Introduction
The liver is an organ consisting of hepatocytes,
stellate cells, endothelial cells and Kupffer cells. It consists of
hepatic lobules as basic units and plays important roles in the
production of bile acids, proteosynthesis, detoxification, glycogen
storage and hematopoiesis. Injured or partially hepatectomized
livers can recover their size and function by enlarging hepatocytes
(1).
During the process of liver development, hepatoblast
cells differentiate from the endoderm and stellate cells, and
endothelial cells differentiate from the mesoderm. The construction
of the liver in the fetus is induced by the interaction among cells
through hormones and/or cytokines. Once a fertilized egg implants
in the uterus, the ectoderm, endoderm and mesoderm differentiate
from the inner clump of cells. The endoderm forms the foregut under
the control of the forkhead box A2 transcription factor and GATA
binding protein 4, and a section of the foregut forms hepatoblast
cells (2,3). Fibroblast growth factor secreted from
the transverse septum and bone morphogenetic protein secreted from
the heart endoderm stimulate endoderm cells of the foregut to
differentiate into hepatoblast cells (4,5).
Hepatocyte growth factor secreted from endothelial cells enhances
the proliferation of hepatoblast cells and the process of liver
development (6,7).
Mature hepatocytes and bile duct epithelial cells
are derived together from hepatoblast cells. The change in the gene
expression of CCAAT enhancer binding protein α, a liver-specific
transcription factor, affects the differentiation into mature
hepatocytes and bile duct epithelial cells (8). In addition, Nodal, which
belongs to the transforming growth factor-β (TGF-β) superfamily, is
involved in the selection of mature hepatocytes or bile duct
epithelial cells for the differentiation (9). In general, TGF-β suppresses the cell
proliferation. The cells destined to be bile duct epithelial cells
stop proliferating due to TGF-β stimulation and form bile duct
cells. By contrast, the cells destined to be hepatocytes continue
to proliferate by avoiding TGF-β stimulation. Functional non-coding
RNAs, such as microRNAs (miRs), have been reported to be involved
in the regulation of the TGF-β pathway. Rogler et al
(10) reported that miR-23b
suppressed the expression of Smad, which functions downstream of
the TGF-β pathway and inactivates the TGF-β pathway. In addition,
Hand et al (11) reported
that miR-30 participated in the formation of bile ducts from
hepatoblast cells in mice. Therefore, it is important to
investigate the gene expression of non-coding RNAs during the
development of the fetal liver to understand the mechanisms of the
liver development.
Natural antisense transcripts (NATs), which are
transcribed from the DNA strand opposite to the sense strand, are
involved in controlling the gene expression in eukaryotes (12). NATs are usually non-coding RNAs. The
study by Matsui et al (13)
reported that NATs participated in maintaining the stability of the
inducible nitric oxide synthase mRNA in the liver. In addition, our
previous study (14) reported that
the expression of NATs was up/downregulated in the regenerating
livers of mice. However, the effects of NAT expression during the
liver development remain unclear.
In the present study, the expression profiles of
NATs were investigated using samples of the developing liver from
mice. The aim of the study was to identify the up/downregulated
NATs during liver development.
Materials and methods
Liver samples
Livers at different developmental stages, i.e.,
embryonic day (E) 14, E17, E19 and newborn (NB), were obtained from
the C57BL/6 mice at the RIKEN BioResource Center (Tsukuba, Ibaraki,
Japan). All the animal experiments were carried out according to
the RIKEN Guidelines for the care and use of experimental animals.
For the RNA analysis, livers at different developmental stages
(each n=3) were collected, frozen immediately in liquid nitrogen
and stored at −80°C until use.
Total RNA extraction and quality
check
Total RNAs from frozen livers were isolated using
Isogen (Nippon Gene, Tokyo, Japan), according to the manufacturer’s
instructions. The quality and concentration of total RNAs were
assessed using the NanoDrop spectrophotometer (NanoDrop
Technologies, Wilmington, DE, USA), according to manufacturer’s
instructions. All the total RNA samples had 260/280 nm absorbance
ratios of 1.8–2.0. The Agilent 2100 Bioanalyzer (Agilent
Technologies, Santa Clara, CA, USA) and the RNA 6000 Nano LabChip
kit (Agilent Technologies) were used to evaluate the RNA integrity.
