Introduction
Osteoarthritis (OA) is the most prevalent joint
disease, and has been considered to be a type of non-inflammatory
arthritis for a number of years (1). The characteristics of OA include pain,
tenderness, limited movement, crepitus and different levels of
inflammation without systemic effects (2). Due to the close association with age,
OA has become one of the major problems in elderly individuals
(3). Articular cartilage
degradation is one of the principal pathological alterations in OA,
and the fate of articular cartilage is determined by articular
chondrocytes (4). Mechanistically,
the imbalance in extracellular matrix (ECM) synthesis and
degradation results in injuries in articular cartilage, including
inflammatory injury. Moreover, apoptosis of chondrocytes has been
suggested to serve an essential role in the pathogenesis of OA
(5). Therefore, chondrocytes, as
the only cell type in articular cartilage (6), have been most commonly used in OA
research to elucidate the pathogenesis of this condition.
Increasing evidence has suggested that non-coding
RNAs participate in OA (7), and
microRNAs (miRNAs/miRs) are one class of non-coding RNAs with about
22 nucleotides (8). Recently,
dysregulation of miRNAs has also been indicated to be associated
with the maintenance of cartilage homeostasis and inflammatory
response during the progression of OA (9), including miR-320, miR-21 and miR-140
(10-12).
miR-200b-3p (miR-200b) has been revealed to be an important
regulator of cell proliferation and motility in various types of
cancer, such as cervical cancer and breast cancer (13-15).
However, although the regulatory mechanism of miR-200b has been
preliminarily identified in cartilage cells from patients with OA,
its potential role in OA has not been widely studied (16).
As key membrane-bound proteins in the surface of
cells residing in the endoplasmic reticulum and Golgi (17), fucosyltransferases (FUTs) have been
reported to participate in miscellaneous biological processes,
including inflammation (15,18).
Accumulating evidence has indicated that FUTs may regulate the
development of rheumatoid arthritis (RA) and juvenile idiopathic
arthritis (19,20), whereas the effect of FUTs in OA
remains not fully understood. FUT4 has been indicated to mediate OA
progression following miR-26a/b targeting (21). The present study examined the
association between FUT4 and miR-200b in injured chondrocytes in
OA.
Lipopolysaccharide (LPS), also known as endotoxin,
can induce inflammatory response both in vitro and in
vivo (22). Numerous
inflammatory cytokines have been indicated to be highly expressed
after LPS treatment, such as interleukin IL-1β, IL-6, IL-8 and
TNF-α (23). Therefore,
LPS-stimulated cell models have been widely used to identify novel
therapeutic targets for the treatment of OA (23-25).
Moreover, miRNAs have been revealed to be implicated in LPS-induced
articular chondrocyte injury in OA (4,24,25).
The present study aimed to investigate the
expression level of miR-200b in cartilage tissues from patients
with OA and its role in an LPS-induced chondrocyte OA model in
vitro. A mechanistic study was also performed to explore the
association between miR-200b and FUT4 in modulating inflammatory
injury in LPS-treated knee articular chondrocytes.
Materials and methods
Patients and tissue specimens
Normal cartilage specimens were obtained from 14
patients with emergency traumatic amputation (8 males and 6
females; average age, 34.6 years) and 14 patients with OA (5 males
and 9 females; average age, 45.3 years) between Jan, 2014 and Dec.
2016 at the Yan'an People's Hospital (Yan'an, China). The average
body height and weight were 165.6 cm and 65.4 kg in normal patients
and 162.3 cm and 62.7 kg in patients with OA. All normal patients
with arthritis, including RA, OA and septic arthritis, were
excluded from the present study. Among the patients with OA,
patients were excluded if they received orthopedic surgery before,
or suffered a fracture, septic arthritis or other serious diseases.
The pathological diagnosis was confirmed by two professor
pathologists, and the diagnostic criteria of knee OA used were in
accordance with the International Classification of Diseases, Tenth
Revision, Clinical Modification (code M17) (26). The samples were isolated form the
knee articular cartilage and subsequently placed into liquid
nitrogen at -73˚C. Written informed consent was obtained
from each patient and the study was approved by the Ethics
Committee of Yan'an People's Hospital. Moreover, the present study
involving human subjects, human material or human data was
performed in accordance with the Declaration of Helsinki.
Cell culture and LPS stimulation
Under aseptic conditions, normal cartilage tissues
and OA cartilages were removed from fibrous connective tissues and
cut into small pieces. After washing with PBS containing 100 U/ml
penicillin and 100 µg/ml streptomycin solution (Gibco; Thermo
Fisher Scientific, Inc.), the cartilage tissues were subjected to
sequential digestion with 0.25% trypsin (Invitrogen; Thermo Fisher
Scientific, Inc.) for 30 min at 37˚C and subsequently
with 0.2% collagenase type II (EMD Millipore) in DMEM (Gibco;
Thermo Fisher Scientific, Inc.) for 10 h at 37˚C.
