Introduction
Several studies have suggested that the microRNAs
(miRNAs or miRs) miR-145-5p and miR-148b-3p are linked to the
development of prostate cancer (PCa) as they have been observed to
be dysregulated during this disease, and their expression levels
differ from patients with benign prostate diseases (1-4).
Furthermore, mutations in the genes that code for these miRNAs and
in nearby regions have been reported, suggesting a possible
explanation for their dysregulation. miR-145-5p has been classified
as a tumor suppressor, specifically in PCa; this miRNA has been
linked to various mechanisms that promote the development of this
pathology (5,6). Some factors have been associated with
the underexpression of this miRNA; for example, single nucleotide
polymorphisms (SNPs) have been suggested as possible causes of
miRNA aberrant expression. In PCa, the rs4705342 polymorphism
stands out; in the Asian population, it has been observed that men
with the TC/CC genotypes have a significantly lower risk of having
PCa than those with the TT genotype. Functionality analyses
demonstrated that the T allele increases protein-binding affinity
and thus reduces promoter activity, resulting in decreased
transcription of the miR-145-5p (7). In recent years, this polymorphism has
been studied in the European population. It has been observed that,
unlike the Asian population, the C allele appears to be a risk
factor in Serbian men (8). These
findings highlight the importance of analyzing mutations to
identify their involvement in regulating this miRNA and PCa
susceptibility.
miR-148b-3p has been observed to be overexpressed in
PCa, both in early and metastatic stages (9). Regarding the regulation of this miRNA
through single nucleotide polymorphisms, Chen et al
(10) observed in 2014 that the
rs11170877 (A>G) polymorphism, specifically the G allele and the
rs12231393 (T>C) allele C, were associated as a protective
factor for gastric cancer in Chinese patients. The rs11170877
variant is located in the exon of COPZ1, the gene that contains or
hosts miR-148b. These two polymorphisms form a haplotype due to
linkage disequilibrium; this haplotype showed a significant
association with this cancer (10).
Despite the interesting results obtained with the rs11170877
polymorphism in this type of cancer, few studies have linked this
SNP to other types of cancer or even to different populations.
Therefore, the present study aimed to identify mutations in
miR-145-5p and miR-148b-3p and regions near them and relate them to
their expression in patients with and without PCa.
Materials and methods
Patient samples
Non-probabilistic convenience sampling was conducted
on prostatic tissue samples collected via transrectal
ultrasound-guided 18G core needle biopsy. The biopsies were
conducted by a single Board-Certified Diagnostic Radiologist with
20 years of experience in the procedure, utilizing the standard
technique, including the sampling of ultrasound-suspicious areas
(nodules). The samples were obtained from the tumor tissue of
patients with a suspicious diagnosis of PCa based on either
positive digital rectal examination or rising prostate specific
antigen (PSA). The collected samples were stored in
Qiazol® (Qiagen GmbH) at 20˚C for later analysis.
The diagnosis and classification of patients with
benign prostatic hyperplasia (BPH) and PCa were performed by a
single Board-Certified Pathologist with 7 years of experience using
histopathology. The inclusion criteria were as follows: Mexican,
≥18 years, histopathological diagnosis of PCa or BPH. The exclusion
criteria included patients with other types of cancer and those who
had received chemotherapy or radiotherapy for any other medical
reason.
The present study consisted of 71 samples: 41 from
patients with PCa diagnosis (PCa group) and 30 from patients
diagnosed with benign prostatic disease (BPD group). The BPD group
included patients with BPH (n=7), prostatitis (n=18), both
hyperplasia and prostatitis (n=4), and intraepithelial neoplasia
(n=1). All patients were recruited from the Alvarez & Arrazola
Radiologists Clinic in Sinaloa, Mexico, from August 2016 to
December 2021. Clinicopathological characteristics including age
(applicable to individuals aged ≥18 years, with an upper limit of
90 years), weight, height, body mass index, PSA and family history
of cancer were collected through direct questionnaires and the
clinical database. An expert pathologist provided the diagnosis and
Gleason score. All patients signed informed consent forms, which
were reviewed and approved by the Alvarez and Arrazola Radiologists
Clinic's Ethics and Research Committee (study approval number
P-3103).
RNA and DNA extraction
Tissue samples were used for RNA extraction. The
prostate tissue in Qiazol was macerated using sterile pistils,
until leaving a homogeneous solution. Total RNA, including miRNAs,
was isolated using the miRNEasy kit (Qiagen) according to the
manufacturer's protocol. DNA extraction was performed using organic
solvents and salt precipitation from the remnants of
Qizol® obtained from RNA extraction using the previously
described methodology of Chomczynski (1993) (11). The concentration of isolated RNA and
DNA was measured with a GENESYS 10S UV-Vis Spectrophotometer
(Thermo Fisher Scientific, Inc.) at 260 nm. It also allows for the
evaluation of RNA and DNA integrity, detection of contaminants, the
analysis of low or atypical concentrations, and warning of invalid
results. The concentration and integrity analysis were performed in
triplicate.
Analysis of polymorphisms in
miRNAs
To design primers for miRNA amplification and
sequencing, the accession numbers NC_00005 and NC_000012
(https://www.ncbi.nlm.nih.gov/nuccore/NC_000005.10
and https://www.ncbi.nlm.nih.gov/nuccore/NC_000012,
respectively) were used for miR-145 and miR-148b, respectively. CLC
Sequence Viewer program (https://resources.qiagenbioinformatics.com/manuals/clcsequenceviewer/current/index.php?manual=Introduction_CLC_Sequence_Viewer.html)
was used to select a region of ~1,000 base pairs spanning the gene
sequence of each selected miRNA to cover the gene and possible
regulatory sequences.
Primer-BLAST Tool from NCBI (https://www.ncbi.nlm.nih.gov/tools/primer-blast/)
was employed to design primers used the sequence selected,
considering the following characteristics: 40-60% G + C,
non-self-complementary and no more than 3˚C (melting temperature
difference) between both primers. The primers selected for
miR-148b-3p were forward 5'-CAGGCTTTAGAAGCCCCTGA-3' and reverse,
5'-GCGCTTAAATGCCGCTTCA-3' with a 986 bp amplicon. And the primers
used for miR-145-5p were forward, 5'-TCTCCAGTAGGTCGTGGACT-3' and
reverse, 5'-CACAAGAGGGCGTTCTGAGT-3', with a 1,000 bp amplicon.