The total RNAs examined were shown as RNA integrity numbers
>8.1. On the basis of the Bioanalyzer’s instructions, total RNAs
obtained were judged to be suitable for microarray analysis.
Microarray analysis
Agilent 44K × 4 mouse sense and antisense
custom-microarray slides [Agilent eArray Design ID, 021137;
produced by Tsukuba GeneTechnology Laboratories (http://www.tsukuba-genetech.com), Ibaraki, Japan]
and Cy3-labeled cDNA were used. The microarray analysis was
performed as described previously (14).
The expression profiles at each stage (E17, E19 and
NB) were compared against those at E14 using GeneSpring GX12
software (Agilent Technologies). The Kruskal-Wallis test was
performed to identify the genes with significant differences at
each time point; P<0.05 was considered to indicate a
statistically significant difference. The upregulated and
downregulated NATs were selected with fold changes (>5.0) at
each time point against E14 and with signal intensities
>100.0.
Strand-specific reverse transcription
quantitative polymerase chain reaction (RT-qPCR)
Strand-specific RT-qPCR was performed using total
RNA obtained from livers at different developmental stages to
confirm the expression patterns of NATs during the liver
development.
First-strand cDNAs derived from NATs or mRNA were
synthesized using a forward or a reverse primer, 18S ribosomal RNA
(rRNA) reverse primer and Reverse Transcriptase (Promega, Madison,
WI, USA), according to the manufacturer’s instructions. The
mixtures were incubated at 50°C for 60 min. The resulting cDNA was
incubated with RNase A at 99°C for 5 min and at 37°C for 60 min to
digest the RNA.
qPCR of NATs or mRNA was performed using
first-strand cDNAs, FastStart Universal SYBR-Green Master (Roche
Diagnostics, Basel, Switzerland) and primer pairs. qPCR analyses
were performed using the StepOne Plus Real-Time PCR system (Life
Technologies, Carlsbad, CA, USA) for 1 min at 95°C, followed by 40
cycles each of 95°C for 15 sec and 60°C for 60 sec, following the
manufacturer’s instructions. Ct values were averaged and normalized
to that of 18S rRNA. The relative expression ratio was determined
by the ΔΔCt comparative threshold method. The results of qPCR are
presented as the mean ± standard error of samples.
Results and Discussion
Previously, several non-coding RNAs have been
reported to be involved in the liver development (15). In the present study, the expression
of NATs was investigated to obtain further information on the
mechanisms occurring during the liver development. The
up/downregulated NATs during the liver development were identified
using mouse sense/antisense microarray slides.
The upregulated NATs of 87 genes were identified
with >5.0-fold changes in E17, E19 or NB compared to those at
E14. In addition, the downregulated NATs of 26 genes were
identified (Table I). These results
indicate that the expression of several NATs changed, suggesting
