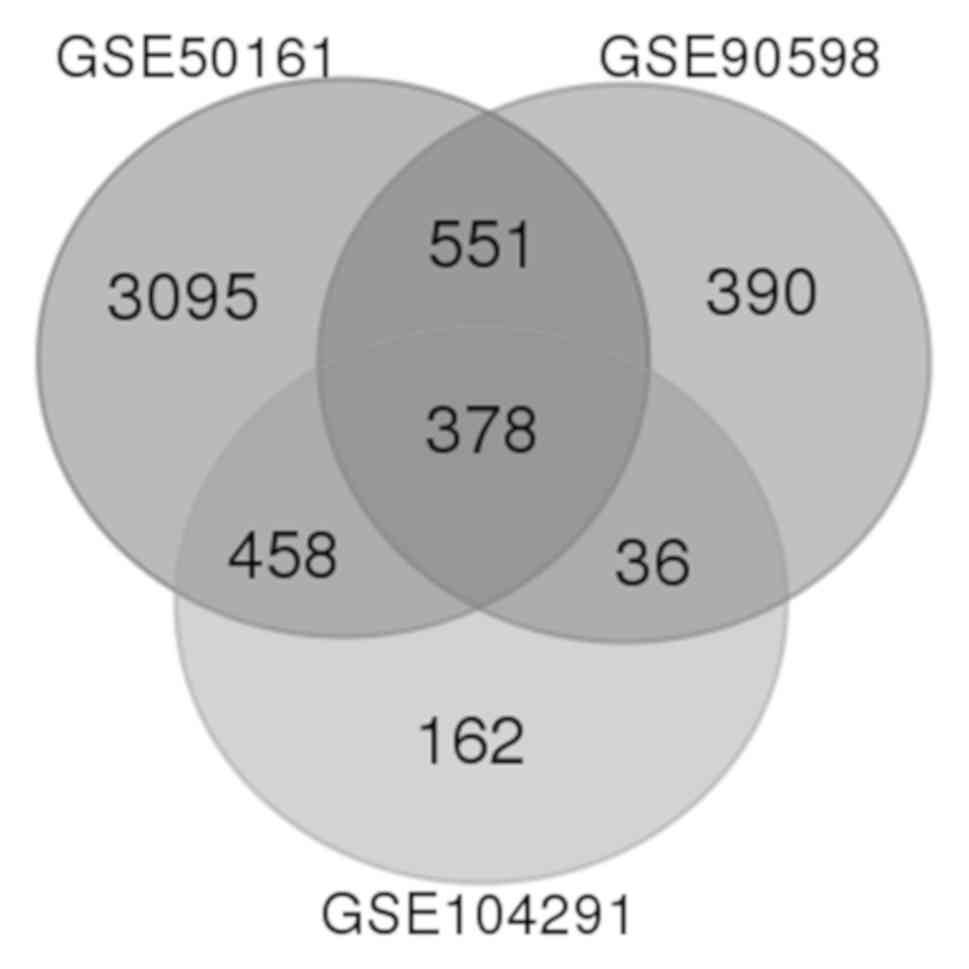
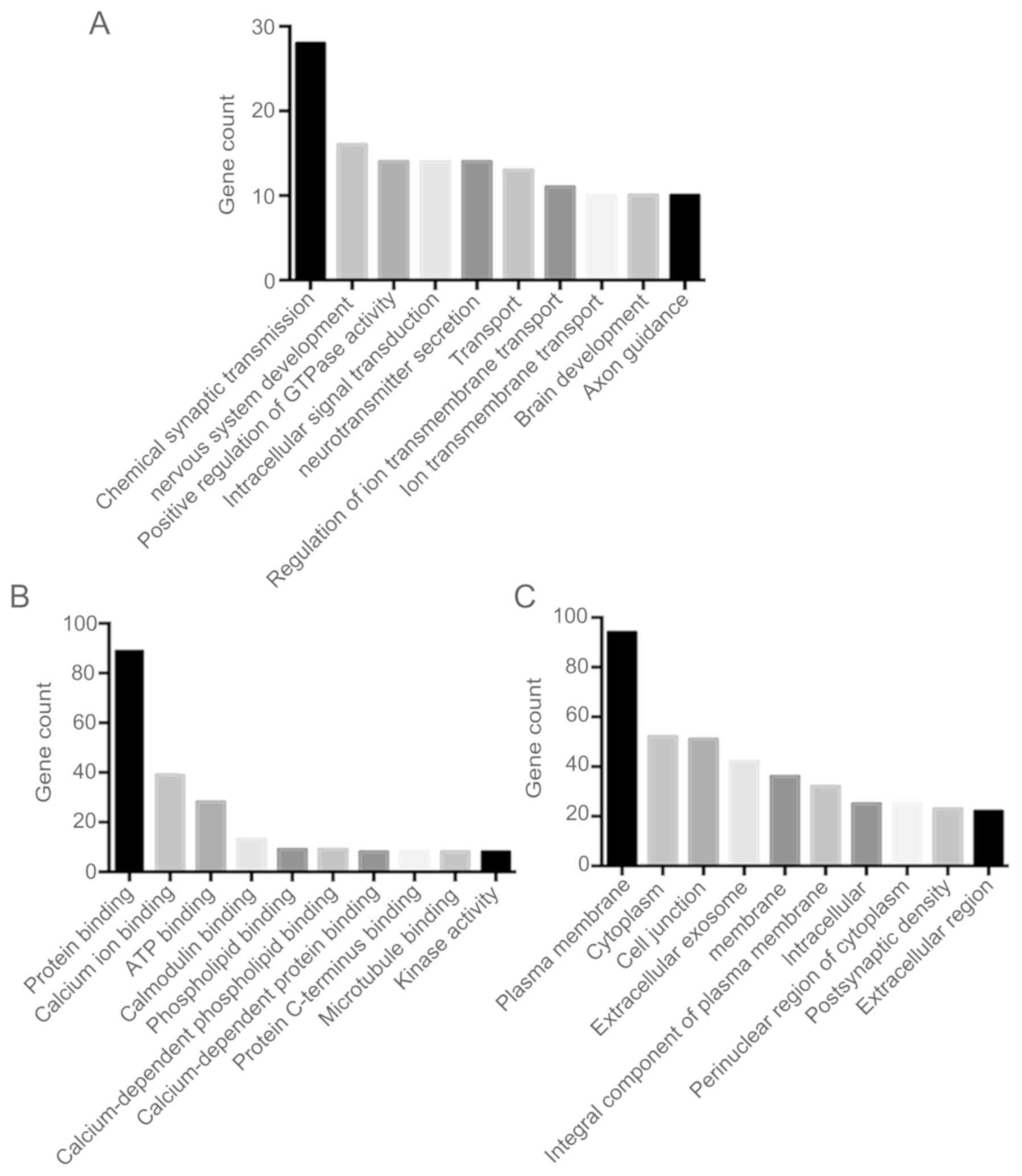
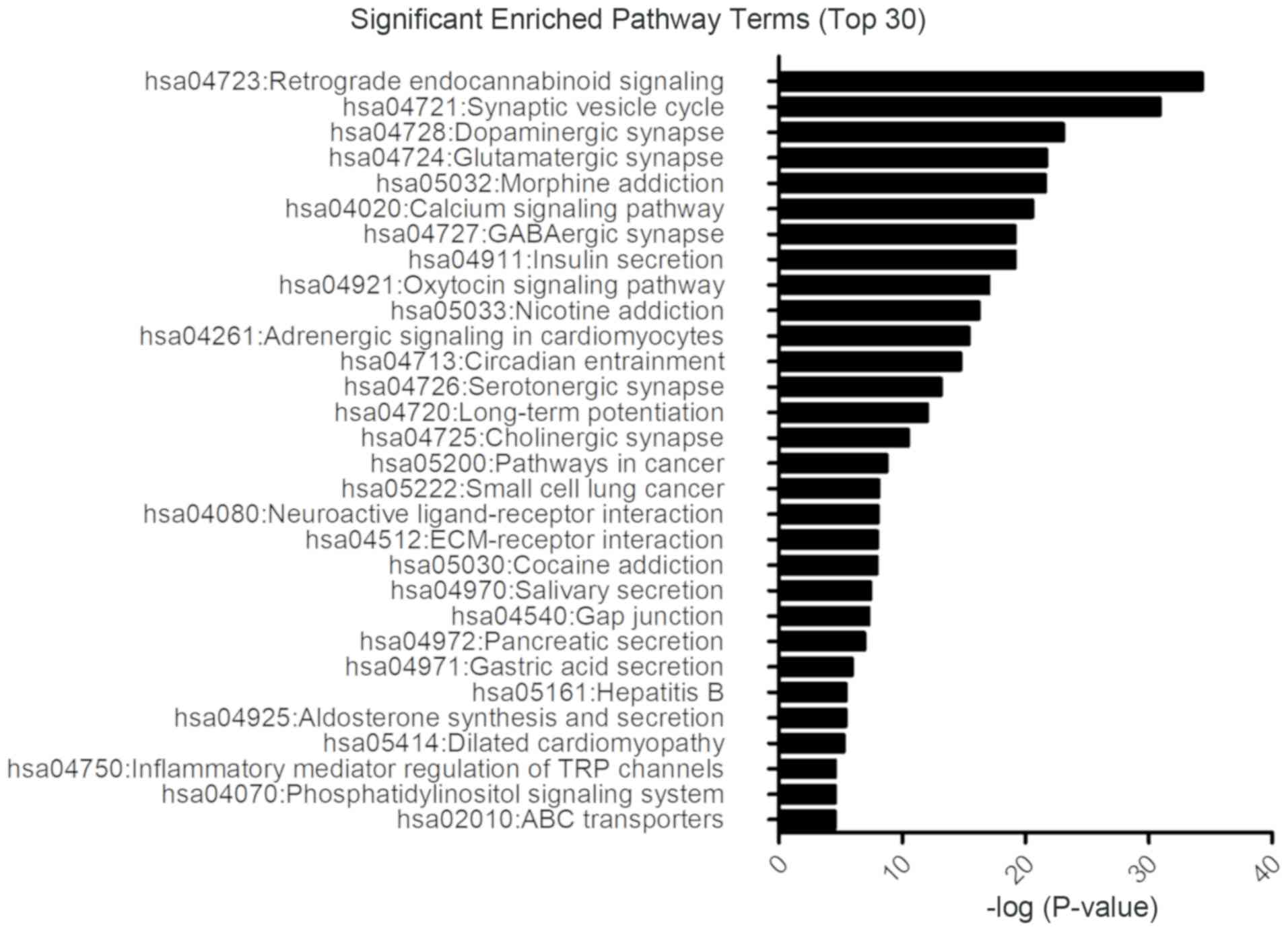
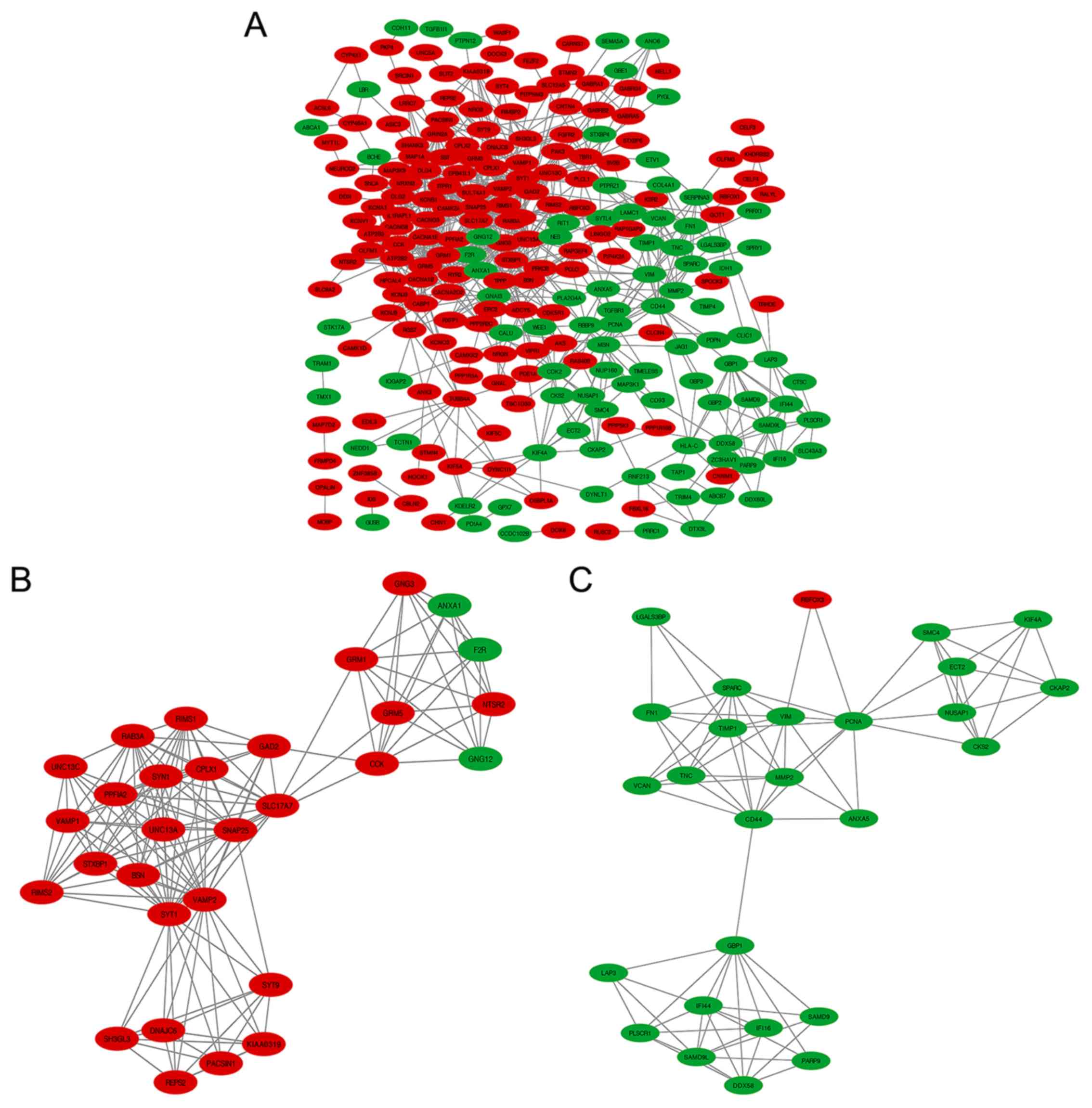
|
1
|
Wen WS, Hu SL, Ai Z, Mou L, Lu JM and Li
S: Methylated of genes behaving as potential biomarkers in
evaluating malignant degree of glioblastoma. J Cell Physiol.
232:3622–3630. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yano S, Miwa S, Kishimoto H, Toneri M,
Hiroshima Y, Yamamoto M, Bouvet M, Urata Y, Tazawa H, Kagawa S, et
al: Experimental curative fluorescence-guided surgery of highly
invasive glioblastoma multiforme selectively labeled with a
killer-reporter adenovirus. Mol Ther. 23:1182–1188. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Stupp R, Taillibert S, Kanner A, Read W,
