, hsa-mir-193b, hsa-mir-182, hsa-mir-17, hsa-mir-150,
hsa-mir-143, hsa-mir-141 |
| AC112721.1 | hsa-mir-424 |
| GDNF-AS1 | hsa-mir-187,
hsa-mir-424, hsa-mir-145, hsa-mir-143 |
| ITPK1-AS1 | hsa-mir-22,
hsa-mir-17, hsa-mir-150, hsa-mir-144, hsa-mir-141 |
 | Table III.lncRNAs that may target specific
miRNAs in right colon cancer. |
Table III.
lncRNAs that may target specific
miRNAs in right colon cancer.
| lncRNAs | miRNAs |
|---|
| LINC00483 | hsa-mir-223,
hsa-mir-21, hsa-mir-215, hsa-mir-192, hsa-mir-183, hsa-mir-182,
hsa-mir-17, hsa-mir-150, hsa-mir-144, hsa-mir-96, hsa-mir-106a |
| LINC00488 | hsa-mir-21,
hsa-mir-215, hsa-mir-192, hsa-mir-98, hsa-mir-144, hsa-mir-96 |
| LPP-AS1 | hsa-mir-338,
hsa-mir-143 |
| COL4A2-AS2 | hsa-mir-338,
hsa-mir-424, hsa-mir-150, hsa-mir-152 |
| ST7-AS2 | hsa-mir-22,
hsa-mir-429, hsa-mir-215, hsa-mir-192, hsa-mir-145 |
| MIR205HG | hsa-mir-22,
hsa-mir-215, hsa-mir-192, hsa-mir-183, hsa-mir-150, hsa-mir-152,
hsa-mir-145, hsa-mir-143, hsa-mir-454, hsa-mir-301b |
| WASIR2 | hsa-mir-338,
hsa-mir-193b, hsa-mir-150 |
| OSBPL10-AS1 | hsa-mir-375,
hsa-mir-182, hsa-mir-424, hsa-mir-145, hsa-mir-96 |
| ERVH48-1 | hsa-mir-338,
hsa-mir-223, hsa-mir-22, hsa-mir-21, hsa-mir-187, hsa-mir-98,
hsa-mir-182, hsa-mir-145, hsa-mir-144, hsa-mir-141, hsa-mir-96,
hsa-mir-454, hsa-mir-301b |
| DSCAM-AS1 | hsa-mir-338,
hsa-mir-150, hsa-mir-143, hsa-mir-141 |
| EGOT | hsa-mir-375,
hsa-mir-21, hsa-mir-183, hsa-mir-424, hsa-mir-143, hsa-mir-141 |
| THOC7-AS1 | hsa-mir-215,
hsa-mir-192, hsa-mir-187 |
| ZBTB20-AS1 | hsa-mir-217,
hsa-mir-152, hsa-mir-454, hsa-mir-301b |
| AC007731.1 | hsa-mir-215,
hsa-mir-192, hsa-mir-183, hsa-mir-150, hsa-mir-152 |
| ITGB5-AS1 | hsa-mir-21,
hsa-mir-193b |
| ARHGEF38-IT1 | hsa-mir-338,
hsa-mir-150, hsa-mir-143 |
| ANO1-AS2 | hsa-mir-375,
hsa-mir-98, hsa-mir-17, hsa-mir-152, hsa-mir-106a |
| C8orf49 | hsa-mir-375,
hsa-mir-338, hsa-mir-32, hsa-mir-429, hsa-mir-17, hsa-mir-424,
hsa-mir-150, hsa-mir-152, hsa-mir-143, hsa-mir-106a, hsa-mir-454,
hsa-mir-301b |
| RMST | hsa-mir-375,
hsa-mir-338, hsa-mir-32, hsa-mir-429, hsa-mir-193b, hsa-mir-182,
hsa-mir-17, hsa-mir-424, hsa-mir-150, hsa-mir-145, hsa-mir-144,
hsa-mir-96, hsa-mir-454, hsa-mir-301b |
| NOVA1-AS1 | hsa-mir-223,
hsa-mir-22, hsa-mir-217 |
| KCNQ1OT1 | hsa-mir-375,
hsa-mir-338, hsa-mir-32, hsa-mir-223, hsa-mir-22, hsa-mir-217,
hsa-mir-429, hsa-mir-193b, hsa-mir-215, hsa-mir-192, hsa-mir-187,
hsa-mir-98, hsa-mir-183, hsa-mir-301b, hsa-mir-454, hsa-mir-106a,
hsa-mir-96, hsa-mir-141, hsa-mir-143, hsa-mir-145, hsa-mir-152,
hsa-mir-150, hsa-mir-182, hsa-mir-17, hsa-mir-424 |
Prediction of lncRNA-targeted
miRNAs
For further investigation, 165 and 227 side-specific
miRNAs that were differentially expressed between tumour tissues
and adjacent tissues from left and right colon cancer,
respectively, were determined. These miRNAs were identified as
side-specific miRNAs. Then, it was determined whether these 165
miRNAs could target the aforementioned 277 RIDElncRNAs in left
colon cancer and whether the 227 miRNAs could target the 314
RIDElncRNAs in right colon cancer. Based on miRcode (http://www.mircode.org/), 22 miRNAs targeting 18
lncRNAs were predicted in left colon cancer (Table II), and there were 27 miRNAs and 21
lncRNAs predicted in the ceRNA of right colon cancer (Table III).
Prediction of miRNA-targeted
mRNAs
First, 2,028 and 2,069 differentially expressed
mRNAs between tumour tissues and adjacent tissues [absolute
log2(fold change)>2, FDR<0.01] in left and right
colon cancer, respectively, were identified. These mRNAs were
identified as side-specific mRNAs (DEmRNAs). The 2,028 and 2,069
DEmRNAs were analysed with DAVID bioinformatics resources.