Sealed 10 µl aliquots of DNA were subsequently sent to the Korean
company MACROGEN for capillary electrophoresis sequencing. Once the
data was obtained, Chromas V2.6.6 (https://technelysium.com.au/wp/chromas/) was used to
clean the sequence, eliminating the ends in which the sequence was
not reliable using the default parameters. The reference sequence
GRCh38.p14 was obtained using the CLC sequence viewer 8.0 program.
Subsequently, the SnackVar v2.4.3(12) and NovoSNP v.3.0.1(13) tools were used to identify mutations.
Both programs are automatically using the default parameters detect
SNVs and indel type mutations by comparing them with a reference
sequence. Once the mutations were identified, only those that
coincided in both programs and those that had a quality score
>40 were included in this analysis.
Reverse transcription (RT) PCR and
relative miRNA expression
RT was performed from 10 ng of total RNA with the
TaqMan Advanced miRNA cDNA Synthesis kit (Applied Biosystems;
Thermo Fisher Scientific, Inc.) in a T100 Thermal Cycler (Bio-Rad
Laboratories, Inc.). To obtain cDNA, the TaqMan Advanced miRNA cDNA
Synthesis kit was used, which consists of four steps as follows: i)
Polyadenylation reaction: First, a reaction mix was prepared where,
for a final volume of 5 µl, 0.5 µl of 10x Poly(A) buffer, 0.5 µl of
ATP, 0.3 µl of Poly A enzyme and 2 µl of miRNAs were added.
Vortexing was briefly performed. The T100 Thermal Cycler (Bio-Rad
Laboratories, Inc.) was programmed as follows: 45 min at 37˚C and
10 min at 65˚C. The next reaction was immediately carried out. ii)
Ligation Reaction: The following reaction mixture was prepared for
a final volume of 15 µl: 3 µl of 5X DNA ligase buffer, 4.5 µl of
50% PEG 8000, 0.6 µl of 25X ligation adapter, 1.5 µl RNA ligase and
0.4 µl of RNase-free water; and then, the 5 µl reaction mixture
obtained in the polyadenylation was added and vortexed briefly. It
was subsequently incubated at 16˚C for 60 min in the same thermal
cycler. The reverse transcription reaction was carried out
immediately afterwards. iii) Reverse transcription (RT) reaction:
The following reaction mixture was prepared for a final volume of
30 µl: 6 µl of 5X RT buffer, 1.2 µl of dNTP mix (25 mM), 1.5 µl of
20X Universal RT primer, 3 µl of 10X RT enzyme mix and 3.3 µl of
RNase-free water. It was mixed by pipetting, then this reaction
mixture was added to the 15 µl of reaction obtained from the
ligation reaction and vortexed briefly. The thermal cycler was
programmed as follows: 15 min at 42˚C and 5 min at 85˚C. For each
of the miRNAs, standard curves of five points of serial dilutions
1:10 were made to obtain the efficiencies from the slope.
Quantitative estimations for miR-145-5p (assay ID: 477916_mir),
miR-148b-3p (assay ID: 477824_mir), and miR-191-5p (assay ID:
477952_mir) were performed using TaqMan Advance miRNA Assays by
Thermo Fisher Scientific, Inc. using a StepOnePlusÔ thermal cycler
(Applied Biosystems). To select the normalizing miRNA, the
RefFinder tool (https://blooge.cn/RefFinder/) was used, which
integrates different algorithms to select normalizers, identifying
miR-191-5p as the most appropriate miRNA for the present study.
Relative expression analysis
To obtain relative miRNA expression of miR-145-5p
and miR-148b-3p, the previously described method by Taylor et
al (2019) (14) was used based
on the ∆∆Cq method (2-∆∆Cq algorithm) (15) and the miR-191-5p was used as
reference gene. First, the average of the Cq of the control samples
was calculated; subsequently, the average Cq obtained was
subtracted individually for each test sample, obtaining a ΔCq, and
then, the RQ was calculated using the formula 2∆ΔCq.
Subsequently, the normalized expression was obtained for each
sample using the formula RQ problem gene/RQ normalizing gene; and
finally, the normalization of the expression per sample was carried
out using the Log2 of each result obtained in the previous
formula.
Statistical analysis
Once the information was collected, the data was
organized into tables and figures. Measures of central tendency
(mean, median, mode), measures of dispersion (standard deviation),
and confidence intervals were estimated for the quantitative
variables. For the qualitative variables, the frequencies and
percentages were estimated, as well as their respective confidence
intervals. To identify differences between means of the different
quantitative variables in patients with and without PCa, the
unpaired Student's t-test and Mann-Whitney U statistics were
performed for parametric and non-parametric variables,
respectively. To find a relationship between qualitative variables,
the Chi-square test was used. For the correlation analyses, the
Pearson's correlation coefficient and the Spearman's test were
used, for parametric and non-parametric variables, respectively.
All the analyses were carried out using the statistical package
SPSS v.20 (IBM Corp.). To determine if the distribution of the
genotypes of each mutation was in Hardy-Weinberg (EHW) equilibrium,
the SNPStats software (http://www.snpstats.net/start.htm) was used.
Similarly, the haplotype frequencies and linkage analysis were
calculated using the same software. P<0.05 was considered to
indicate a statistically significant difference.
Results
miRNAs expression analysis
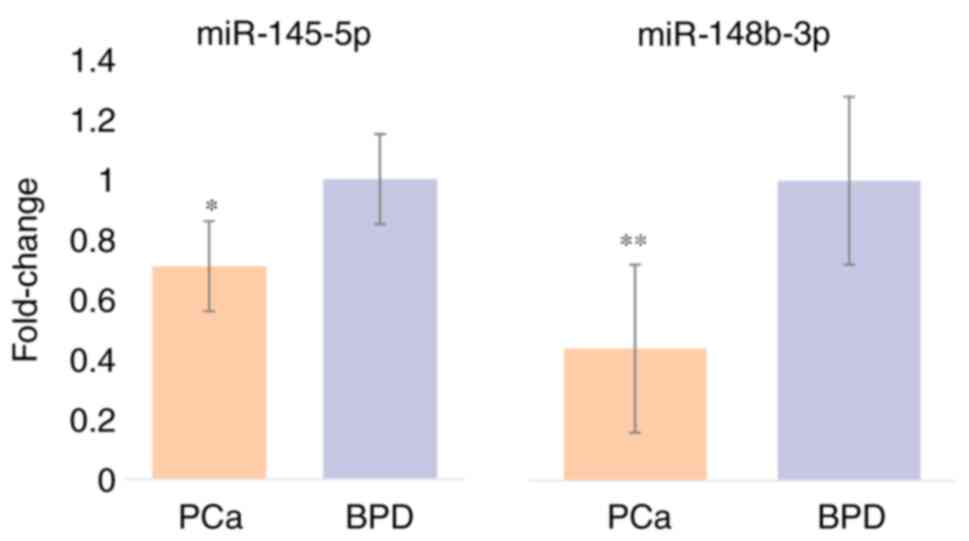
Relative expression of miR-145-5p and miR-148b-3p
was quantified in all samples of BPD and PCa patients (Fig. 1). When analyzing miR-145-5p
expression, an expression factor of 0.71±0.15 was identified in PCa
patients compared with BPD patients, showing a significant
difference (P=0.033), and miR-148b-3p was underexpressed in PCa
patients with an expression factor of 0.44±0.28 with a
statistically significant difference between groups (P=0.001).