Following filtration with filters (100 and 40 µm; JingAn
Biological), the cells in medium were centrifuged at 400 x g for 15
min at room temperature, and subsequently resuspended in fresh
culture media supplemented with 10% FBS (Gibco; Thermo Fisher
Scientific, Inc.). The cartilage cells and HEK 293T (293T) (cat.
no. CRL-3216; American Type Culture Collection) cells were cultured
in DMEM (Gibco; Thermo Fisher Scientific, Inc.) containing 10% FBS
(Gibco; Thermo Fisher Scientific, Inc.) in humidified air with 5%
(v/v) CO2 at 37˚C. The first passage
chondrocytes were obtained after cell cultivation for 10 days.
All experiments were performed on cells between the
first and third passage stimulated with LPS (MilliporeSigma), and
the cells were treated with 0.25% trypsin (Invitrogen; Thermo
Fisher Scientific, Inc.) for 1 min at 37˚C between
passages. LPS was dissolved in ultrapure water at a stock
concentration of 5 mg/ml according to the manufacturer's
instructions. For LPS stimulation, chondrocytes were incubated in
serum-free DMEM containing LPS at 1, 5 and 10 µg/ml for 48 h at
37˚C. The blank group was incubated in serum-free DMEM
without LPS.
Cell Counting Kit (CCK)-8 assay
The viability of the chondrocytes was determined
using CCK-8 kit (Dojindo Molecular Technologies, Inc.). In brief,
the chondrocytes were seeded into 96-well plates (Corning, Inc.) at
a density of 1x104 cells/well for 24 h, and subsequently
treated with 0, 1, 5 and 10 µg/ml LPS for 48 h as aforementioned.
The cells were cultured with 20 µl CCK-8 solution (5 g/l) in PBS
for another 2 h, and the optical density at 450 nm was measured on
a microplate reader. Each group was assayed in triplicate.
Flow cytometry
For cell apoptosis analysis, LPS-treated
chondrocytes were seeded into 6-well plates (Corning, Inc.) at a
density of 1x105 cells/well for 24 h and were
subsequently analyzed with flow cytometry using an Annexin V-FITC
apoptosis detection kit (Beyotime Institute of Biotechnology). The
apoptotic cells were labelled according to the manufacturer's
protocol. In brief, the adherent and floating cells were harvested
and washed twice with PBS three times. Subsequently, 100 µl cells
of each group were stained in a binding buffer containing Annexin
V-FITC and PI for 30 min at 4˚C in the dark.
Fluorescence was analyzed with CytoFLEX LX flow cytometer (Beckman
Coulter, Inc.) using CytExpert software (version 2.0; Beckman
Coulter, Inc.). The blank group was treated with 0 µg/ml LPS, and
the control groups were transfected with miR-NC mimic or
co-transfected with miR-200b mimic and pcDNA. For cell surface
marker detection, cartilage cells on passage 1 were collected by
0.25% trypsin (Invitrogen; Thermo Fisher Scientific, Inc.) for 1
min at 37˚C, and washed with PBS three times; 1x107
cells were resuspended in the binding buffer, followed by
incubation with monoclonal antibodies against CD44 (cat. no.
AF0105; 1:100; Beyotime Institute of Biotechnology) and CD151 (cat.
no. FAB1884P; 1:100; R&D Systems China Co., Ltd.) conjugated to
FITC or phycoerythrin for 30 min at room temperature in the dark.
After washing with PBS once, these cells were analyzed with a
FACScan flow cytometer (Becton-Dickinson and Company), using
CellQuest software (version 5.1; Becton-Dickinson and Company). The
results were obtained from three independent experiments.
ELISA
ELISA was performed to measure the concentration of
IL-1β, IL-6 and TNF-α that were released in the culture
supernatants. Chondrocytes without and with transfection were
seeded into 24-well plates (Corning, Inc.) for 24 h. After LPS (0,
1, 5 and 10 µg/ml) stimulation, the culture supernatants were
collected to measure the inflammatory factor levels according to
the manufacturer's instructions of each kit. The ELISA kits used
were as follows: Human IL-1β (cat. no. ab100562), human IL-6 (cat.