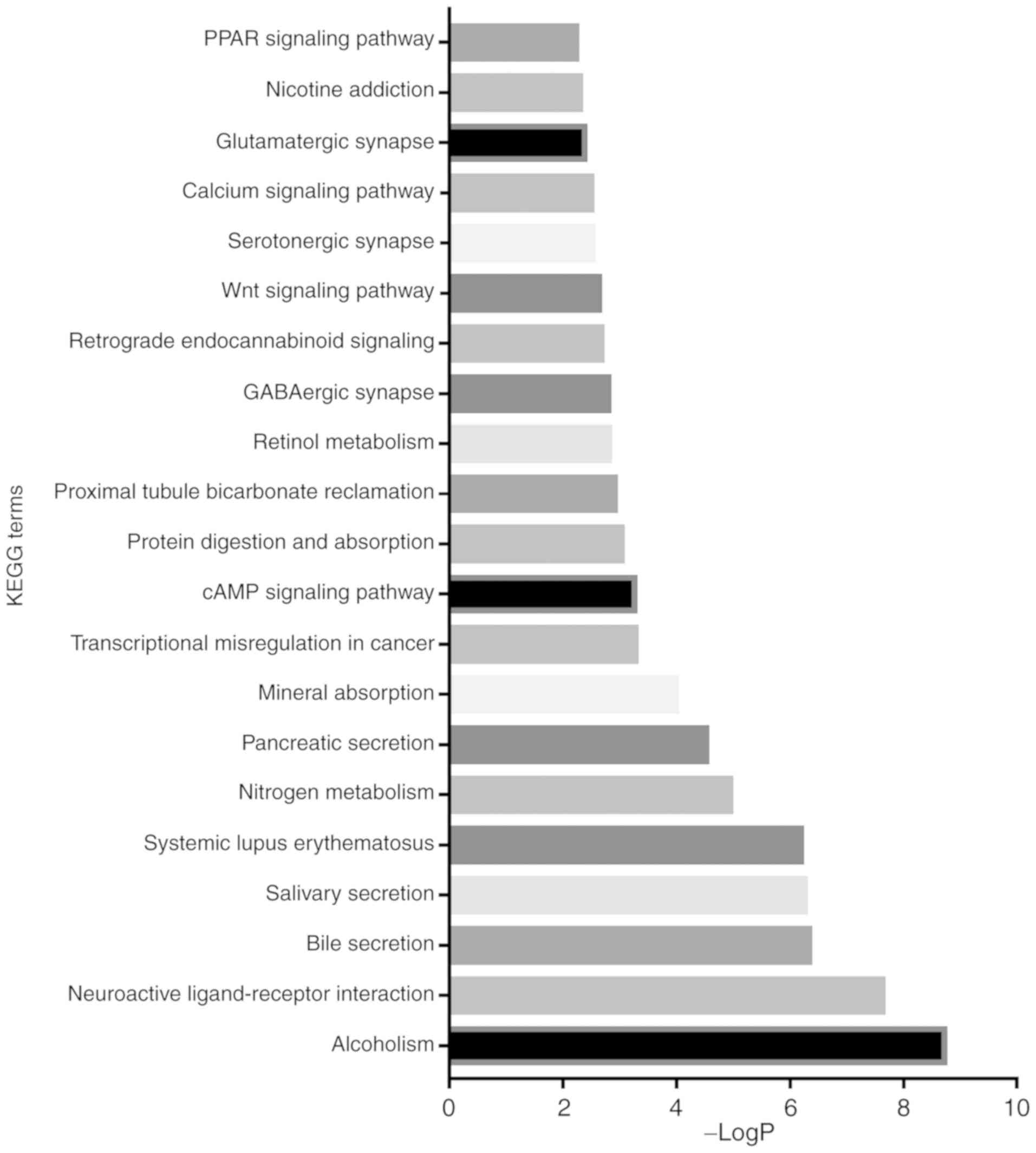
According to the P-values, the top 21 KEGG pathways of the DEmRNAs
were revealed (Figs. 3 and 4). Among these pathways, the
transcriptional dysregulation in cancer, cAMP, Wnt and PPAR
signalling pathways, which were related to cancer-associated
pathways, were revealed to be more important in left colon cancer.
However, the chemical carcinogenesis pathway was more involved in
right colon cancer. For further investigation, the miRNAs included
in the ceRNA network were then used to predict the targeted mRNAs
using miRanda, TargetScan and miRDB. The intersection of the
predicted mRNAs and DEmRNAs was obtained. Finally, 57 mRNAs were
included in the left colon cancer ceRNA network, and 55 were
included in the right (Tables IV
and V).
 | Table IV.miRNAs that may target specific mRNAs
in left colon cancer. |
Table IV.
miRNAs that may target specific mRNAs
in left colon cancer.
| miRNAs | mRNAs |
|---|
| hsa-mir-141 | PHLPP2, ELAVL4,
MACC1, KIAA1549, EPHA7 |
| hsa-mir-143 | COL1A1,
SERPINE1 |
| hsa-mir-144 | GRIK3, TBX18 |
| hsa-mir-145 | SERPINE1 |
| hsa-mir-150 | HILPDA, SLC7A11,
ZNF460, EREG |
| hsa-mir-152 | NPTX1, BMP3,
KLF4 |
| hsa-mir-17 | E2F1, FOXQ1, CADM2,
FJX1, CFL2, SALL3, FAM129A, SMOC1, CLIP4, FAM46C, ANKRD33B |
| hsa-mir-182 | CHL1, NPTX1,
TCEAL7, FOXF2, ULBP2 |
| hsa-mir-183 | KIF5C |
| hsa-mir-192 | TCF7 |
| hsa-mir-193b | PLAU, DCAF7 |
| hsa-mir-21 | EDIL3, OSR1,
ATP2B4, TGFBI |
| hsa-mir-215 | TCF7 |
| hsa-mir-22 | RGS2 |
| hsa-mir-32 | UGP2, CCDC113,
PHLPP2, ATP2B4 |
| hsa-mir-375 | ELAVL4 |
| hsa-mir-424 | PSAT1, PHLPP2,
ZNRF3, TPM2, TMEM100, AXIN2, CBX2 |
| hsa-mir-454 | RBM20, SALL3, CFL2,
SMOC1 |
| hsa-mir-96 | ALK, TRIB3 |
| hsa-mir-98 | TRIM71, CPA4,
IGF2BP3, HAND1, SLC5A6, PRSS22, IGF2BP1 |
 | Table V.miRNAs that may target specific mRNAs
in right colon cancer. |
Table V.
miRNAs that may target specific mRNAs
in right colon cancer.