Identification of mutations
A total of 18 mutations were identified in the
analyzed sequence for miR-148b-3p in 71 study samples (Table I). Different types of mutations were
observed, which were single nucleotide variations (SNVs), deletions
(Del), an insertion (Ins), and a duplication (Dup). Among the
mutations obtained, 12 had already been registered, while the
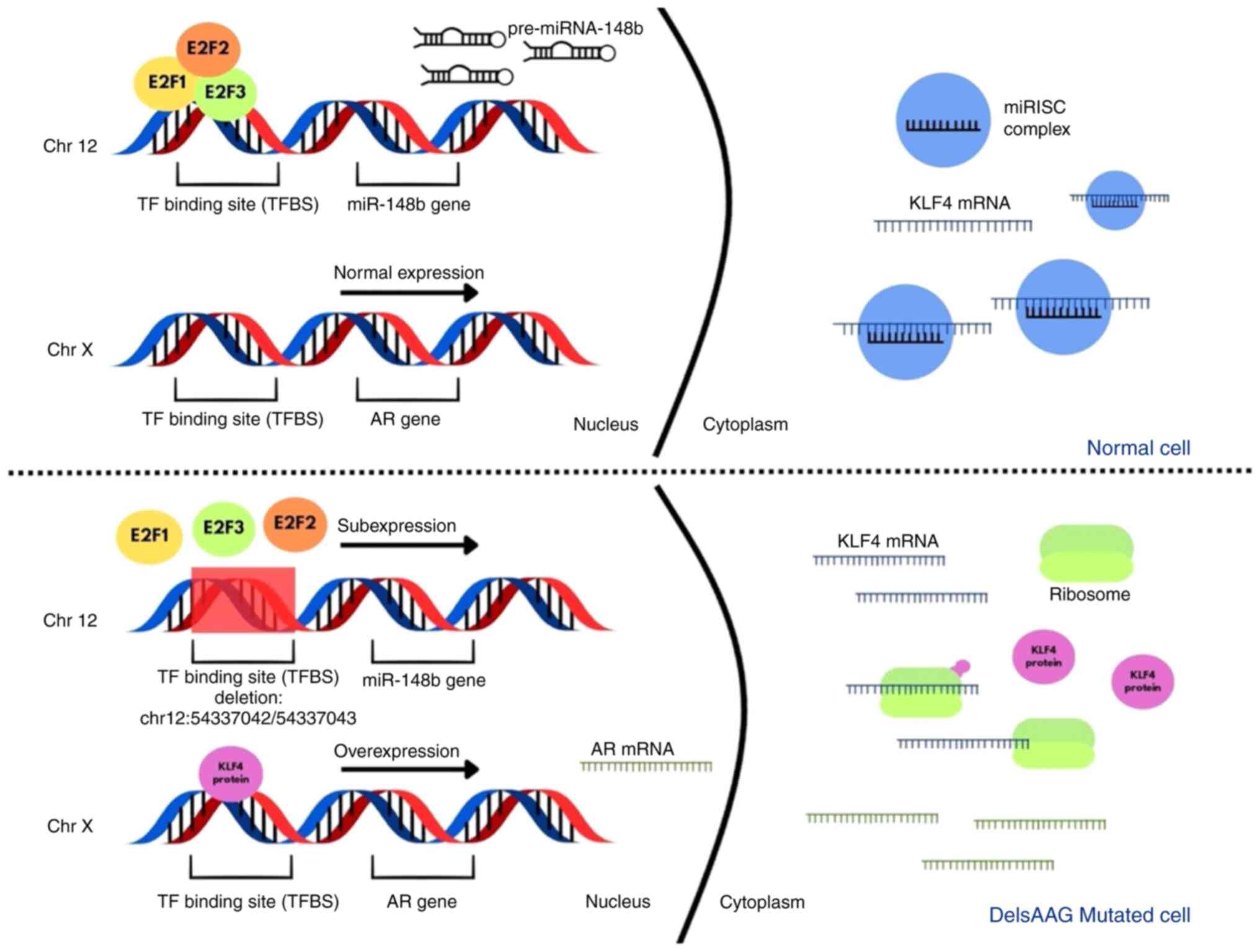
remaining six had not. In position chr12:54337042/54337043, there
were four different deletions, two of which were previously
registered (rs1465330647 and rs1187801243), and two which have not
been reported in any database (Fig.
2). The rs1465330647 mutation is the deletion of AAAGAA, the
rs1187801243 mutation is the loss of AAGAA, and the other two
unrecorded mutations comprise the deletion of AAAGA in one case and
AAGAAG in the other. As they are very similar to each other and
have a low frequency separately, they were grouped into a single
variable called DelsAAG. To analyze the relationship of these
mutations with the expression of miR-148b-3p, those with the
highest frequency of the mutated allele were selected: DupCT
(rs59761210), SNV G/T (rs1381668656), SNV A/C (NA), DelG
(rs368666376), SNV G/C (rs56818309) and DelsAAG (Table I). Mutation analysis for miR-145-5p
was only performed on 44 of the 71 study samples since the
remaining samples could not be sequenced. Of the 44 samples, 21
were from the PCa and 23 from the BPD groups. Mutation analysis for
miR-145-5p was performed, where 62 mutations were identified, of
which 24 had previously been reported. Of the 62 mutations, 59 were
single nucleotide variations (SNV), two deletions (Del), and one
duplication (Dup). Most of the mutations had a low frequency of the
mutated allele; for this reason, only the rs353291 mutation,
consisting of a T/C SNV with a frequency of 0.18 of the mutated
alleles in the total population, was chosen.