no. ab46027) and human TNF-α ELISA kit (cat. no. ab46087), all from
Abcam. Three independent experiments in triplicate were
performed.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA from OA cartilages and LPS-treated
chondrocytes was isolated using TRIzol® reagent (Thermo
Fisher Scientific, Inc.). Briefly, 300 ng total RNA was used to
synthesize cDNA at 42˚C for 15 min using All-in-one
MasterMix (Applied Biological Materials Inc.) for mRNA and
Bestar™ qPCR RT kit for miRNA (DBI Bioscience). The
amplification of cDNA was performed using SYBR® Premix
Ex Taq Master Mix (Takara Bio, Inc.) or Bestar® SYBR
Green qPCR Master Mix (DBI Bioscience) on the ABI PRISM 7500
Real-time PCR System (Applied Biosystems; Thermo Fisher Scientific,
Inc.). The thermocycling conditions were 40 cycles of
95˚C for 15 sec, 60˚C for 60 sec and
95˚C for 15 sec. RNA expression was calculated using the
2-ΔΔCq method (27). All
primers were synthesized by Shanghai GenePharma Co., Ltd. and are
listed in Table I. All experiments
were performed at least in triplicate. The data are presented as
fold change relative to control levels. GAPDH (for mRNA) or U6 (for
miRNA) were used for normalization.
 | Table ISequences of primers used in reverse
transcription-quantitative PCR. |
Table I
Sequences of primers used in reverse
transcription-quantitative PCR.
| Gene | Primer sequence
(5'-3') |
|---|
| miR-200b | F:
GCTGCTGAATTCCATCTAATTTCCAAAAG |
| | R:
TATTATGGATCCGCCCCCAGGGCAATGGG |
| FUT4 | F:
TCCTACGGAGAGGCTCAG |
| | R:
TCCTCGTAGTCCAACACG |
| U6 | F:
AACGCTTCACGAATTTGCGT |
| | R:
CTCGCTTCGGCAGCACA |
| GAPDH | F:
GTCAACGGATTTGGTCTGTATT |
| | R:
AGTCTTCTGGGTGGCAGTGAT |
Cell transfection
The primary chondrocyte were seeded into six-well
plates (Corning, Inc.) at a density of 5x104 cells/well
for 24 h prior to transfection, and 293T cells were pre-seeded in
24-well plate at a density of 5x103 cells/well. For
overexpression purposes, the coding domain sequence of FUT4 was
inserted into pcDNA3.1 vector (Invitrogen; Thermo Fisher
Scientific, Inc.), and equal amount of empty vector was used as
negative control. miR-200b mimic and its negative control miR-NC
mimic were purchased from Shanghai GenePharma Co., Ltd.. For
knockdown purposes, anti-miR-200b and anti-miR-NC were provided by
Shanghai GenePharma Co., Ltd.. Cell transfection was performed
using Lipofectamine® 3000 reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) in serum-free DMEM at 37˚C for
6 h, and the culture medium was replaced with complete cell culture
DMEM medium for 48 h prior to further analysis. Briefly, 2 µg
vectors and 30 nM oligonucleotides were transfected into cells in
six-well plate, and 20 ng vectors and 20 nM oligonucleotides were
transfected in a 24-well plate. After transfection for 48 h,
chondrocytes were treated with 0 or 10 µg/ml LPS at 37˚C
for another 48 h.
Dual-luciferase reporter assay
Prediction of potential targets was performed using
TargetScan software (version 7.2; http://www.targetscan.org/vert_72/). The putative
binding sites of hsa-miR-200b in the 3'-untranslated region of FUT4
(FUT4 3'-UTR) were mutated (FUT4-MUT). Subsequently, the wild-type
FUT4 3'-UTR (FUT4-WT) and FUT4-MUT were cloned into pMIR-Luciferase
Reporter Vector (Invitrogen; Thermo Fisher Scientific, Inc.). 293T
cells (cat. no. CRL-3216; American Type Culture Collection;
1x104 cells/well) in the logarithmic growth phase were
plated in 24-well plates (Corning, Inc.), followed by
co-transfection with 20 nM miR-200b/NC mimic and 20 ng FUT4-WT/MUT
using Lipofectamine® 3000 reagent (Invitrogen; Thermo
Fisher Scientific, Inc.). After transfection for 48 h, the cells
were collected to measure the relative luciferase activity using
the Dual-Luciferase® Reporter Assay System (Promega
Corporation) with comparison with Renilla luciferase. All
transfections were performed in triplicate.
RNA immunoprecipitation (RIP)
Magna RIP™ RNA-binding protein
immunoprecipitation kit (EMD Millipore; cat. no. 17-700) was used
for RIP assay in chondrocyte extracts after transfection with
miR-200b/NC mimic. Immunoprecipitation of mRNAs were bound to 5 µl
argonaute-2 (Ago2; cat. no. 03-110; EMD Millipore) or IgG (control;
03-198; EMD Millipore) antibody. The immunoprecipitated mRNAs were
isolated using TRIzol reagent (Thermo Fisher Scientific, Inc.) and
FUT4 mRNA expression was measured via RT-qPCR. All procedures were
performed according to the manufacturer's protocol.