| miRNAs | mRNAs |
|---|
| hsa-mir-106a | CFL2, FAM129A,
FJX1, CADM2, FOXQ1 |
| hsa-mir-141 | KIAA1549, PHLPP2,
ELAVL4, EPHA7, DPY19L1 |
| hsa-mir-143 | COL1A1,
SERPINE1 |
| hsa-mir-144 | GRIK3, KCNQ5 |
| hsa-mir-145 | SERPINE1 |
| hsa-mir-150 | ZNF460, PDCD4,
SLC7A11, HILPDA, EREG |
| hsa-mir-152 | KLF4, NPTX1,
BMP3 |
| hsa-mir-17 | FOXQ1, CADM2, FJX1,
FAM129A, CFL2, SLC16A9, CYBRD1 |
| hsa-mir-182 | ULBP2, CHL1, NPTX1,
FOXF2 |
| hsa-mir-183 | PDCD4 |
| hsa-mir-192 | GRHL1 |
| hsa-mir-193b | PLAU |
| hsa-mir-21 | PDCD4, TGFBI,
JPH1 |
| hsa-mir-217 | DACH1 |
| hsa-mir-223 | ECT2, EPB41L3 |
| hsa-mir-32 | PAX9, PBLD, MIER3,
PHLPP2, UGP2 |
| hsa-mir-338 | NOVA1 |
| hsa-mir-375 | ELAVL4 |
| hsa-mir-424 | TPM2, TMEM100,
PHLPP2, CBX2, E2F7, PSAT1, AXIN2 |
| hsa-mir-429 | PMAIP1 |
| hsa-mir-454 | CFL2, RBM20 |
| hsa-mir-96 | TRIB3 |
| hsa-mir-98 | RGS16, IGF2BP3,
TRIM71, PRSS22, HAND1, IGF2BP1, FMO4, CPA4 |
According to the relationship between RNAs revealed
by Tables II–V, lncRNA-miRNA-mRNA ceRNA networks were
constructed. Cytoscape 3.0 was used to draw the ceRNA network. In
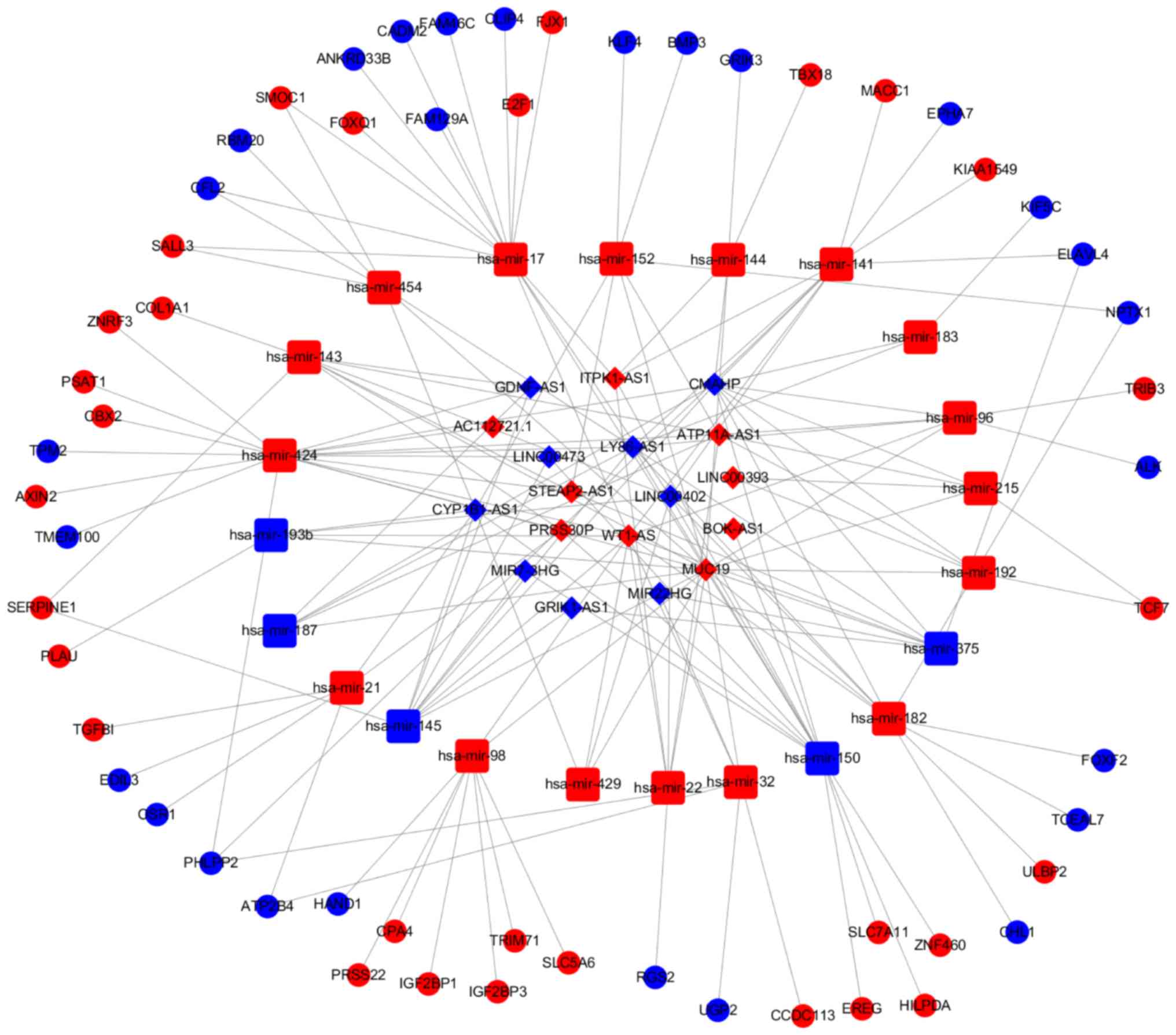
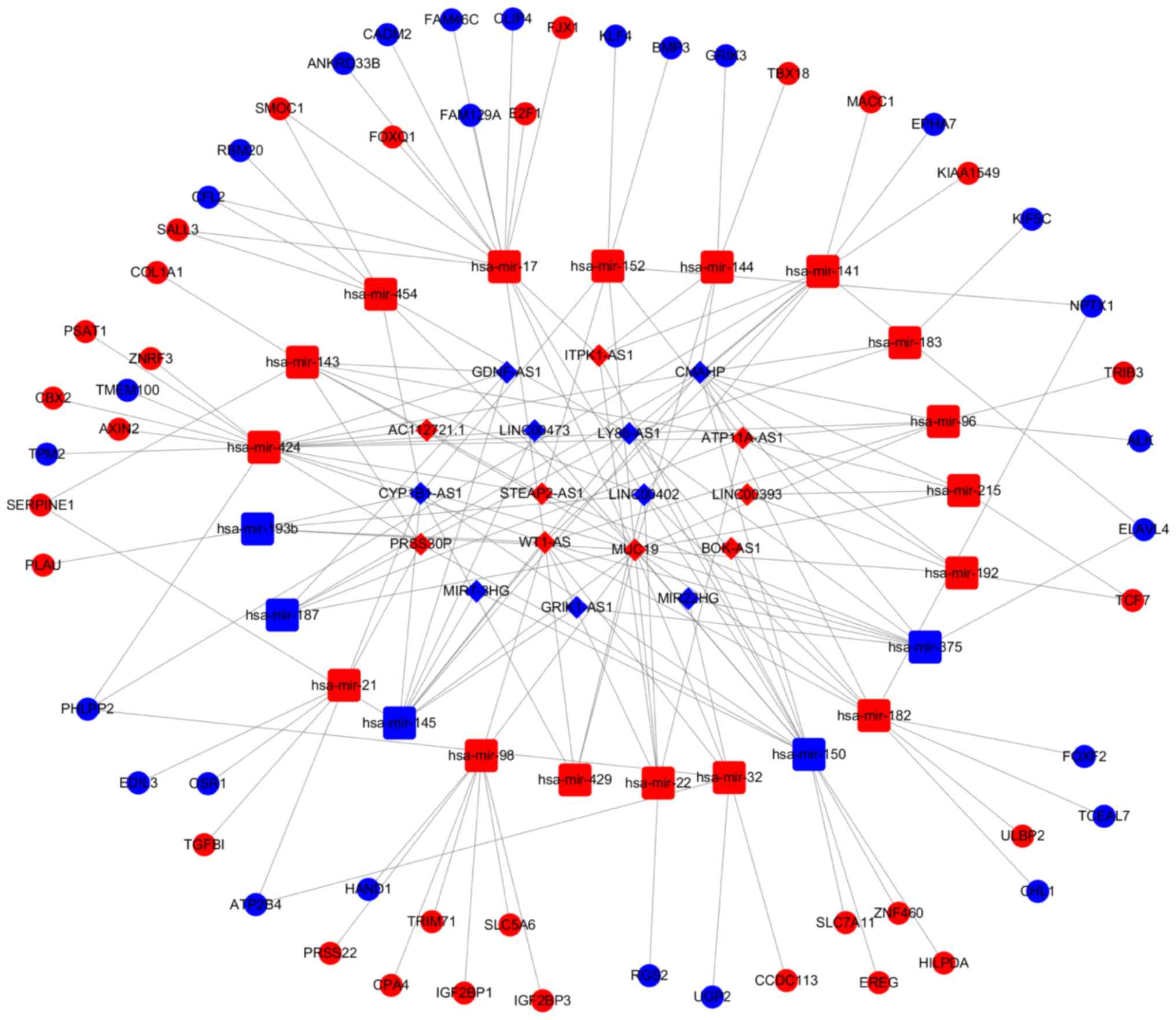
our results, there were 18 lncRNAs, 22 miRNAs and 57 mRNAs included
in the left colon cancer ceRNA network and 21 lncRNAs, 27 miRNAs
and 55 mRNAs included in the right ceRNA network (Figs. 5 and 6).
Key lncRNAs and clinical feature
association
The clinical features of the key lncRNAs in the
ceRNA network were then further analysed. The clinical features,
including sex, tumour grade, TNM stage and lymphatic metastasis
status, were provided by TCGA data sets. In total 15 lncRNAs were
revealed, including 3 in left colon cancer and 12 in right colon
cancer, which were significantly related to clinical features
(Table VI). The results revealed
that CYP1B1-AS1, LPP-AS1, MIR205HG, DSCAM-AS1, RMST and NOVA1-AS1
were related to age, LPP-AS1, WASIR2, OSBPL10-AS1 and DSCAM-AS1