important roles during the liver development.
 | Table IIdentification of up- or downregulated
natural antisense transcripts during liver development by
microarray analyses. |
Table I
Identification of up- or downregulated
natural antisense transcripts during liver development by
microarray analyses.
| Gene regulation | Gene symbol |
|---|
| Up (87 genes) | Syt10, Fgg,
Serpina1a, Rbp4, Acaa2, Ttll10, Acadm, Scp2, Igfbp2, Elovl5,
Angptl3, Serpina1c, Cyp2c67, Arhgef10l, Tmprss9, Itih3, Cpb2, Ahsg,
H2-Q1, Ly6e, Crot, Csad, Fga, Cyp2c40, Lyz1, Cfb, Abhd2, Serpina1b,
F2, Olfr91, Serpina1d, Acat1, H2-T23, Selenbp2, Itsn1, Camp, Phyh,
Zfp62, Lamp2, Serpinc1, Elovl2, Mpc1, Apoe, Pygl, F10, Pate4,
LOC641235, Fgb, Spata21, Vtn, Lgals9, Fads2, B2 m, Trf, Fabp1, Hdc,
Il18, P4hb, Proc, Pm20d2, Fbxw5, Apof, C3, Fah, Pzp, Ech1, Bdh1,
Kng1, Sepp1, Apoa2, Gcat, H2-T3, Sdr42e1, Nudt7, Apoh, Pah, Crp,
Apoa1, Kcnab3, Ctsl, Adh1, Mypop, Lpl, Alb, Fgfbp1, S100a9 and
Acad11 |
| Down (26 genes) | Slc25a37, Rpl4,
Pklr, Cbln2, Ccl5, Car2, Pa2g4, Cpox, Ermap, Ptcd1, Hemgn, Ncl,
Was, Scn4b, Gpr150, Sypl, Klf1, Prdx2, Nop56, Tfrc, Cdk13, Agap3,
Asns, Yod1, Rhag and Magoh |
Several NATs described above were validated by the
strand-specific RT-qPCR to verify the expression of NATs obtained
by microarray analyses. The expression of fibrinogen α chain
(Fga), fibrinogen β chain (Fgb), fibrinogen γ chain
(Fgg), coagulation factor II (F2), fatty acid binding
protein 1 (Fabp1), complement component 3 (C3),
apolipoprotein A-I (Apoa1) and protein C (Proc) NATs
was examined and these were observed as upregulated candidate NATs
in the progression of the liver development. As shown in Fig. 1, the expression of Fga,
Fgb and Fgg, known as the fibrinogen family,
increased in E17 and/or E19 compared to that in E14. This result
indicates that these NATs may play important roles in E17 and/or
E19 stages. F2, known as thrombin, is a blood coagulation
factor [Online Mendelian Inheritance in Man (OMIM): 176930] and
Proc inhibits the blood coagulation (OMIM: 612283). The
expression of F2 and Proc NATs increased 3.4-fold and
2.5-fold, respectively, in E17 compared to that in E14 and
subsequently decreased. This result indicates that F2 and
Proc NATs may function only during the E17 stage.
Fabp1 is involved in the transport and metabolism of fatty
acids by binding to free fatty acids (OMIM: 134650). C3 is a
protein that mediates immunoreactions and acts during the
bacteriolysis by binding to the bacterial surface (OMIM: 120700).
The increased levels of Fabp1 and C3 NATs had two
phases: One peak was observed at E17 and another was observed at
NB, suggesting that Fabp1 and C3 NATs may be involved
in the regulation of gene expression at E17 following birth and may
indicate further high expression in the adult liver. The albumin
(Alb) and alcohol dehydrogenase 1 (Adh 1) genes shown
in Table I were involved in liver