Western blotting
Total protein from LPS-treated chondrocytes was
extracted using RIPA lysis buffer (Beyotime Institute of
Biotechnology), and protein concentration was determined using a
BCA Protein Assay kit (Pierce; Thermo Fisher Scientific, Inc.). A
total of 20 µg of proteins/lane were resolved on SDS-PAGE (12%
gel), and transferred onto PVDF membranes (EMD Millipore). The
membranes were blocked with 5% nonfat milk for 1 h at
25˚C, and subsequently incubated with primary antibodies
overnight at 4˚C. The following primary antibodies were
supplied by Abcam: Anti-FUT4 (1:500; cat. no. ab181461) and
anti-β-actin (1:1,000; cat. no. ab8227). After incubation with
HRP-conjugated secondary antibodies against mouse (cat. no. A0216;
1:1,000; Beyotime Institute of Biotechnology) or rabbit (cat. no.
A0208; 1:1,000; Beyotime Institute of Biotechnology) at room
temperature for 1.5 h, the bands were visualized using an ECL kit
(EMD Millipore) according to standard protocols. β-actin was used
as an internal standard to normalize protein levels analyzed on
Image-Pro Plus software (version 6.0; Media Cybernetics, Inc.).
Western blotting of the same proteins was performed in
triplicate.
Statistical analysis
Data are presented as the mean ± SD from three
independent experiments. Statistical significance was determined
using independent Student's t-test for comparisons between two
groups and one-way ANOVA followed by Tukey's post hoc test for
comparisons among multiple groups. Data analysis was performed
using GraphPad Prism v6.0 (GraphPad Software Inc.). P<0.05 was
considered to indicate a statistically significant difference.
Results
LPS-induced inflammatory injury in
knee articular chondrocytes in vitro
In the present study, patients with OA and control
patients were recruited for knee cartilage tissue isolation. OA
cartilages were used for the separation of OA chondrocytes
(Fig. S1), while normal
chondrocytes were isolated from normal cartilages, followed by LPS
treatment for the establishment of the OA model. Firstly, the
cultured cells from normal cartilages were analyzed using surface
markers of chondrocytes (CD44 and CD151) via flow cytometry. The
results indicated that 83.2% of the cells were
CD44+/CD151+ (Fig. S2), suggesting that the majority of
the cells were chondrocytes. Subsequently, a cell model of
LPS-induced chondrocyte injury in vitro was established and
verified. Chondrocytes were exposed to LPS, and cell behaviors were
measured. As presented in Fig. 1A,
5 and 10 µg/ml LPS induced a significant decrease in cell viability
compared with control cells, while the apoptosis rate was increased
from 6.9 to 16.7% (5 µg/ml LPS) and 26.3% (10 µg/ml LPS) (Fig. 1B and C). The release of IL-1β, IL-6 and TNF-α
was gradually upregulated by LPS in a concentration-dependent
manner (Fig. 1D-F). Taken together,
these results indicated that LPS reduced cell viability, but
promoted apoptosis and pro-inflammatory factor secretion in
chondrocytes in a dose-dependent manner, suggesting that LPS may
mimic the inflammatory injury of OA in knee articular chondrocytes
in vitro. Moreover, OA chondrocytes isolated from patients
with OA also exhibited increased apoptosis and inflammatory
response compared with control chondrocytes (Fig. S3A-E).
miR-200b expression is downregulated
in OA cartilage and LPS-treated chondrocytes
miR-200b expression was detected using RT-qPCR, and
it was revealed that the level of miR-200b was lower in cartilage
samples isolated from patients with OA compared with that in normal
patient samples (Fig. 2A).
Consistently, LPS treatment altered miR-200b expression level in
chondrocytes, as 5 and 10 µg/ml LPS resulted in a significant
decrease in miR-200b level compared with that in the blank group (0
µg/ml LPS; Fig. 2B). These data
indicated that miR-200b expression was downregulated in patients
with OA and LPS-stimulated chondrocytes.
FUT4 expression is directly modulated
by miR-200b
Recently, FUT4 has been reported to be aberrantly
upregulated in OA cartilage tissues (19), therefore we hypothesized that FUT4
may serve an important role in knee articular chondrocytes. FUT4
was predicted by TargetScan to be a potential target of miR-200b,
and the potential binding sites are presented in Fig. 3A. To examine this hypothesis,
dual-luciferase reporter assay and RIP assay were subsequently
conducted in miRNA mimic-transfected 293T cells and primary
chondrocytes, respectively. Transfection of miR-200b mimic resulted
in overexpression of miR-200b in 293T cells (Fig. S4A), which significantly reduced the
luciferase activity of FUT4-WT, but not that of FUT4-MUT compared
with miR-NC (Fig. 3B). In
chondrocytes, miR-200b mimic also induced higher expression levels
of miR-200b compared with miR-NC (Fig.