were related to sex, AC112721.1, DSCAM-AS1, ARHGEF38-IT1, C8orf49
and KCNQ1OT1 were related to lymphatic metastasis, LINC00402 and
ZBTB20-AS1 were related to tumour grade, and AC112721.1, DSCAM-AS1,
ARHGEF38-IT1, C8orf49 and KCNQ1OT1 were related to TNM stage
(Table VI).
 | Table VI.Correlation between key lncRNAs
involved in the ceRNA network and their clinical features. |
Table VI.
Correlation between key lncRNAs
involved in the ceRNA network and their clinical features.
| Comparisons | Left colon
cancer | Right colon
cancer |
|---|
| Age (<60 vs.
>60 years) | CYP1B1-AS1 | LPP-AS1, MIR205HG,
DSCAM-AS1, RMST, NOVA1-AS1 |
| Sex (female vs.
male) |
| LPP-AS1, WASIR2,
OSBPL10-AS1, DSCAM-AS1 |
| Lymphatic
metastasis (no vs. yes) | AC112721.1 | DSCAM-AS1,
ARHGEF38-IT1, C8orf49, KCNQ1OT1 |
| TNM staging system
(T1+T2 vs. T3+T4) | LINC00402 | ZBTB20-AS1 |
| Tumour stage (stage
I, II vs. stage III, IV) | AC112721.1 | DSCAM-AS1,
ARHGEF38-IT1, C8orf49, KCNQ1OT1 |
| MSI status (MSI-H
vs. other status) |
| LINC00483,
LINC00488, WASIR2, DSCAM-AS1, NOVA1-AS1 |
Furthermore, the prognostic characteristics of
RIDElncRNAs from both sides were detected. Based on the univariate
Cox proportional hazards regression model, the association of the
overall survival of patients with the expression level of
RIDElncRNAs was analysed. Finally, 20 lncRNAs from left colon
cancer that were significantly associated with overall survival
(log-rank P<0.05) (Fig. S1 and
Table VII) were revealed, and 25
lncRNAs from right colon cancer were determined to be related to
overall survival (log-rank P<0.05) (Fig. S2 and Table VII). Multivariate Cox regression
analysis was then performed with these lncRNAs. Fifteen lncRNAs
from the left colon cancer were revealed to be independent factors
of survival time and 12 in the right colon cancer. The results are
presented in Table VIII.
 | Table VII.Kaplan-Meier survival analysis for
lncRNAs that were associated with overall survival. |
Table VII.