metabolism, indicating that these NATs may have important roles in
the fetal and adult liver. In addition, the sense transcripts
(mRNAs) of the genes described above were confirmed to be
upregulated during liver development (Fig. 1). These findings indicate that these
NATs and mRNAs are co-expressed in the developmental stages and may
participate in the regulation of gene expression, such as the
maintenance of the mRNA stability (13).
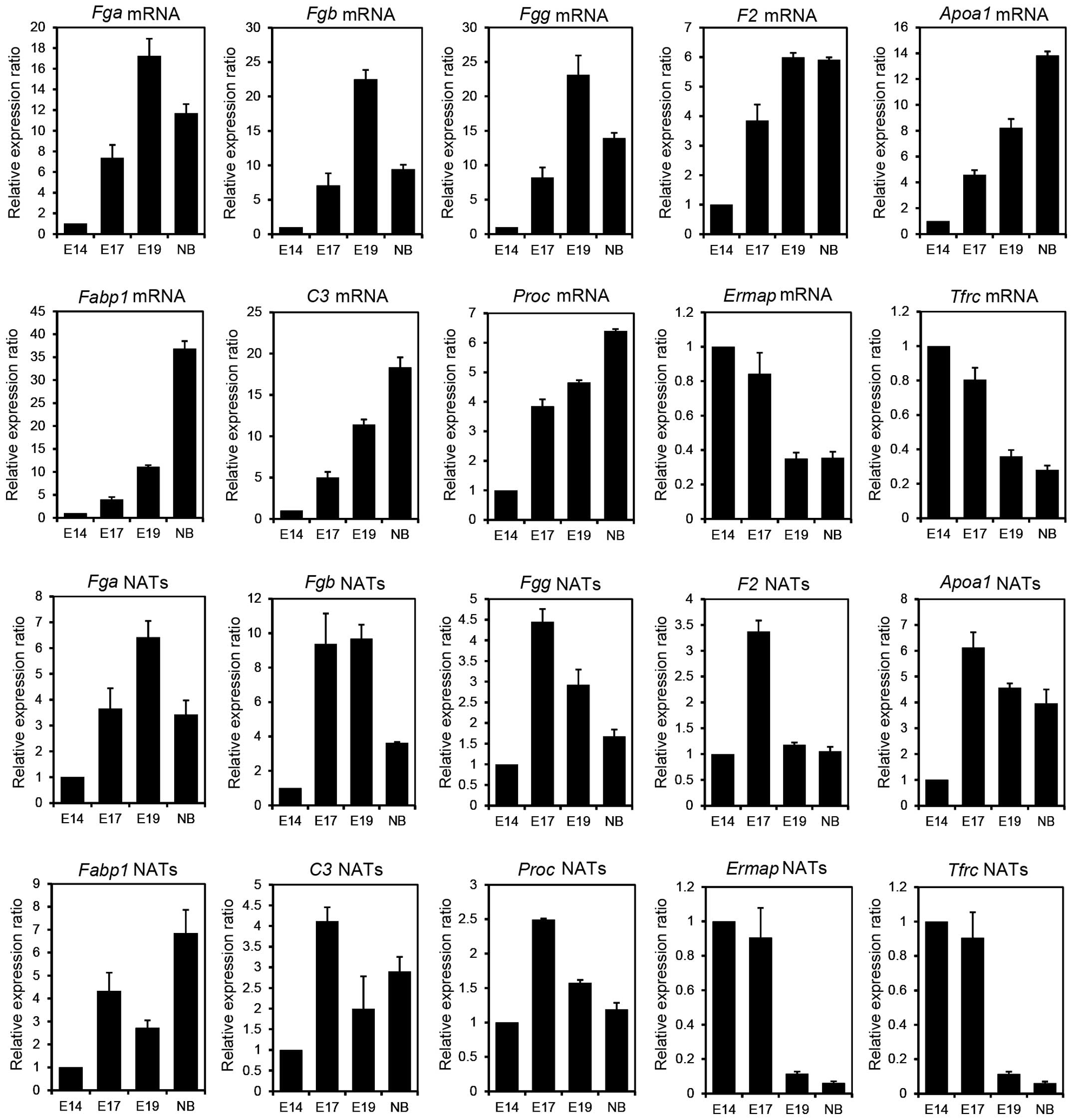 | Figure 1A comparison of natural antisense
transcript (NAT) and mRNA expression at various liver developmental
stages by strand-specific reverse transcription-quantitative
polymerase chain reaction (RT-qPCR). First-strand cDNAs derived
from NATs or mRNA were synthesized using a forward primer and a
reverse primer (Table II).
Strand-specific RT-qPCR was performed to investigate the values of
the expressed NATs. The Ct values obtained were averaged and
normalized to that of 18S ribosomal RNA. The relative expression
ratio was determined by the ΔΔCt comparative threshold method and
calculated using the values at embryonic day (E) 14 as 1.0. qPCR
results are presented as the mean ± standard error of samples (each
n=3). NB, newborn; Fga, fibrinogen α chain; Fgb,
fibrinogen β chain; Fgg, fibrinogen γ chain; F2,
coagulation factor II; Apoa1, apolipoprotein A-I;
Fabp1, fatty acid binding protein 1; C3, complement
component 3; Proc, protein C; Ermap, erythroblast
membrane-associated protein; Tfrc, transferrin receptor. |
By contrast, strand-specific RT-qPCR was performed
to confirm the expression of erythroblast membrane-associated
protein (Ermap), transferrin receptor (Tfrc) p90 and
CD71 as downregulated candidate NATs. Ermap functions as a
cell-adhesion or a receptor molecule of erythroid cells (OMIM:
609017). Tfrc is an essential gene that imports iron into
erythroid cells (OMIM: 190010). As shown in Fig. 1, the expression of Ermap and
Tfrc NATs was increased up to the E17 stage but showed
markedly decreased expression following the E19 stage. In addition,
the NATs of hemogen (Hemgn), which regulate the
proliferation and differentiation of hematopoietic cells (OMIM:
610715), were downregulated during liver development (Table I). These results indicate that
decreased NATs of Ermap, Hemgn and Tfrc during
liver development may be involved in the regulation of the
hematopoietic function.
In our previous study (14), we showed that several NATs,
including Fga, Fgb, Fgg, inter-α-trypsin
inhibitor heavy chain 3 (Itih3) and cathepsin L
(Ctsl), were upregulated when the NAT expression changes
were examined during liver regeneration using 70% partial
hepatectomized mice. Notably, Fga, Fgb, Fgg,
Itih3 and Ctsl NATs were upregulated during liver
development in the present study (Table
I). This result indicates that these NATs may act in the
dramatic functional conversion of the liver, such as during the
liver regeneration and development.