S4B), which resulted in an enrichment of FUT4 mRNA in RIP-Ago2
immunoprecipitated complexes (Fig.
3C). Moreover, following co-transfection of miR-200b mimic with
either FUT4-WT or FUT4-MUT, miR-200b expression was highly induced,
and FUT4 mRNA expression level was reduced compared with
miR-NC-transfected cells (Fig. S4C
and D). Therefore, these results
suggested a direct binding relationship between miR-200b and
FUT4.
 | Figure 3FUT4 is negatively regulated by
miR-200b via direct binding. (A) The putative binding sites of
miR-200b in FUT4 3'-UTR were predicted, and luciferase reporter
vectors containing the FUT4-WT or FUT4-MUT were constructed. (B)
Luciferase assay in 293T cells co-transfected with FUT4-WT or
FUT4-MUT and miR-200b mimic or miR-NC. (C) Reverse
transcription-quantitative PCR following RIP assay was used to
detect the mRNA expression level of FUT4 in RIP-Ago2 and
IgG-RIP-IgG in chondrocytes transfected with miR-200b or miR-NC.
*P<0.05 vs. miR-NC. miR-200b, microRNA-200b-3p; FUT4,
fucosyltransferase 4; NC, negative control; RIP, RNA
immunoprecipitation; WT, wild-type, MUT, mutant; UTR, untranslated
region; Ago2, argonaute-2. |
FUT4 is upregulated in injured
cartilage tissues and chondrocytes, and modulated by miR-200b
Subsequently, the expression of FUT4 was examined
using both RT-qPCR and western blotting. The expression of FUT4 at
the mRNA (Fig. 4A) and protein
level (Fig. 4B) was upregulated in
patients with OA compared with control patients, as well as in
LPS-treated chondrocytes compared with control cells (Fig. 4C and D). Moreover, overexpression of miR-200b
via mimic transfection reduced and knockdown of miR-200b via
anti-miR-200b transfection increased the FUT4 protein expression
level in chondrocytes (Fig. 4E and
F, respectively). The transfection
efficiency of miR-200b mimic and anti-miR-200b is presented in
Fig. S4B. These results
demonstrated that FUT4 was highly expressed in injured cartilage
tissues and chondrocytes and it was negatively modulated by
miR-200b.
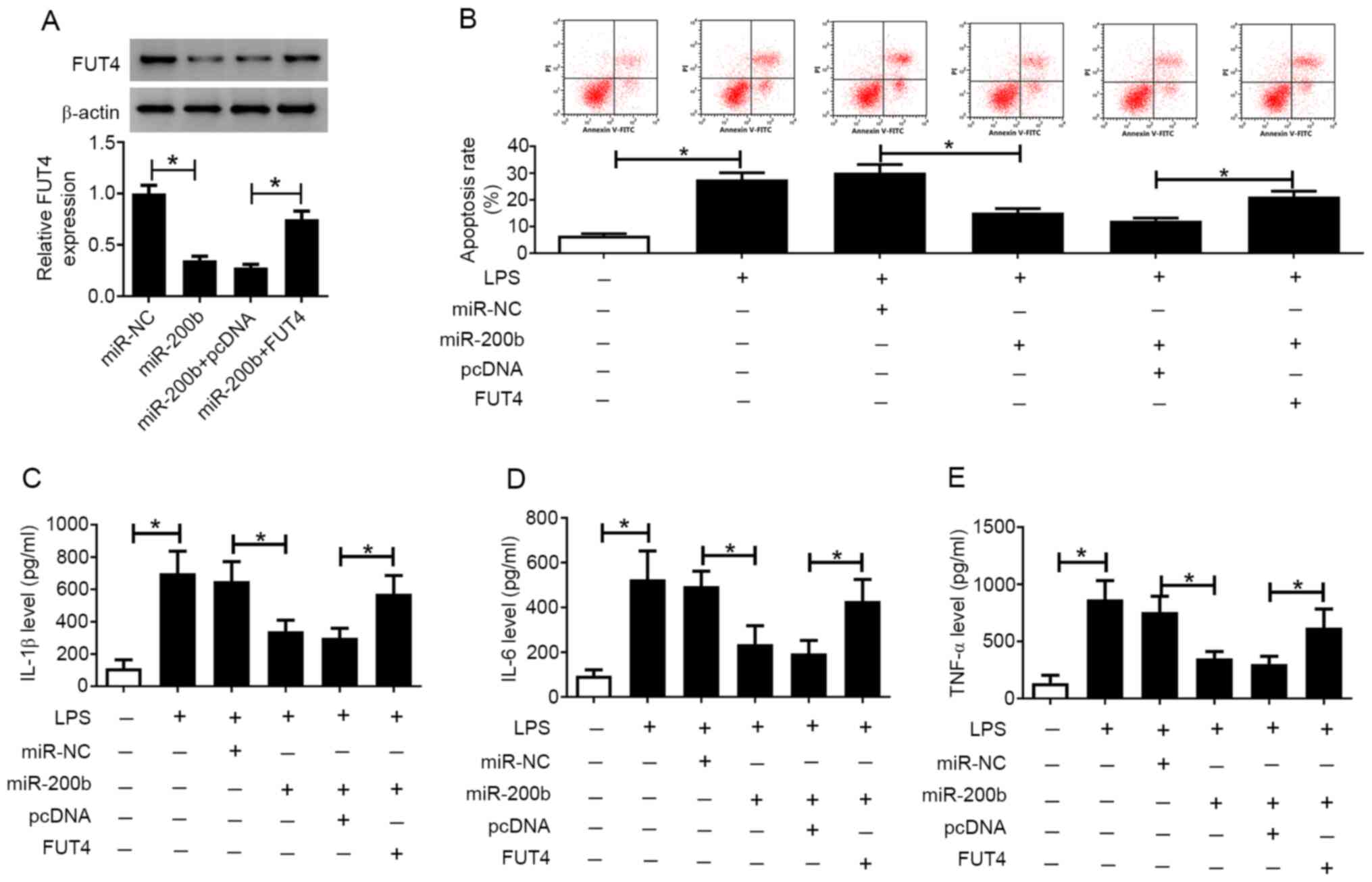
Restoring FUT4 levels partially
reverses the miR-200b-induced protective effect in LPS-induced
inflammatory injury in chondrocytes in vitro
The roles of miR-200b and FUT4 in the LPS-induced OA
model in chondrocytes were explored. The pcDNA-FUT4 vector was used
to overexpress FUT4 in chondrocytes, and chondrocytes were