Kaplan-Meier survival analysis for
lncRNAs that were associated with overall survival.
| lncRNAs from the
left | P-value | lncRNAs from the
right | P-value |
|---|
| AC019118.4 | 0.003 | AC003991.3 | 0.005 |
| CTB-181H17.1 | 0.01 | AC011288.2 | 0.003 |
| CTD-2308L22.1 | 0.04 | AC012531.25 | 0.01 |
| FGF13-AS1 | 0.006 | B4GALT1-AS1 | 0.0005 |
| HORMAD2-AS1 | 0.03 | CTC-428G20.6 | 0.02 |
| IGBP1-AS2 | 0.03 | CTC-573N18.1 | 0.05 |
| LINC01990 | 0.02 | CTD-2591A5.2 | 0.05 |
| RP1-10C16.1 | 0.03 | CTD-3064M3.7 | 0.05 |
| RP11-108K3.1 | 0.04 | LINC00513 | 0.005 |
| RP11-205M3.3 | 0.03 | LINC01630 | 0.03 |
| RP11-281P23.2 | 0.002 | LINC01633 | 0.008 |
| RP11-342A23.2 | 0.01 | RP11-126H7.4 | 0.04 |
| RP11-354P11.4 | 0.02 | RP11-138A9.1 | 0.04 |
| RP11-475B2.1 | 0.03 | RP11-138A9.2 | 0.01 |
| RP11-515O17.2 | 0.01 | RP11-157F20.3 | 0.03 |
| RP11-674E16.4 | 0.02 | RP11-277P12.20 | 0.05 |
| RP11-686O6.1 | 0.04 | RP11-304L19.12 | 0.03 |
| RP11-713P17.3 | 0.01 | RP11-495P10.5 | 0.002 |
| RP3-453C12.15 | 0.04 | RP11-619I22.1 | 0.03 |
| RP6-114E22.1 | 0.02 | RP11-661A12.9 | 0.04 |
|
|
| RP11-67K19.3 | 0.05 |
|
|
| RP1-29C18.10 | 0.05 |
|
|
| RP4-676L2.1 | 0.05 |
|
|
| RP4-811H24.9 | 0.05 |
|
|
| SSTR5-AS1 | 0.03 |
 | Table VIII.Results of multivariate cox
regression analysis. |
Table VIII.
Results of multivariate cox
regression analysis.
| lncRNAs from the
left | β | OR (95% CI) | P-value | lncRNAs from the
right | β | OR (95% CI) | P-value |
|---|
| RP6-114E22.1 | −1.037 | 0.354
(0.158–0.793) | 0.012 | AC003991.3 | 0.676 | 1.966
(1.142–3.387) | 0.015 |
| RP3-453C12.15 | −0.515 | 0.597
(0.273–1.306) | 0.197 | AC011288.2 | −0.706 | 0.494
(0.286–0.851) | 0.011 |
| RP11-713P17.3 | −0.886 | 0.412
(0.188–0.904) | 0.027 | AC012531.25 | −0.672 | 0.511
(0.287–0.908) | 0.022 |
| RP11-686O6.1 | 0.809 | 2.245
(1.055–4.778) | 0.036 | B4GALT1-AS1 | 0.879 | 2.408
(1.347–4.305) | 0.003 |
| RP11-674E16.4 | 0.684 | 1.982
(0.899–4.369) | 0.09 | CTC-428G20.6 | 0.413 | 1.511
(0.869–2.628) | 0.144 |
| RP11-515O17.2 | 1.404 | 4.072
(1.591–10.424) | 0.003 | CTC-573N18.1 | −0.543 | 0.581
(0.335–1.007) | 0.053 |
| RP11-475B2.1 | −0.931 | 0.394
(0.179–0.865) | 0.02 | CTD-2591A5.2 | −0.708 | 0.492
(0.280–0.866) | 0.014 |
| RP11-354P11.4 | 1.049 | 2.854
(1.252–6.506) | 0.013 | CTD-3064M3.7 | −0.541 | 0.582
(0.336–1.009) | 0.054 |
| RP11-342A23.2 | −0.916 | 0.400
(0.182–0.877) | 0.022 | LINC00513 | 0.641 | 1.898
(1.079–3.337) | 0.026 |
| RP11-281P23.2 | 1.023 | 2.783
(1.213–6.384) | 0.016 | LINC01630 | 0.462 | 1.587
(0.896–2.811) | 0.113 |
| RP11-205M3.3 | 0.79 | 2.204
(1.016–4.780) | 0.045 | LINC01633 | −0.702 | 0.496
(0.285–0.863) | 0.013 |
| RP11-108K3.1 | −0.545 | 0.580
(0.255–1.318) | 0.193 | RP11-126H7.4 | −0.372 | 0.689
(0.398–1.194) | 0.184 |
| RP1-10C16.1 | −0.921 | 0.398
(0.180–0.879) | 0.023 | RP11-138A9.1 | 0.421 | 1.524
(0.887–2.619) | 0.127 |
| LINC01990 | −0.761 | 0.467
(0.208–1.051) | 0.066 | RP11-138A9.2 | 0.728 | 2.070
(1.150–3.729) | 0.015 |
| IGBP1-AS2 | 0.737 | 2.089
(0.904–4.828) | 0.085 | RP11-157F20.3 | −0.495 | 0.610
(0.349–1.064) | 0.081 |
| HORMAD2-AS1 | −1.334 | 0.263
(0.114–0.607) | 0.002 | RP11-277P12.20 | −0.478 | 0.620
(0.358–1.073) | 0.088 |
| FGF13-AS1 | −1.154 | 0.315
(0.143–0.695) | 0.004 | RP11-304L19.12 | −0.47 | 0.625
(0.365–1.069) | 0.086 |
| CTD-2308L22.1 | −1.085 | 0.338
(0.147–0.774) | 0.01 | RP11-495P10.5 | −0.693 | 0.500
(0.282–0.886) | 0.017 |
| CTB-181H17.1 | −1.03 | 0.357
(0.160–0.795) | 0.012 | RP11-619I22.1 | 0.578 | 1.783
(1.048–3.035) | 0.033 |
| AC019118.4 | −0.991 | 0.371
(0.170–0.810) | 0.013 | RP11-661A12.9 | −0.32 | 0.726
(0.417–1.265) | 0.259 |
|
|
|
|
| RP11-67K19.3 | 0.655 | 1.926
(1.103–3.363) | 0.021 |
|
|
|
|
| RP1-29C18.10 | −0.455 | 0.634
(0.370–1.086) | 0.097 |
|
|
|
|
| RP4-676L2.1 | 0.505 | 1.656
(0.924–2.969) | 0.09 |
|
|
|
|
| RP4-811H24.9 | −0.385 | 0.681
(0.394–1.176) | 0.168 |
|
|
|
|
| SSTR5-AS1 | 0.615 | 1.850
(1.061–3.225) | 0.03 |
Subsequently, to confirm the reliability of the
bioinformatics results, 2 key lncRNAs (LINC00402 from the left side
and KCNQ1OT1 from the right side) were randomly selected from the
networks and their RNA expression level was determined in 58 paired
colon cancer tissues from left colon cancer and right colon cancer.