 | Table IMutations identified for
miR-148b-3p. |
Table I
Mutations identified for
miR-148b-3p.
| Mutation | Position | ID | Mutated allele
frequency |
|---|
| SNV G/T | 54336876 | NA | 0.01 |
| SNV G/A | 54337007 | NA | 0.01 |
| DupCT | 54337015 | rs59761210 | 0.15a |
| SNV C/G | 54337017 | rs555306172 | 0.01 |
| SNV G/T | 54337018 | rs1381668656 | 0.11a |
| SNV A/C | 54337021 | NA | 0.10a |
| SNV A/C | 54337042 | rs1953883052 | 0.02 |
| SNV A/C | 54337043 | rs918405335 | 0.01 |
| DelAAAGA | 54337042 | NA | 0.01 |
| DelAAAGAA | 54337042 | rs1465330647 | 0.08 |
| DelAAGAA | 54337043 | rs1187801243 | 0.02 |
| DelAAGAAG | 54337043 | NA | 0.09 |
| DelG | 54337045 | rs368666376 | 0.12a |
| InsA | 54337045 | NA | 0.01 |
| SNV G/A | 54337045 | rs879146327 | 0.01 |
| SNV A/C | 54337046 | rs1048873442 | 0.01 |
| SNV A/C | 54337047 | rs1481050082 | 0.02 |
| SNV G/C | 54336970 | rs56818309 | 0.27a |
|
DelsAAGb |
54337042/54337043 | NA | 0.21a |
Analysis by inheritance model
Subsequently, an analysis of the possible
inheritance models of these six mutations of miR-148-3p, was
carried out according to the response variable that groups patients
in PCa and BPD using the SNPStat.net
software. Of the six mutations, only DelsAAG was significant for
the codominant (P=0.03, AIC=95.4), dominant (P=0.01, AIC=94.7), and
over dominant (P=0.02, AIC=94.9) models. To conduct subsequent
analyses using genotypes and alleles, the inheritance model was
selected according to the lowest value of the Akaike Information
Criterion (AIC) obtained. For the rs59761210 mutation, the dominant
model was selected; for the rs368666376 mutation, the recessive
model, for the rs56818309 mutation, the over dominant model; and
for the DelsAAG mutation, the dominant model. In the case of the
rs1381668656 and SNV A/C mutations, the analysis by different
inheritance models was not obtained because the mutated homozygous
genotype was not observed in the study groups; hence the codominant
inheritance model was selected. In the case of miR-145-5p for the
rs353291 mutation, none of the inheritance models were
statistically significant; therefore, the model with the lowest AIC
value was chosen (over dominant model, AIC=62.2).
Genotypic and allele frequencies and
their association with PCa
The genotype and allele frequencies were calculated
for each of the selected mutations, and both genotypes and alleles
were associated with PCa. The allele and genotypic frequencies of
the rs59761210, rs1381668656, rs368666376, rs56818309 and SNV A/C
mutations are in Hardy-Weinberg equilibrium (P=0.052, P=0.59,
P=0.98, P=0.36 and P=0.44, respectively); in the case of the
DelsAAG mutation, this was not observed in Hardy-Weinberg
equilibrium (P=0.001). The DelsAAG mutation was significantly
related to the presence of PCa (P=0.014), wherein a dominant model,
both mutated homozygotes and heterozygotes, were related to the
pathology, unlike normal homozygotes. Likewise, the analysis by
alleles revealed that patients who carry the DelsAAG mutation are
4.16 (P=0.017) times more likely to develop PCa; the other
mutations were not observed to be related to the pathology
(Table II). The rs353291 mutation
demonstrated a frequency of the heterozygous genotype of 33.3% in
patients with PCa, and 13% in the BPD group. The genotype analysis
found no statistically significant relationship between the
genotypes and the presence of PCa. The mutated allele presents a
frequency of 0.21 in the PCa group and 0.15 in the BPD group; no
significant relationship between the mutated allele and the
pathology was observed (P=0.11) (Table III).
 | Table IIAllele and genotypic frequencies of
miR-148b-3p mutations. |
Table II
Allele and genotypic frequencies of
miR-148b-3p mutations.
| Mutation | Inheritance
model | Genotype | Group BPD | Group PCa | RM (95% CI) | P-value |
|---|
| dupCT
rs59761210 | Dominant | CT/CT | 21 (70%) | 32 (78%) | 0.66
(0.22-1.92) | 0.44 |
| | | CT/CTCT | 7 (23%) | 7 (17%) | | |
| | | CTCT/CTCT | 2 (7%) | 2 (5%) | | |
| | | CTCT | 0.18 | 0.13 | 0.66
(0.22-1.92) | 0.44 |
| | | G/G | 22 (73%) | 33 (81%) | | |
| SNP G/T
rs1381668656 | Codominant | G/T | 8 (27%) | 8 (19%) | 0.67
(0.22-2.04) | 0.48 |
| | | T/T | - | - | | |
| | | T | 0.13 | 0.1 | 0.72
(0.33-1.56) | 0.41 |
| | | G/G | 22 (74%) | 33 (80%) | | |
| DelG
rs368666376 | Recessive |
G/G- | 7 (23%) | 8 (20%) | 0.00 (0.00-NA) | 0.19 |
| | |
G-/G- | 1 (3%) | 0 (0%) | | |
| | | G- | 0.13 | 0.1 | 0.41
(0.31-0.55) | 0.24 |
| | | G/G | 13 (44%) | 23 (56%) | | |
| SNV G/C
rs56818309 | Overdominant | G/C | 16 (53%) | 16 (39%) | 0.56
(0.22-1.45) | 0.23 |
| | | C/C | 1 (3%) | 2 (5%) | | |
| | | C | 0.3 | 0.24 | 0.56
(0.22-1.45) | 0.23 |
| | | A/A | 23 (77%) | 36 (88%) | | |
| SNV A/C
54337021 | Codominant | A/C | 7 (23%) | 5 (12%) | 0.46
(0.13-1.61) | 0.22 |
| | | C/C | - | - | | |
| | | C | 0.12 | 0.06 | 0.49
(0.15-1.63) | 0.24 |
| | | AAG/AAG | 26 (87%) | 25 (61%) | | |
| DelsAAG | Dominant |
AAG/AAG- | 1 (3%) | 9 (22%) | 4.16
(1.22-14.17) | 0.014 |
| | |
AAG-/AAG- | 3 (10%) | 7 (17%) | | |
| | |
AAG- | 0.12 | 0.28 | 4.16
(1.22-14.17) | 0.017 |
 | Table IIIAllele and genotypic frequencies of
the rs353291 mutation. |
Table III
Allele and genotypic frequencies of
the rs353291 mutation.