transfected with miR-200b mimic alone or together with pcDNA-FUT4
vector or empty vector. FUT4 mRNA level was significantly
upregulated in cells transfected with pcDNA-FUT4 compared with
those transfected with the empty vector (Fig. S4E). The downregulation of FUT4
induced by miR-200b overexpression was attenuated by transfection
with pcDNA-FUT4 (Fig. 5A). After
cell treatment with 10 µg/ml LPS for 48 h, the apoptosis rate was
decreased from 30.5 to 16.9% in chondrocytes transfected with
miR-200b mimic compared with those transfected with miR-NC, while
it was increased to 22.8% in cells transfected with pcDNA-FUT4
(Fig. 5B). Moreover, the high level
of IL-1β, IL-6 and TNF-α in LPS-treated chondrocytes was decreased
by miR-200b mimic transfection, while this suppression was reversed
following FUT4 overexpression (Fig.
5C-E). These data demonstrated that miR-200b protected
chondrocytes from LPS-induced inflammatory injury in vitro
by inhibiting FUT4.
Discussion
OA is a degenerative joint disease and its effective
treatments primarily include the relief of pain and inflammation
(28). One key pathological feature
of OA is articular cartilage degeneration, which is largely
attributed to the apoptosis of chondrocytes (29). Statistics have revealed that ~10% of
the population suffer from OA, especially the elderly (30). At present, the available treatments
are often accompanied by severe side effects. Therefore, there is a
requirement to develop therapies for OA by identifying novel target
genes using patients with OA tissues and cells, as well as multiple
stimuli-induced OA models. In the present study, chondrocytes were
isolated from healthy patients and patients with OA, and normal
chondrocytes were subjected to LPS stimulation.
LPS-induced inflammatory injury in chondrocytes has
been widely studied (23,25). LPS has been reported to result in
multiple organ damage by stimulating pro-inflammatory cytokine
secretion and inflammation-related signaling pathways (23). For example, Jia et al
(4) revealed that LPS stimulation
effectively attenuated cell viability, and increased apoptosis and
the production of IL-1β, IL-6 and TNF-α in knee articular
chondrocytes ex vivo compared with LPS-untreated
chondrocytes. Emerging evidence has indicated the involvement of
non-coding RNAs, including miRNAs and long non-coding RNAs in ECM
degeneration and chondrocyte reduction in LPS-stimulated
chondrogenic ATDC5 cells (31,32).