The bioinformatics results revealed that LINC00402 presented lower
expression in left colon cancer tissues than adjacent non-tumour
tissues, and KCNQ1OT1 was significantly expressed higher in tumour
tissues. Fig. 7 revealed that the
qRT-PCR results were consistent with the bioinformatics results.
The relationship between the expression of the 2 lncRNAs and
clinicopathological characteristics was then further analysed. The
results revealed that LINC00402 was related to TNM staging
(Table IX), and KCNQ1OT1 was
significantly associated with lymphatic metastasis and tumour stage
(Table IX), which were almost the
same as the bioinformatics results (Table VI).
 | Table IX.Expression of lncRNAs related to
clinical features according to the clinicopathological
characteristics of patients. |
Table IX.
Expression of lncRNAs related to
clinical features according to the clinicopathological
characteristics of patients.
|
|
| LINC00402 |
|
|---|
|
|
|
|
|
|---|
|
Characteristics | No. | Low group | High group | P-value |
|---|
| Age (years) |
|
|
| 0.581 |
|
<60 | 20 | 11 | 9 |
|
|
≥60 | 38 | 18 | 20 |
|
| Sex |
|
|
| 0.597 |
|
Female | 26 | 12 | 14 |
|
|
Male | 32 | 17 | 15 |
|
| Lymphatic
metastasis |
|
|
| 0.185 |
| No | 33 | 14 | 19 |
|
|
Yes | 25 | 15 | 10 |
|
| TNM staging
system |
|
|
| 0.03 |
|
T1+T2 | 22 | 7 | 15 |
|
|
T3+T4 | 36 | 22 | 14 |
|
| Tumor stage |
|
|
| 0.293 |
| Stage
I, II | 30 | 13 | 17 |
|
| Stage
III, IV | 28 | 16 | 12 |
|
| MSI status |
|
|
| 0.066 |
|
MSI-H | 14 | 5 | 9 |
|
| Other
status | 44 | 22 | 12 |
|
|
|
|
|
KCNQ1OT1 |
|
|
|
|
|
|
|
Characteristics | No. | Low
group | High
group | P-value |
|
| Age (years) |
|
|
| 0.279 |
|
<60 | 22 | 9 | 13 |
|
|
≥60 | 36 | 20 | 16 |
|
| Sex |
|
|
| 0.430 |
|
Female | 27 | 12 | 15 |
|
|
Male | 31 | 17 | 14 |
|
| Lymphatic
metastasis |
|
|
| 0.007 |
| No | 36 | 23 | 13 |
|
|
Yes | 22 | 6 | 16 |
|
| TNM staging
system |
|
|
| 0.426 |
|
T1+T2 | 25 | 11 | 14 |
|
|
T3+T4 | 33 | 18 | 15 |
|
| Tumour stage |
|
|
| 0.033 |
| Stage
I, II | 34 | 21 | 13 |
|
| Stage
III, IV | 24 | 8 | 16 |
|
| MSI status |
|
|
| 0.557 |
|
MSI-H | 16 | 9 | 7 |
|
| Other
status | 42 | 20 | 22 |
|
Discussion
In terms of clinical behaviour and response to
therapy, colorectal cancer (CRC) is a heterogeneous disease
(15,16). Patients can benefit from this
heterogeneity when stratified for their response to therapeutic
strategies. However, CRC development involves the complex
transformation of molecular events of which we lack enough
knowledge. CRC still has a high incidence and mortality. It is our
hope that the study of the molecular difference between left and
right side colon cancer will help with the exploration of CRC
heterogeneity.
In the present study, we determined side-specific
lncRNAs, miRNAs and mRNAs based on the differential expression
between tumour tissues and adjacent non-tumour tissues in the two
sides. Through KEGG analysis, we analysed the pathways that the
side-specific mRNAs may be involved in. Combining the
bioinformatics resources, we established ceRNA networks with
side-specific DEmRNAs, DEmiRNAs and RIDElncRNAs. We then further
analysed the clinical features of the key lncRNAs belonging to the
ceRNA network. Side-specific RIDElncRNAs were further analysed to
determine whether they were correlated with overall survival. To
check the reliability of the bioinformatics results, we randomly
selected 2 key lncRNAs (LINC00402 from the left and KCNQ1OT1 from
the right) and determined their expression by qRT-PCR.
There were several cancer-related roles in the
RIDElncRNA group from the two sides. For example, MIR22HG was
reported to suppressed hepatocellular and endometrial carcinoma
(17,18) and linc00483 was reported to promoted
gastric cancer (19). We conducted
univariate and multivariate Cox regression analyses and found that
15 lncRNAs from the left and 12 lncRNAs from the right were found
to be independent factors of survival time (Table VIII). In KEGG analysis, we
identified the top 21 pathways of the DEmRNAs (Figs. 3 and 4). After removing the same KEGG terms, the
results revealed that different cancer-related pathways were
involved in the two sides, i.e., the transcriptional dysregulation
in cancer, cAMP, Wnt and PPAR signalling pathways were more
important in the left colon cancer, and the chemical carcinogenesis
pathway played a more important role in the right colon cancer.
Some of these cancer-related pathways, such as cAMP, Wnt and PPAR
signalling pathways, have been reported to play important roles in
the CRC. Lu et al reported that the cAMP pathway could be
activated to inhibit angiogenesis and vasculogenic mimicry in CRC
(20). As previous studies reported
(21,22), the Wnt pathway could reduce
apoptosis, stimulate cell proliferation and promote metastasis in
CRC. Zarkou et al revealed that lncRNAs can modulate the WNT
pathway by affecting gene expression through diversified
mechanisms, from the transcriptional to the post-translational
level (23). As for the PPAR
signalling pathway, it has been revealed to be inhibited in CRC
(24). However, few studies have
examined the performance of these pathways in left or right colon
cancer.