Steinberg D, Lhermitte B, Toms S, Idbaih A, Ahluwalia MS, Fink K,
et al: Effect of tumor-treating fields plus maintenance
temozolomide vs maintenance temozolomide alone on survival in
patients with glioblastoma: A randomized clinical trial. JAMA.
318:2306–2316. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Stupp R, Mason WP, van den Bent MJ, Weller
M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn
U, et al: Radiotherapy plus concomitant and adjuvant temozolomide
for glioblastoma. N Engl J Med. 352:987–996. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ostrom QT, Bauchet L, Davis FG, Deltour I,
Fisher JL, Langer CE, Pekmezci M, Schwartzbaum JA, Turner MC, Walsh
KM, et al: The epidemiology of glioma in adults: A ‘state of the
science’ review. Neuro Oncol. 16:896–913. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zeng T, Li L, Zhou Y and Gao L: Exploring
long noncoding RNAs in glioblastoma: Regulatory mechanisms and
clinical potentials. Int J Genomics. 2018:28959582018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Filbin MG and Suvà ML: Gliomas genomics
and epigenomics: Arriving at the start and knowing it for the first
time. Annu Rev Pathol. 11:497–521. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Stupp R, Hegi ME, Gilbert MR and
Chakravarti A: Chemoradiotherapy in malignant glioma: Standard of
care and future directions. J Clin Oncol. 25:4127–4136. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Delgado-López PD and Corrales-García EM:
Survival in glioblastoma: A review on the impact of treatment
modalities. Clin Transl Oncol. 18:1062–1071. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lim M, Xia Y, Bettegowda C and Weller M:
Current state of immunotherapy for glioblastoma. Nat Rev Clin
Oncol. 15:422–442. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chinot OL, Wick W, Mason W, Henriksson R,
Saran F, Nishikawa R, Carpentier AF, Hoang-Xuan K, Kavan P, Cernea
D, et al: Bevacizumab plus radiotherapy-temozolomide for newly
diagnosed glioblastoma. N Engl J Med. 370:709–722. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Furnari FB, Cloughesy TF, Cavenee WK and
Mischel PS: Heterogeneity of epidermal growth factor receptor
signalling networks in glioblastoma. Nat Rev Cancer. 15:302–310.