For instance, overexpression of miR-125b was revealed to inhibit
the LPS-induced inflammatory injury by suppressing MIP-1α
expression and NF-κB and JNK signaling activation (4). On the contrary, high expression of
miR-146a has been indicated to aggravate the LPS-induced
inflammatory injury by downregulating CXCR4 and inhibiting the
PI3K/AKT and Wnt/β-catenin signaling pathways (24). In the present study, the expression
levels and the role of miR-200b in an OA model of inflammatory
injury were investigated. The results indicated that miR-200b was
downregulated in OA cartilage tissues and LPS-stimulated
chondrocytes, which was in accordance with the finding of Wu et
al (16). They proposed that
miR-200b might function as a repair factor for OA cartilage,
because its upregulation suppressed the degradation of ECM in OA
cartilage cells ex vivo, as indicated by the lower MMP and
higher collagen II levels in miR-200b mimic-transfected cells
compared with negative control-transfected cells (16). Similarly, the present study
demonstrated a suppressive role of miR-200b in apoptosis and the
inflammatory response in LPS-treated chondrocytes in vitro.
However, the role of miR-200b in ECM degradation requires further
investigation.
miR-200b is considered to function as a regulatory
factor of cell proliferation and motility in a number of types of
cancer. For example, the epithelial-mesenchymal transition and
tumor growth of glioma were suppressed by miR-200b overexpression
via downregulating ERK5 expression (33). Moreover, miR-200b downregulation by
low p73 has been indicated to promote androgen independence in
prostate cancer cells (34).
Nevertheless, few studies have investigated the potential functions
of miR-200b in OA pathogenesis. Additional evidence is required to
verify the role of miR-200b in vivo.
In the present study, overexpression of FUT4 was
revealed to inhibit the miR-200-mediated protective effect in knee
articular chondrocyte injury in vitro. DNA
(cytosine-5)-methyltransferase 3A and FUT4 have been identified as
downstream target genes of miR-200b (16). FUT4 belongs to the
fucosyltransferase family, which is a group of fucosylation
synthases (21). It has been
reported that FUTs are associated with signal transduction,
inflammation, tumor progression and metastasis. For example, FUTs
have been reported to mediate multidrug resistance in human
hepatocellular carcinoma via the PI3K/AKT signaling pathway
(18). In arthritis, FUT1 and FUT7
were upregulated in RA synovial fibroblast cells and fluid,
respectively (19,20). FUT1 and FUT2 have been demonstrated
to mediate angiogenesis and inflammatory cell adhesion in RA.
However, to the best of our knowledge, FUTs in OA have not been
widely studied with the exception of one study. According to the
results of Hu et al (21),
the expression profile of FUT genes in healthy and OA human
cartilage tissues was unraveled, and the expression level of FUT4,
FUT1, FUT2 and FUT3 was notably increased in OA tissues. According
to the findings of the present study, FUT4 expression at the mRNA
and protein level was increased in OA cartilage tissues and
LPS-treated chondrocytes compared with control tissues and cells,
respectively. Functionally, overexpression of FUT4
attenuated the miR-200b-induced suppressive effects on the
apoptosis rate and the expression level of IL-1β, IL-6 and TNF-α in
LPS-treated chondrocytes.
In consideration of the protective role of miR-200b
overexpression in injured chondrocytes, including OA chondrocytes
ex vivo (16) and
LPS-stimulated chondrocytes in vitro (from the results of
the current study), miR-200b may be a cartilage repair gene in OA.
The findings of the current study suggested that miR-200b may be a
potential candidate to improve chondrocyte viability in knee OA by
affecting apoptosis and inflammatory response of injured
chondrocytes, as well as ECM degeneration, which is indicated by a
previous study (35). In the
future, additional studies should be performed to explore the
signaling pathways of the miR-200b/FUT4 axis involved in the
progression of chondrocyte injury, such as the NF-κB (21) and MAPK/ERK (16) pathway.
Collectively, the present study demonstrated that
miR-200b was downregulated in cartilage tissues from patients with
OA, and upregulating miR-200b attenuated LPS-induced apoptosis and
inflammatory response in knee articular chondrocytes in
vitro via targeting FUT4.
Supplementary Material
Flow chart of the present study. FUT4,
fucosyltransferase 4; miR-200b, microRNA-200b-3p; LPS,
lipopolysaccharide.
Expression of surface markers in
chondrocytes. CD44 and CD151 antibodies were conjugated FITC or to
phycoerythrin. The percentage of CD44+/CD151+
cells was calculated. *P<0.05.
Inflammatory injury in knee articular
chondrocytes isolated from patients with OA. Knee articular
chondrocytes were isolated from tissue samples of patients with OA
and normal controls. (A and B) The apoptosis rate was determined
using flow cytometry. The production of (C) IL-1β, (D) IL-6 and (E)
TNF-α was evaluated by ELISA. *P<0.05 vs. control.
OA, osteoarthritis.