By constructing the ceRNA network, our research
focused on the potentially different mechanisms of distal and
proximal colon cancers. Several previous studies have already
reported the interactions between RNAs in the ceRNA network. For
example, MIR22HG, an lncRNA from the left ceRNA network, was
reported to interact with miR-141-3p and therefore inhibited
endometrial carcinoma cell proliferation (18). Additionally, KCNQ1OT1, which
belonged to the right ceRNA network, modulated CCNE2 by sponging
miR-145 in BRCA (25). Another
study reported that KCNQ1OT1 could ameliorate particle-induced
osteolysis by inhibiting miR-21a-5p (26). These previous studies strongly
demonstrated that our analysis was reliable. Therefore, there may
be some internal contact between lncRNA/miRNA/mRNA in the
progression and development of CRC. Based on significant
differences in lncRNA, miRNA and mRNA expression data, a ceRNA
network was constructed by bioinformatics prediction and
correlation analysis. The relationship between the key lncRNAs and
clinical features was then further analysed. In the left ceRNA
network, 3 key lncRNAs (CYP1B1-AS1, LINC00402, and AC112721.1) were
confirmed to be related to clinical features. AC112721.1 has been
reported to be correlated with bladder cancer patient survival. In
the right ceRNA network, 12 key lncRNAs (LINC00483, LPP-AS1,
MIR205HG, WASIR2, OSBPL10-AS1, DSCAM-AS1, ZBTB20-AS1, ARHGEF38-IT1,
C8orf49, RMST, NOVA1-AS1, and KCNQ1OT1) were identified to be
associated with clinical features. Among these 12 lncRNAs,
LINC00483 was reported to regulate proliferation and apoptosis in
gastric cancer (19); Di Agostino
et al reported that MIR205HG led to unrestrained
proliferation in head and neck squamous cell carcinoma by depleting
miR-590-3p (27); DSCAM-AS1 could
interact with miR-137 to enhance tamoxifen resistance in breast
cancer (28). Microarray data
revealed that lncRNA RMST was differentially expressed in cervical
cancer, and the in vitro assay revealed that RMST played the
role of tumour suppressor in TNBC by inhibiting cell proliferation,
invasion and migration (29); some
studies reported that KCNQ1OT1 played an important role in multiple
malignancies by interacting with several miRNAs, such as
miR-140-5p, miR-384b, miR-145, miR-211-5p, miR-7-5p and miR-504
(25,30–34).
However, more research still needs to be carried out to confirm and
understand the relationship between these RNAs and clinical
features.
Our analysis has ramifications for the ceRNA network
in colon cancer, and some results were confirmed by qRT-PCR.
However, there are still several limitations in our study. First,
the present study was derived only from the data of The Cancer
Genome Atlas (TCGA) database, and the conclusion is relatively
preliminary and requires validation from large-scale clinical
trials. Second, our confirmation experiment is limited to qRT-PCR
of two lncRNAs, and further research is also required on the
functions of key lncRNAs in vivo and in vitro, and
the relationship of expression and function in the RNAs also
requires further validation.
In conclusion, our research aimed to detect the
difference between left and right colon cancer. By constructing
ceRNA networks and analysing the relationship between the key
lncRNAs and the clinical features, the putatively different
mechanism of the two sides and the relationship among these 3 types
of RNAs was partially revealed. The present study may further our
insight into the difference between left and right colon cancer at
the genetic level.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank the faculty and
staff at the Department of General Surgery (The First Affiliated
Hospital of Nanjing Medical University, Jiangsu, China) for
providing language and technology support.
Funding
The present study was funded by Jiangsu Key Medical
Discipline-General Surgery (grant no. ZDXKA2016005).
Availability of data and materials
The data sets used and/or analysed during the
present study are available from the corresponding author on
reasonable request.
Authors' contributions
YS conceptualized and designed the research. WQ and
YF performed the experiments, analysed and interpreted the results,
made the figures and wrote the manuscript. JL, WP and QG performed
the experiments and analysed the data. JL, ZZ and DJ provided the
patient tissues. QG, ZZ and DJ also helped design the experimental
studies and edited the manuscript. QW and DZ interpreted the
results and wrote the manuscript. All authors read and approved the
manuscript and agree to be accountable for all aspects of the
research in ensuring that the accuracy or integrity of any part of
the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
The present study was approved by the Research
Ethics Committee of The First Affiliated Hospital of Nanjing
Medical University, and written informed consent was obtained from
all patients prior to enrolment in the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CRC
|
colorectal cancer
|
|
lncRNAs
|
long non-coding RNAs
|
|
DElncRNAs
|
differentially expressed lncRNAs
|
|
RIDElncRNAs
|
DElncRNAs after removing the elements
of the intersection of the left and right colon cancer
|
|
miRNAs
|
microRNAs
|
|
KEGG
|
Kyoto Encyclopaedia of Genes and
Genomes
|
|
ceRNAs
|
competing endogenous RNAs
|
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bufill JA: Colorectal cancer: Evidence for
distinct genetic categories based on proximal or distal tumor
location. Ann Intern Med. 113:779–788. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gervaz P, Bucher P and Morel P: Two
colons-two cancers: Paradigm shift and clinical implications. J
Surg Oncol. 88:261–266. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Distler P and Holt PR: Are right- and
left-sided colon neoplasms distinct tumors? Dig Dis. 15:302–311.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Glebov OK, Rodriguez LM, Nakahara K,
Jenkins J, Cliatt J, Humbyrd CJ, DeNobile J, Soballe P, Simon R,
Wright G, et al: Distinguishing right from left colon by the
pattern of gene expression. Cancer Epidemiol Biomarkers Prev.
12:755–762. 2003.PubMed/NCBI
|
|
6
|
Iacopetta B: Are there two sides to
colorectal cancer? Int J Cancer. 101:403–408. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sana J, Faltejskova P, Svoboda M and Slaby
O: Novel classes of non-coding RNAs and cancer. J Transl Med.
10:1032012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pan W, Liu L, Wei J, Ge Y, Zhang J, Chen
H, Zhou L, Yuan Q, Zhou C and Yang M: A functional lncRNA HOTAIR
genetic variant contributes to gastric cancer susceptibility. Mol
Carcinog. 55:90–96. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Huang C, Cao L, Qiu L, Dai X, Ma L, Zhou
Y, Li H, Gao M, Li W, Zhang Q, et al: Upregulation of H19 promotes
invasion and induces epithelial-to-mesenchymal transition in
esophageal cancer. Oncol Lett. 10:291–296. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zheng HT, Shi DB, Wang YW, Li XX, Xu Y,
Tripathi P, Gu WL, Cai GX and Cai SJ: High expression of lncRNA
MALAT1 suggests a biomarker of poor prognosis in colorectal cancer.