2015. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ludwig K and Kornblum HI: Molecular
markers in glioma. J Neurooncol. 134:505–512. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yan Y, Xu Z, Li Z, Sun L and Gong Z: An
insight into the increasing role of LncRNAs in the pathogenesis of
gliomas. Front Mol Neurosci. 10:532017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sturm D, Bender S, Jones DT, Lichter P,
Grill J, Becher O, Hawkins C, Majewski J, Jones C, Costello JF, et
al: Paediatric and adult glioblastoma: Multiform (epi)genomic
culprits emerge. Nat Rev Cancer. 14:92–107. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Vogelstein B, Papadopoulos N, Velculescu
VE, Zhou S, Diaz LA Jr and Kinzler KW: Cancer genome landscapes.
Science. 339:1546–1558. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Long H, Liang C, Zhang X, Fang L, Wang G,
Qi S, Huo H and Song Y: Prediction and analysis of key genes in
glioblastoma based on bioinformatics. Biomed Res Int.
2017:76531012017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Huang SW, Ali ND, Zhong L and Shi J:
MicroRNAs as biomarkers for human glioblastoma: Progress and
potential. Acta Pharmacol Sin. 39:1405–1413. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tan SY, Sandanaraj E, Tang C and Ang BT:
Biobanking: An important resource for precision medicine in
glioblastoma. Adv Exp Med Biol. 951:47–56. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Guo Y, Bao Y, Ma M and Yang W:
Identification of key candidate genes and pathways in colorectal
cancer by integrated bioinformatical analysis. Int J Mol Sci.
18:E7222017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tanwar MK, Gilbert MR and Holland EC: Gene
expression microarray analysis reveals YKL-40 to be a potential
serum marker for malignant character in human glioma. Cancer Res.
62:4364–4368. 2002.PubMed/NCBI
|
|
22
|
Kunkle B, Yoo C and Roy D: Discovering
gene-environment interactions in glioblastoma through a
comprehensive data integration bioinformatics method.
Neurotoxicology. 35:1–14. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li W, Li K, Zhao L and Zou H:
Bioinformatics analysis reveals disturbance mechanism of MAPK
signaling pathway and cell cycle in Glioblastoma multiforme. Gene.
547:346–350. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Griesinger AM, Birks DK, Donson AM, Amani
V, Hoffman LM, Waziri A, Wang M, Handler MH and Foreman NK:
Characterization of distinct immunophenotypes across pediatric
brain tumor types. J Immunol. 191:4880–4888. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gulluoglu S, Tuysuz EC, Sahin M, Kuskucu
A, Kaan Yaltirik C, Ture U, Kucukkaraduman B, Akbar MW, Gure AO,
Bayrak OF and Dalan AB: Simultaneous miRNA and mRNA transcriptome
profiling of glioblastoma samples reveals a novel set of OncomiR
candidates and their target genes. Brain Res. 1700:199–210. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bady P, Diserens AC, Castella V, Kalt S,
Heinimann K, Hamou MF, Delorenzi M and Hegi ME: DNA fingerprinting
of glioma cell lines and considerations on similarity measurements.
Neuro Oncol. 14:701–711. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sciuscio D, Diserens AC, van Dommelen K,
Martinet D, Jones G, Janzer RC, Pollo C, Hamou MF, Kaina B, Stupp
R, et al: Extent and patterns of MGMT promoter methylation in
glioblastoma- and respective glioblastoma-derived spheres. Clin
Cancer Res. 17:255–266. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Barrett T, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, Holko M, et al: NCBI GEO: Archive for functional
genomics data sets-update. Nucleic Acids Res. 41:D991–D995. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
He WQ, Gu JW, Li CY, Kuang YQ, Kong B,
Cheng L, Zhang JH, Cheng JM and Ma Y: The PPI network and clusters
analysis in glioblastoma. Eur Rev Med Pharmacol Sci. 19:4784–4790.
2015.PubMed/NCBI
|
|
30
|
Li Y, Min W, Li M, Han G, Dai D, Zhang L,
Chen X, Wang X, Zhang Y, Yue Z and Liu J: Identification of hub
genes and regulatory factors of glioblastoma multiforme subgroups
by RNA-seq data analysis. Int J Mol Med. 38:1170–1178. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wei B, Wang L, Du C, Hu G, Wang L, Jin Y
and Kong D: Identification of differentially expressed genes
regulated by transcription factors in glioblastomas by
bioinformatics analysis. Mol Med Rep. 11:2548–2554. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Huang da W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Franceschini A, Szklarczyk D, Frankild S,
Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C
and Jensen LJ: STRING v9.1: Protein-protein interaction networks,
with increased coverage and integration. Nucleic Acids Res.