In conclusion, several NATs were revealed to be
up/downregulated during liver development. Certain NATs were
co-expressed with mRNA of the same genes. Therefore, NATs may be
involved in the co-regulation with these mRNAs. The fetal liver
functions mainly as a hematopoietic organ. During the developmental
progress, the hematopoietic function declines and the function
conversion is subsequently induced from a hematopoietic organ to a
metabolic and synthesizing organ. NATs may be involved in this
functional conversion of the fetal liver. Further studies are
required to understand the mechanisms of these NATs during the
liver development.
Acknowledgements
I am grateful to Ms. Noriko Hiraiwa (RIKEN
BioResource Center) for offering the samples. The present study was
supported in part by the Grants-in-Aid from the Ministry of
Education, Culture, Sports, Science and Technology of Japan and the
Hirosaki University Grant for Exploratory Research by Young
Scientists.
References
|
1
|
Higgins GM and Anderson RM: Experimental
pathology of the liver: restoration of the liver of the white rat
following partial surgical removal. Arch Pathol. 12:186–202.
1931.
|
|
2
|
Bossard P and Zaret KS: GATA transcription
factors as potentiators of gut endoderm differentiation.
Development. 125:4909–4917. 1998.PubMed/NCBI
|
|
3
|
Cirillo LA, Lin FR, Cuesta I, Friedman D,
Jarnik M and Zaret KS: Opening of compacted chromatin by early
developmental transcription factors HNF3 (FoxA) and GATA-4. Mol
Cell. 9:279–289. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jung J, Zheng M, Goldfarb M and Zaret KS:
Initiation of mammalian liver development from endoderm by
fibroblast growth factors. Science. 284:1998–2003. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rossi JM, Dunn NR, Hogan BL and Zaret KS:
Distinct mesodermal signals, including BMPs from the septum
transversum mesenchyme, are required in combination for
hepatogenesis from the endoderm. Genes Dev. 15:1998–2009. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Schmidt C, Bladt F, Goedecke S, et al:
Scatter factor/hepatocyte growth factor is essential for liver
development. Nature. 373:699–702. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ishikawa KS, Masui T, Ishikawa K and
Shiojiri N: Immunolocalization of hepatocyte growth factor and its
receptor (c-Met) during mouse liver development. Histochem Cell
Biol. 116:453–462. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shiojiri N, Takeshita K, Yamasaki H and
Iwata T: Suppression of C/EBP alpha expression in biliary cell
differentiation from hepatoblasts during mouse liver development. J
Hepatol. 41:790–798. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Vincent SD, Dunn NR, Hayashi S, Norris DP
and Robertson EJ: Cell fate decisions within the mouse organizer
are governed by graded Nodal signals. Genes Dev. 17:1646–1662.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rogler CE, Levoci L, Ader T, Massimi A,
Tchaikovskaya T, Norel R and Rogler LE: MicroRNA-23b cluster
microRNAs regulate transforming growth factor-beta/bone
morphogenetic protein signaling and liver stem cell differentiation
by targeting Smads. Hepatology. 50:575–584. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hand NJ, Master ZR, Eauclaire SF,
Weinblatt DE, Matthews RP and Friedman JR: The microRNA-30 family
is required for vertebrate hepatobiliary development.
Gastroenterology. 136:1081–1090. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Faghihi MA and Wahlestedt C: Regulatory
roles of natural antisense transcripts. Nat Rev Mol Cell Biol.
10:637–643. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Matsui K, Nishizawa M, Ozaki T, et al:
Natural antisense transcript stabilizes inducible nitric oxide
synthase messenger RNA in rat hepatocytes. Hepatology. 47:686–697.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chiba M, Yasue H and Ohkohchi N: Gene
expression profiling of sense and antisense transcripts in liver
regeneration by microarray analysis. Biomed Rep. 1:383–388.
2013.PubMed/NCBI
|
|
15
|
Tzur G, Levy A, Meiri E, et al: MicroRNA
expression patterns and function in endodermal differentiation of
human embryonic stem cells. PLoS One. 3:e37262008. View Article : Google Scholar : PubMed/NCBI
|