Int J Clin Exp Pathol. 7:3174–3181. 2014.PubMed/NCBI
|
|
11
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The rosetta stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Missiaglia E, Jacobs B, D'Ario G, Di Narzo
AF, Soneson C, Budinska E, Popovici V, Vecchione L, Gerster S, Yan
P, et al: Distal and proximal colon cancers differ in terms of
molecular, pathological, and clinical features. Ann Oncol.
25:1995–2001. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Snaebjornsson P, Jonasson L, Jonsson T,
Moller PH, Theodors A and Jonasson JG: Colon cancer in Iceland-a
nationwide comparative study on various pathology parameters with
respect to right and left tumor location and patients age. Int J
Cancer. 127:2645–2653. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Budinska E, Popovici V, Tejpar S, D'Ario
G, Lapique N, Sikora KO, Di Narzo AF, Yan P, Hodgson JG, Weinrich
S, et al: Gene expression patterns unveil a new level of molecular
heterogeneity in colorectal cancer. J Pathol. 231:63–76. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sadanandam A, Lyssiotis CA, Homicsko K,
Collisson EA, Gibb WJ, Wullschleger S, Ostos LC, Lannon WA,
Grotzinger C, Del Rio M, et al: A colorectal cancer classification
system that associates cellular phenotype and responses to therapy.
Nat Med. 19:619–625. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang DY, Zou XJ, Cao CH, Zhang T, Lei L,
Qi XL, Liu L and Wu DH: Identification and functional
characterization of long non-coding RNA MIR22HG as a tumor
suppressor for hepatocellular carcinoma. Theranostics. 8:3751–3765.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cui Z, An X, Li J, Liu Q and Liu W: LncRNA
MIR22HG negatively regulates miR-141-3p to enhance DAPK1 expression
and inhibits endometrial carcinoma cells proliferation. Biomed
Pharmacother. 104:223–228. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li D, Yang M, Liao A, Zeng B, Liu D, Yao
Y, Hu G, Chen X, Feng Z, Du Y, et al: Linc00483 as ceRNA regulates
proliferation and apoptosis through activating MAPKs in gastric
cancer. J Cell Mol Med. 15:136612018.
|
|
20
|
Lu PW, Li L, Wang F and Gu YT: Effects of
long non-coding RNA HOST2 on cell migration and invasion by
regulating MicroRNA let-7b in breast cancer. J Cell Biochem.
119:4570–4580. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Macleod RJ: CaSR function in the
intestine: Hormone secretion, electrolyte absorption and secretion,
paracrine non-canonical Wnt signaling and colonic crypt cell
proliferation. Best Pract Res Clin Endocrinol Metab. 27:385–402.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Basu S, Haase G and Ben-Ze'ev A: Wnt
signaling in cancer stem cells and colon cancer metastasis.
F1000Res. 5:F10002016. View Article : Google Scholar
|
|
23
|
Zarkou V, Galaras A, Giakountis A and
Hatzis P: Crosstalk mechanisms between the WNT signaling pathway
and long non-coding RNAs. Noncoding RNA Res. 3:42–53. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lecarpentier Y, Claes V, Vallee A and
Hébert JL: Interactions between PPAR gamma and the canonical
wnt/beta-catenin pathway in type 2 diabetes and colon cancer. PPAR
Res. 2017:58790902017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Feng W, Wang C, Liang C, Yang H, Chen D,
Yu X, Zhao W, Geng D, Li S, Chen Z and Sun M: The dysregulated
expression of KCNQ1OT1 and its interaction with downstream factors
miR-145/CCNE2 in breast cancer cells. Cell Physiol Biochem.
49:432–446. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gao X, Ge J, Li W, Zhou W and Xu L: LncRNA
KCNQ1OT1 ameliorates particle-induced osteolysis through inducing
macrophage polarization by inhibiting miR-21a-5p. Biol Chem.
399:375–386. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Di Agostino S, Valenti F, Sacconi A,
Fontemaggi G, Pallocca M, Pulito C, Ganci F, Muti P, Strano S and
Blandino G: Long non-coding MIR205HG depletes Hsa-miR-590-3p
leading to unrestrained proliferation in head and neck squamous
cell carcinoma. Theranostics. 8:1850–1868. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ma Y, Bu D, Long J, Chai W and Dong J:
LncRNA DSCAM-AS1 acts as a sponge of miR-137 to enhance tamoxifen
resistance in breast cancer. J Cell Physiol. 234:2880–2894. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang L, Liu D, Wu X, Zeng Y, Li L, Hou Y,
Li W and Liu Z: Long non-coding RNA (LncRNA) RMST in
triple-negative breast cancer (TNBC): Expression analysis and
biological roles research. J Cell Physiol. 233:6603–6612. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang X, Wang M, Sun H, Zhu T and Wang X:
Downregulation of LINC00894-002 contributes to tamoxifen resistance
by enhancing the TGF-β signaling pathway. Biochemistry. 83:603–611.
2018.PubMed/NCBI
|
|
31
|
Shen C, Kong B, Liu Y, Xiong L, Shuai W,
Wang G, Quan D and Huang H: YY1-induced upregulation of lncRNA
KCNQ1OT1 regulates angiotensin II-induced atrial fibrillation by
modulating miR-384b/CACNA1C axis. Biochem Biophys Res Commun.
505:134–140. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang S, Ma H, Zhang D, Xie S, Wang W, Li
Q, Lin Z and Wang Y: LncRNA KCNQ1OT1 regulates proliferation and
cisplatin resistance in tongue cancer via miR-211-5p mediated
Ezrin/Fak/Src signaling. Cell Death Dis. 9:7422018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Fan S, Fan C, Liu N, Huang K, Fang X and
Wang K: Downregulation of the long non-coding RNA ZFAS1 is
associated with cell proliferation, migration and invasion in
breast cancer. Mol Med Rep. 17:6405–6412. 2018.PubMed/NCBI
|
|
34
|
Li J, Wang W, Xia P, Wan L, Zhang L, Yu L,
Wang L, Chen X, Xiao Y and Xu C: Identification of a five-lncRNA
signature for predicting the risk of tumor recurrence in patients
with breast cancer. Int J Cancer. 143:2150–2160. 2018. View Article : Google Scholar : PubMed/NCBI
|