41:D808–D815. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chen WJ, Tang RX, He RQ, Li DY, Liang L,
Zeng JH, Hu XH, Ma J, Li SK and Chen G: Clinical roles of the
aberrantly expressed lncRNAs in lung squamous cell carcinoma: A
study based on RNA-sequencing and microarray data mining.
Oncotarget. 8:61282–61304. 2017.PubMed/NCBI
|
|
36
|
Scardoni G, Petterlini M and Laudanna C:
Analyzing biological network parameters with CentiScaPe.
Bioinformatics. 25:2857–2859. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Anaya J: OncoLnc: Linking TCGA survival
data to mRNAs, miRNAs, and lncRNAs. Peer J Computer Sci. 2:e672016.
View Article : Google Scholar
|
|
38
|
Iser IC, Pereira MB, Lenz G and Wink MR:
The epithelial- to-mesenchymal transition-like process in
glioblastoma: An updated systematic review and in silico
investigation. Med Res Rev. 37:271–313. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Alshabi AM, Vastrad B, Shaikh IA and
Vastrad C: Identification of crucial candidate genes and pathways
in glioblastoma multiform by bioinformatics analysis. Biomolecules.
9:E2012019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Caragher SP, Hall RR, Ahsan R and Ahmed
AU: Monoamines in glioblastoma: Complex biology with therapeutic
potential. Neuro Oncol. 20:1014–1025. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Cheng YC, Tsai WC, Sung YC, Chang HH and
Chen Y: Interference with PSMB4 expression exerts an anti-tumor
effect by decreasing the invasion and proliferation of human
glioblastoma cells. Cell Physiol Biochem. 45:819–831. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Turkowski K, Brandenburg S, Mueller A,
Kremenetskaia I, Bungert AD, Blank A, Felsenstein M and Vajkoczy P:
VEGF as a modulator of the innate immune response in glioblastoma.
Glia. 66:161–174. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Li C, Chan J, Haeseleer F, Mikoshiba K,
Palczewski K, Ikura M and Ames JB: Structural insights into
Ca2+-dependent regulation of inositol
1,4,5-trisphosphate receptors by CaBP1. J Biol Chem. 284:2472–2481.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Oz S, Tsemakhovich V, Christel CJ, Lee A
and Dascal N: CaBP1 regulates voltage-dependent inactivation and
activation of Ca(V)1.2 (L-type) calcium channels. J Biol Chem.
286:13945–13953. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Dixit AB, Banerjee J, Srivastava A,
Tripathi M, Sarkar C, Kakkar A, Jain M and Chandra PS: RNA-seq
analysis of hippocampal tissues reveals novel candidate genes for
drug refractory epilepsy in patients with MTLE-HS. Genomics.
107:178–188. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Mubaraki MA, Hafiz TA, Al-Quraishy S and
Dkhil MA: Oxidative stress and genes regulation of cerebral malaria
upon Zizyphus spina-christi treatment in a murine model. Microb
Pathog. 107:69–74. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Chen ML, Chen YC, Peng IW, Kang RL, Wu MP,
Cheng PW, Shih PY, Lu LL, Yang CC and Pan CY: Ca2+
binding protein-1 inhibits Ca2+ currents and exocytosis in bovine
chromaffin cells. J Biomed Sci. 15:169–181. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhao J, Zhu X, Shrubsole MJ, Ness RM,
Hibler EA, Cai Q, Long J, Chen Z, Jiang M, Kabagambe EK, et al:
Interactions between calcium intake and polymorphisms in genes
essential for calcium reabsorption and risk of colorectal neoplasia
in a two-phase study. Mol Carcinog. 56:2258–2266. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Sun P, Shrubsole MJ, Ness RM, Cai Q, Long
J, Edwards T, Chen Z, Zhu X, Deng X, Luo J, et al: Calcium intake,
CABP1 polymorphisms, and the risk of colorectal adenoma: Results
from Tennessee Colorectal Polyp Study. Cancer Res. 71 (Suppl
8):S37632011.
|
|
50
|
Zhang Y, Xu J and Zhu X: A 63 signature
genes prediction system is effective for glioblastoma prognosis.
Int J Mol Med. 41:2070–2078. 2018.PubMed/NCBI
|
|
51
|
Delfino KR, Serão NV, Southey BR and
Rodriguez-Zas SL: Therapy-, gender- and race-specific microRNA
markers, target genes and networks related to glioblastoma
recurrence and survival. Cancer Genomics Proteomics. 8:173–183.
2011.PubMed/NCBI
|