| Mutation | Inheritance
model | Genotype | Group BPD | Group PCa | RM (95% CI) | P-value |
|---|
| SNV T/C
rs353291 | | T/T | 18 (78.3%) | 13 (61.9%) | | |
| | | | | | 3.33
(0.73-15.17) | 0.11 |
| | Sobredominant | T/C | 3 (13%) | 7 (33.3%) | | |
| | | C/C | 2 (8.7%) | 1 (4.8%) | 3.33
(0.73-15.17) | 0.11 |
| | | C | 0.15 | 0.21 | | |
Linkage disequilibrium and analysis by
haplotypes
Linkage disequilibrium analysis was performed to
identify possible haplotypes, where the set of mutations of
rs368666376, rs56818309, and DelsAAG was found to be in linkage
disequilibrium. In total, three haplotypes (haplotypes I, II and
III) were obtained from the set of mutations, with ‘haplotype I’
having the normal alleles. An analysis was performed in which each
haplotype was linked to the presence of PCa. However, no
statistically significant association was found. In the case of
miR-145-5p, since only one mutation was studied, this analysis was
not performed.
Relationship of mutations with miRNAs
expression
The gene expression of miR-148b-3p between the
genotypes and alleles of each mutation was compared and was
observed that there is a statistically significant difference
between the expression of the DelsAAG mutation genotypes (P=0.004),
specifically between the normal homozygous genotypes with a mean
expression of -0.35 and heterozygous with a mean of -2.10 (P=0.003)
(Table IV). Similarly, this
analysis compared alleles, where it was identified that the
patients carrying the mutated allele of the DelsAAG mutation had a
mean of -1.53. In the case of non-carriers, a mean of -0.36 was
found, with a statistically significant difference (P=0.005). The
remaining mutations were not observed with significant differences
in the analyses by genotypes or by alleles (Table V). To identify a relationship
between the rs353291 mutation and the expression of miR-145-5p,
mean difference analyses were performed between genotypes and
alleles. It was observed that the patients with the heterozygous
genotype had a mean miR-145-5p expression of -0.38, however, no
statistically significant difference was observed when compared
with the other genotypes. The allele analysis revealed that
patients carrying the mutated allele had a mean expression of -0.38
and non-carrier patients had a mean expression of -0.17; however,
no statistically significant difference was observed between groups
(P=0.47).
 | Table IVAnalysis of the mean difference in
the expression of miR-148b-3p between the genotypes of the
mutations in the study population. |
Table IV
Analysis of the mean difference in
the expression of miR-148b-3p between the genotypes of the
mutations in the study population.
| Mutation | Genotype | N | Average | Standard error | P-value |
|---|
| dupCT
rs59761210 | CT/CT | 53 | -0.88 | 0.23 | |
| | CT/CTCT | 14 | -0.88 | 0.30 | 0.17 |
| | CTCT/CTCT | 4 | 0.41 | 0.71 | |
| SNP G/T
rs1381668656 | G/G | 55 | -0.54 | 0.23 | |
| | G/T | 16 | -0.73 | 0.33 | 0.69 |
| | T/T | 0 | - | - | |
| DelG
rs368666376 | G/G | 56 | -0.71 | 0.21 | |
| |
G/G- | 14 | -0.66 | 0.49 | 0.9 |
| |
G-/G- | 1 | - | - | |
| SNV G/C
rs56818309 | G/G | 36 | -1.03 | 0.31 | |
| | G/C | 32 | -0.34 | 0.23 | 0.18 |
| | C/C | 3 | -0.27 | 0.29 | |
| SNV A/C
54337021 | A/A | 59 | -0.69 | 0.21 | |
| | A/C | 12 | -0.66 | 0.44 | 0.95 |
| | C/C | 0 | - | - | |
| DelsAAG | AAG/AAG | 51 | -0.36 | 0.19 | |
| |
AAG/AAG- | 10 | -2.10 | 0.75 | 0.004
(0.03a) |
| |
AAG-/AAG- | 10 | -0.95 | 0.33 | |
 | Table VAnalysis of the mean difference in
the expression of miR-148b-3p between the alleles of the mutations
in the study population. |
Table V
Analysis of the mean difference in
the expression of miR-148b-3p between the alleles of the mutations
in the study population.
| Mutation | Allele | N | Average | Standard error | P-value |
|---|
| dupCT
rs59761210 | Carrier | 18 | -0.13 | 0.28 | 0.046 |
| | Normal | 53 | -0.88 | 0.23 | |
| SNP G/T
rs1381668656 | Carrier | 33 | -0.53 | 0.22 | 0.53 |
| | Normal | 109 | -0.73 | 0.16 | |
| DelG
rs368666376 | Carrier | 1 | 0.03 | - | 0.65 |
| | Normal | 70 | -0.7 | 0.19 | |
| SNV G/C
rs56818309 | Carrier | 32 | -0.34 | 0.23 | 0.09 |
| | Normal | 39 | -0.98 | 0.28 | |
| SNV A/C
54337021 | Carrier | 12 | -0.66 | 0.44 | 0.95 |
| | Normal | 130 | -0.7 | 0.14 | |
| DelsAAG | Carrier | 20 | -1.53 | 0.42 | 0.005 |
| | Normal | 51 | -0.36 | 0.19 | |
Discussion
miRNAs are small non-coding RNAs and are part of the
epigenetic machinery that can regulate the expression of genes;
therefore, increasingly altered mechanisms are identified due to
the malfunction of these miRNAs, for example, it has been observed
that several miRNAs dysregulated in PCa, are related to the
regulatory T cell marker FOXP3, suggesting a relationship between
immune cells, miRNAs and PCa (16).
Therefore, the identification of dysregulated miRNAs could have
implications both in understanding the pathology and in identifying
possible targets for the detection, prognosis and treatment of
various cancers (17,18).
The dysregulation of the expression of a miRNA can
be due to mutations in its coding genes or in their regulatory
regions; for example, SNPs in miR-146a and in miR-100 have been
identified that act as a factor of bad and an improved prognosis in
PCa. In addition, it has been mentioned that these polymorphisms
could alter miRNA function (19).
Furthermore, it has been observed that mutations can affect the
biogenesis of the mature miRNA while it is in the ‘seed’ region of
the passenger strand, thus decreasing its expression (20,21).
Finally, mutations in the promoter regions can also cause changes
in the expression of miRNAs, which have been observed in the
miR-143/145 cluster associated with PCa (22).
In the case of miR-145-5p, several mutations were
identified; among them, the one with the highest frequency was the
rs353291 mutation. This mutation is located in an intronic region
of the CARMN gene, 450 bp downstream of miR-145-5p. According to
the NCBI database, this SNV has a mutated allele frequency of
C=0.40 and normal allele frequency of T=0.60 in the global
population. In the present study, the frequencies in the general
population were C=0.18 and T=0.82, which are remarkably similar to
those reported in the African and Afro-American population (C=0.23
and T=0.77) and to those reported in Latin Americans (C=0.30 and
T=0.70). The Asian population is the most distinct from the
frequencies observed in the present study (C=0.41 and T=0.59)
(23).
The present results indicated that the rs353291
mutation is unrelated to CaP or miR-145-5p underexpression. At
present, there is only one study that analyzes the relationship of
this miR-145-5p SNV with cancer in an Australian population, which
observed that the mutated allele was significantly related to
breast cancer; additionally, it was suggested that this SNV could
be part of the gene regulation mechanisms of miR-145-5p. It should
be noticed, however, that they did not analyze miRNA expression
(24). The dysregulation of this
miRNA in CaP is probably caused by molecular mechanisms other than
mutations close to the miRNA, such as DNA methylation. This is
consistent with previous research; Zaman et al (2010)
(25), reported that the promoter
of this miRNA was hypermethylated in PCa cell lines. Similarly,
previous studies have shown that methylation plays a role in
regulating miR-145-5p expression, which contributes to tumor growth
and response to treatment (26,27).
Furthermore, Xue et al (2015) (28), observed that miR-145-5p targets the
DNMT3b methyltransferase, one of its mechanisms as a tumor
suppressor miRNA. In addition, they observed that DNMT3b leads to
hypermethylation of the miRNA promoter, with feedback regulation
between the two. To the best of the authors' knowledge, the only
study analyzing the regulation of this miRNA by epigenetic
mechanisms in tissue from patients with PCa is that of Suh et
al (2011) (29) in the United
States population, who observed a relationship between the
underexpression of this miRNA and the hypermethylation in its
promoter region. This suggested that epigenetic mechanisms in the
regulation of miR-145-5p may be the most common cause of its gene
expression dysregulation.
At present, the mechanisms that lead to the
dysregulation of miR-148b-3p in PCa have not been studied. The
present study identified a set of deletions with a start site at
chr12:54337042/54337043, which were unified and called DelsAAG.
This mutation was related to the presence of PCa; likewise, it was
associated with the underexpression of miR-148b-3p. These deletions
are located in a particular region, specifically in the first
intron of the COPZ1 gene, in the sequence comprising
chr12:54337020–54337044, there is a set of 25 adenines in tandem,
behaving as a pure or perfect microsatellite, and as previously
reported, this type of repeated sites are hot spots for mutations,
as they are susceptible to the generation of deletions and
insertions, being highly polymorphic, having mutation rates ranging
from 103 and 106 for each cell generation, thus this could explain
the existence of the mutations in the analyzed site (30-32).
According to the GeneCards database (33), this mutation is in a region that
functions as a proximal intragenic enhancer (ID: GH12J054336),
which could be a key cis-acting regulator promoting the expression
of miR-148b-3p (34). Similarly,
according to the ENSEMBL database, this mutation is found within a
binding site for the transcription factors E2F1, E2F2 and E2F3
(chr:54337040-54337055) (35);
previously these transcription factors have been identified to
regulate the expression of other miRNAs, such as miRNAs 17 and 20,
by binding to their promoter regions and promoting their expression
(36,37). This is consistent with previous
studies, as ~50% of the variants or mutations in transcription
factor binding regions (RUFT) are in intronic regions (38). Previously, some RUFT mutations in
Pac have been reported to modify their affinity and be involved in
pathology development. For example, Huang et al (2014)
(39), reported that an SNV
increases the binding affinity of HOXB13, resulting in the
overexpression of RFX6, leading to proliferation, migration and
invasion of prostate tumor cells. Likewise, the rs7077275
polymorphism increases the binding of CTFC, which leads to the
overexpression of CTBP2, decreasing apoptosis, and increasing tumor
growth in PCa (40). Eickhardt
et al (41) reported in 2016
that deletions are more disruptive variations than SNVs, leading to
a more significant modification in the affinity of the
transcription factors for the affected sequence. The aforementioned
description indicated that the DelsAAG mutation could affect the
binding of transcription factors, resulting in the absence or
reduction of miR-148b-3p transcription.
This miRNA is directly involved in molecular
mechanisms related to the development of PCa. Tomeva et al
(2022) (42), observed that
miR-148b-3p was underexpressed in tumors that occur at the androgen
receptor (AR). Feng et al (43), in 2019 demonstrated that this miRNA
was underexpressed in tumor tissue of patients with PCa. In
addition, they observed that the 3'-untranlated region of the KLF4
gene contains the conserved miR-148b-3p binding site, making it a
potential target for this miRNA binding, which was verified through
their in vitro experiments (43). KLF4 is a transcription factor that
can regulate the expression of various genes and that has been
previously observed to be overexpressed in PCa (44). This transcription factor has an
important implication in the prostate, since Siu et al
(2016) (45), demonstrated a
reciprocal relationship between KLF4 and the AR, where AR can
induce the expression of this transcription factor. KLF4 can then
bind to the AR promoter generating its transcription. In turn, AR
activity is commonly altered in PCa, which promotes the evolution
and progression of this cancer (46,47).
Therefore, it was proposed by the auhtors that the mutation near
miR-148b-3p, which includes a set of deletions in a binding site
for transcription factors, promotes miRNA underexpression, which
means that it cannot regulate the expression of KLF4 increasing its
expression and, as a result, leading to the overexpression of the
AR, enhancing the proliferation and progression of PCa (Fig. 3).
In conclusion, mutations in regions near the gene
that codes for a miRNA may be related to its expression and be part
of the regulation mechanisms in a pathogenic event such as PCa.
Thus, the present study becomes the first to propose a method for
regulating the expression of miR-148b-3p in PCa, involving
deletions in a transcription factor binding site.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the Autonomous
University of Sinaloa (grant no. PROFAPI PRO_A3_001) and the
Polytechnic University of Sinaloa internal resource.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
FBH and FLO performed the statistical analysis,
critically reviewed literature findings and drafting the
manuscript. NGM, MAA and EAM collected the sample, conceived and
designed the study and revised the manuscript. FBH and NGM confirm
the authenticity of all the raw data. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
The present study was approved (approval no. P-3103)
by the Alvarez and Arrazola Radiologists Clinic's Ethics and
Research Committee (Mazatlán, Mexico). Written informed consent was
obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Xiao J, Gong AY, Eischeid AN, Chen D, Deng
C, Young CY and Chen XM: miR-141 modulates androgen receptor
transcriptional activity in human prostate cancer cells through
targeting the small heterodimer partner protein. Prostate.
72:1514–1522. 2012.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Sun Q, Zhao X, Liu X, Wang Y, Huang J,
Jiang B, Chen Q and Yu J: miR-146a functions as a tumor suppressor
in prostate cancer by targeting Rac1. Prostate. 74:1613–1621.
2014.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Bergez-Hernández F, Arámbula-Meraz E,
Alvarez-Arrazola M, Irigoyen-Arredondo M, Luque-Ortega F,
Martínez-Camberos A, Cedano-Prieto D, Contreras-Gutiérrez J,
Martínez-Valenzuela C and García-Magallanes N: Expression Analysis
of miRNAs and their potential role as biomarkers for prostate
cancer detection. Am J Mens Health.
16(15579883221120989)2022.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Arámbula-Meraz E, Bergez-Hernández F,
Leal-León E, Romo-Martínez E, Picos-Cárdenas V, Luque-Ortega F,
Romero-Quintana J, Alvarez-Arrazola M and García-Magallanes N:
Expression of miR-148b-3p is correlated with overexpression of
biomarkers in prostate cancer. Genet Mol Biol.
43(e20180330)2020.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Xu WX, Liu Z, Deng F, Wang DD, Li XW, Tian
T, Zhang J and Tang JH: MiR-145: A potential biomarker of cancer
migration and invasion. Am J Transl Res. 11:6739–6753.
2019.PubMed/NCBI
|
|
6
|
Jin W, Fei X, Wang X, Song Y and Chen F:
Detection and prognosis of prostate cancer using blood-based
biomarkers. Mediators Inflamm. 2020(8730608)2020.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Chu H, Zhong D, Tang J, Li J, Xue Y, Tong
N, Qin C, Yin C, Zhang Z and Wang M: A functional variant in
miR-143 promoter contributes to prostate cancer risk. Arch Toxicol.
90:403–414. 2016.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Kotarac N, Dobrijevic Z, Matijasevic S,
Savic-Pavicevic D and Brajuskovic G: Analysis of association of
potentially functional genetic variants within genes encoding
miR-34b/c, miR-378 and miR-143/145 with prostate cancer in Serbian
population. EXCLI J. 18:515–529. 2019.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Watahiki A and Wang Y, Morris J, Dennis K,
O'Dwyer HM, Gleave M, Gleave M, Gout PW and Wang Y: MicroRNAs
associated with metastatic prostate cancer. PLoS One.
6(e24950)2011.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Chen X, Wang G, Lu X, Gao P, Song Y, Sun
J, Li A, Xu Y, Xu H and Wang Z: Polymorphisms and haplotypes of the
miR-148/152 family are associated with the risk and
clinicopathological features of gastric cancer in a Northern
Chinese population. Mutagenesis. 29:401–407. 2014.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Chomczynski P: A reagent for the
single-step simultaneous isolation of RNA, DNA and proteins from
cell and tissue samples. Biotechniques. 15:532–4, 536-7.
1993.PubMed/NCBI
|
|
12
|
Kim YG, Kim MJ, Lee JS, Lee JA, Song JY,
Cho SI, Park SS and Seong MW: SnackVar: An open-source software for
sanger sequencing analysis optimized for clinical use. J Mol Diagn.
23:140–148. 2021.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Rijk PD and Del-Favero J: novoSNP3:
Variant detection and sequence annotation in resequencing projects.
Methods Mol Biol. 396:331–344. 2007.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Taylor SC, Nadeau K, Abbasi M, Lachance C,
Nguyen M and Fenrich J: The Ultimate qPCR experiment: Producing
publication quality, reproducible data the first time. Trends
Biotechnol. 37:761–774. 2019.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Akalin I, Erol B, Aslan E, Ozkanli SS,
Efiloglu O, Yildirim S, Caskurlu T, Yildirim A and Karaman MI: A
new promising pathway in aggressive prostate cancer:
Treg/mir-let8c/lin28b. Arch Esp Urol. 75:459–466. 2022.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Liu S, Zhao Y, Zhao Y and Tang X: The
prognostic value of miR-487a in clear cell renal cell carcinoma and
its influence on cell biological behavior. Arch Esp Urol.
75:346–353. 2022.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Wang Z, Zhu X, Zhai H, Wang Y and Hao G:
Integrated analysis of mRNA-single nucleotide polymorphism-microRNA
interaction network to identify biomarkers associated with prostate
cancer. Front Genet. 13(922712)2022.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Camargo JA, Lopes RE, Ferreira GFD, Viana
NI, Guimaraes V, Leite KRM, Nahas WC, Srougi M, Antunes AA and Reis
ST: The role of single nucleotide polymorphisms of miRNAs 100 and
146a as prognostic factors for prostate cancer. Int J Biol Markers.
36:50–56. 2021.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Xu B, Feng NH, Li PC, Tao J, Wu D, Zhang
ZD, Tong N, Wang JF, Song NH, Zhang W, et al: A functional
polymorphism in Pre-miR-146a gene is associated with prostate
cancer risk and mature miR-146a expression in vivo. Prostate.
70:467–472. 2010.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Huang S, Cui H, Lou Z, Wang X, Chen L, Xie
Z, Hehir M, Yao X, Ren Y, Cen D and Weng G: Association of
rs3787016 in Long Non-coding RNAs POLR2E and rs2910164 in
MiRNA-146a with prostate cancer: A systematic review and
meta-analysis. Iran J Public Health. 47:623–632. 2018.PubMed/NCBI
|
|
22
|
Zhao CG, Xu HB and Xu B: The rs4705342
gene mutation in the promoter region of the miR-143/145 cluster
associated with the risk of prostate cancer in the Chinese Han
population. Zhonghua Nan Ke Xue. 25:696–702. 2019.PubMed/NCBI(In Chinese).
|
|
23
|
Sherry ST, Ward MH, Kholodov M, Baker J,
Phan L, Smigielski EM and Sirotkin K: dbSNP: The NCBI database of
genetic variation. Nucleic Acids Res. 29:308–311. 2001.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Chacon-Cortes D, Smith RA, Haupt LM, Lea
RA, Youl PH and Griffiths LR: Genetic association analysis of miRNA
SNPs implicates MIR145 in breast cancer susceptibility. BMC Med
Genet. 16(107)2015.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Zaman MS, Chen Y, Deng G, Shahryari V, Suh
SO, Saini S, Majid S, Liu J, Khatri G, Tanaka Y and Dahiya R: The
functional significance of microRNA-145 in prostate cancer. Br J
Cancer. 103:256–264. 2010.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Bellissimo T, Ganci F, Gallo E, Sacconi A,
Tito C, De Angelis L, Pulito C, Masciarelli S, Diso D, Anile M, et
al: Thymic Epithelial Tumors phenotype relies on miR-145-5p
epigenetic regulation. Mol Cancer. 16(88)2017.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Donzelli S, Mori F, Bellissimo T, Sacconi
A, Casini B, Frixa T, Roscilli G, Aurisicchio L, Facciolo F,
Pompili A, et al: Epigenetic silencing of miR-145-5p contributes to
brain metastasis. Oncotarget. 6:35183–35201. 2015.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Xue G, Ren Z, Chen Y, Zhu J, Du Y, Pan D,
Li X and Hu B: A feedback regulation between miR-145 and DNA
methyltransferase 3b in prostate cancer cell and their responses to
irradiation. Cancer Lett. 361:121–127. 2015.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Suh SO, Chen Y, Zaman MS, Hirata H,
Yamamura S, Shahryari V, Liu J, Tabatabai ZL, Kakar S, Deng G, et
al: MicroRNA-145 is regulated by DNA methylation and p53 gene
mutation in prostate cancer. Carcinogenesis. 32:772–778.
2011.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Nesta AV, Tafur D and Beck CR: Hotspots of
human mutation. Trends Genet. 37:717–729. 2021.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Gemayel R, Cho J, Boeynaems S and
Verstrepen KJ: Beyond junk-variable tandem repeats as facilitators
of rapid evolution of regulatory and coding sequences. Genes
(Basel). 3:461–480. 2012.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Vieira ML, Santini L, Diniz AL and Munhoz
Cde F: Microsatellite markers: What they mean and why they are so
useful. Genet Mol Biol. 39:312–328. 2016.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Fishilevich S, Nudel R, Rappaport N, Hadar
R, Plaschkes I, Iny Stein T, Rosen N, Kohn A, Twik M, Safran M, et
al: GeneHancer: Genome-wide integration of enhancers and target
genes in GeneCards. Database (Oxford). 2017(bax028)2017.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Peng Y and Zhang Y: Enhancer and
super-enhancer: Positive regulators in gene transcription. Animal
Model Exp Med. 1:169–179. 2018.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Cunningham F, Allen JE, Allen J,
Alvarez-Jarreta J, Amode MR, Armean IM, Austine-Orimoloye O, Azov
AG, Barnes I, Bennett R, et al: Ensembl 2022. Nucleic Acids Res.
50:D988–D995. 2022.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Sylvestre Y, De Guire V, Querido E,
Mukhopadhyay UK, Bourdeau V, Major F, Ferbeyre G and Chartrand P:
An E2F/miR-20a autoregulatory feedback loop. J Biol Chem.
282:2135–2143. 2007.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Woods K, Thomson JM and Hammond SM: Direct
regulation of an oncogenic micro-RNA cluster by E2F transcription
factors. J Biol Chem. 282:2130–2134. 2007.PubMed/NCBI View Article : Google Scholar
|
|
38
|
de Santiago I, Liu W, Yuan K, O'Reilly M,
Chilamakuri CS, Ponder BA, Meyer KB and Markowetz F: BaalChIP:
Bayesian analysis of allele-specific transcription factor binding
in cancer genomes. Genome Biol. 18(39)2017.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Huang Q, Whitington T, Gao P, Lindberg JF,
Yang Y, Sun J, Väisänen MR, Szulkin R, Annala M, Yan J, et al: A
prostate cancer susceptibility allele at 6q22 increases RFX6
expression by modulating HOXB13 chromatin binding. Nat Genet.
46:126–135. 2014.PubMed/NCBI View
Article : Google Scholar
|
|
40
|
Zhang P, Tillmans LS, Thibodeau SN and
Wang L: Single-Nucleotide polymorphisms sequencing identifies
candidate functional variants at prostate cancer risk loci. Genes
(Basel). 10(547)2019.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Eickhardt E, Als TD, Grove J, Boerglum AD
and Lescai F: Estimating the functional impact of INDELs in
transcription factor binding sites: A Genome-Wide Landscape.
Bioinformatics, 2016 Jun. Available from: http://biorxiv.org/lookup/doi/10.1101/057604.
|
|
42
|
Tomeva E, Switzeny OJ, Heitzinger C, Hippe
B and Haslberger AG: Comprehensive approach to distinguish patients
with solid tumors from healthy controls by combining androgen
receptor mutation p. H875Y with Cell-Free DNA methylation and
circulating miRNAs. Cancers (Basel). 14(462)2022.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Feng F, Liu H, Chen A, Xia Q, Zhao Y, Jin
X and Huang J: miR-148-3p and miR-152-3p synergistically regulate
prostate cancer progression via repressing KLF4. J Cell Biochem.
120:17228–17239. 2019.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Wei LZ, Wang YQ, Chang YL, An N, Wang X,
Zhou PJ, Zhu HH, Fang YX and Gao WQ: Imbalance of a KLF4-miR-7
auto-regulatory feedback loop promotes prostate cancer cell growth
by impairing microRNA processing. Am J Cancer Res. 8:226–244.
2018.PubMed/NCBI
|
|
45
|
Siu MK, Suau F, Chen WY, Tsai YC, Tsai HY,
Yeh HL and Liu YN: KLF4 functions as an activator of the androgen
receptor through reciprocal feedback. Oncogenesis.
5(e282)2016.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Desai K, McManus JM and Sharifi N:
Hormonal therapy for prostate cancer. Endocr Rev. 42:354–373.
2021.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Auchus RJ and Sharifi N: Sex Hormones and
Prostate Cancer. Annu Rev Med. 71:33–45. 2020.PubMed/NCBI View Article : Google Scholar
|