Expression of miR-200b and FUT4 in
transfected cells. RT-qPCR was used to detect miR-200b expression
level in (A) 293T cells transfected with miR-200b or miR-NC, (B)
Chondrocytes transfected with miR-NC, miR-200b, anti-miR-NC or
anti-miR-200b and (C) 293T cells co-transfected with miR-200b/NC
and FUT4-WT/MUT. RT-qPCR was used to detect FUT4 mRNA expression
level in (D) 293T cells co-transfected with miR-200b/NC and
FUT4-WT/MUT and (E) Chondrocytes transfected with pcDNA-FUT4 or
pcDNA. *P<0.05 vs. the respective control group.
FUT4, fucosyltransferase 4; NC, negative control; RT-qPCR, reverse
transcription-quantitative PCR; miR-200b, microRNA-200b-3p; WT,
wild-type; MUT, mutant.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
SW and ZZ conceived the study, designed the
methodology and performed the final analysis. YL and SW performed
the experiments, data curation, validation and investigation. ZZ
performed GraphPad software analysis. YL wrote the original draft.
SW reviewed and edited the manuscript. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
The present study was conducted in accordance with
the Declaration of Helsinki and was approved by the Ethics
Committee of Yan'an Peoples Hospital (Yan'an, China; February 15,
2019). Written informed consent was obtained from each patient.
Patient consent for publication
Written informed consent for publication was
obtained from each patient.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
van den Bosch MHJ: Inflammation in
osteoarthritis: Is it time to dampen the alarm(in) in this
debilitating disease? Clin Exp Immunol. 195:153–166.
2019.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Loeser RF, Collins JA and Diekman BO:
Ageing and the pathogenesis of osteoarthritis. Nat Rev Rheumatol.
12:412–420. 2016.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Greene MA and Loeser RF: Aging-related
inflammation in osteoarthritis. Osteoarthritis Cartilage.
23:1966–1971. 2015.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Jia J, Wang J, Zhang J, Cui M, Sun X, Li Q
and Zhao B: MiR-125b inhibits LPS-induced inflammatory injury via
targeting MIP-1α in chondrogenic cell ATDC5. Cell Physiol Biochem.
45:2305–2316. 2018.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Kourtis A, Adamopoulos PG, Papalois A,
Iliopoulos DC, Babis GC and Scorilas A: Quantitative analysis and
study of the mRNA expression levels of apoptotic genes BCL2, BAX
and BCL2L12 in the articular cartilage of an animal model of
osteoarthritis. Ann Transl Med. 6(243)2018.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Chen Y, Sun Y, Pan X, Ho K and Li G: Joint
distraction attenuates osteoarthritis by reducing secondary
inflammation, cartilage degeneration and subchondral bone aberrant
change. Osteoarthritis Cartilage. 23:1728–1735. 2015.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Cong L, Zhu Y and Tu G: A bioinformatic
analysis of microRNAs role in osteoarthritis. Osteoarthritis
Cartilage. 25:1362–1371. 2017.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Jeffries MA, Donica M, Baker LW, Stevenson
ME, Annan AC, Humphrey MB, James JA and Sawalha AH: Genome-wide DNA
methylation study identifies significant epigenomic changes in
osteoarthritic cartilage. Arthritis Rheumatol. 66:2804–2815.
2014.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Li B, Bai L, Shen P, Sun Y, Chen Z and Wen
Y: Identification of differentially expressed microRNAs in knee
anterior cruciate ligament tissues surgically removed from patients
with osteoarthritis. Int J Mol Med. 40:1105–1113. 2017.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Song J, Ahn C, Chun CH and Jin EJ: A long
non-coding RNA, GAS5, plays a critical role in the regulation of
miR-21 during osteoarthritis. J Orthop Res. 32:1628–1635.
2014.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Yang R, Zhang D, Yu K, Sun L, Yang J, Zhao
C, Li X and Chen Y: Detection of miR-22, miR-140 and bone
morphogenetic proteins (BMP)-2 expression levels in synovial fluid
of osteoarthritis patients before and after arthroscopic
debridement. Med Sci Monit. 24:863–868. 2018.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Zhang HX, Sun C, Yu HC, Song B and Pan ZX:
Targeted inhibition of β-catenin by miR-320 and decreased MMP-13
expression in suppressing chondrocyte collagen degradation. Eur Rev
Med Pharmacol Sci. 22:5828–5835. 2018.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Feng B, Wang R and Chen L: Review of
miR-200b and cancer chemosensitivity. Biomed Pharmacother.
66:397–402. 2012.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Zeng F, Xue M, Xiao T, Li Y, Xiao S, Jiang
B and Ren C: MiR-200b promotes the cell proliferation and
metastasis of cervical cancer by inhibiting FOXG1. Biomed
Pharmacother. 79:294–301. 2016.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Zheng Q, Cui X, Zhang D, Yang Y, Yan X,
Liu M, Niang B, Aziz F, Liu S, Yan Q and Liu J: miR-200b inhibits
proliferation and metastasis of breast cancer by targeting
fucosyltransferase IV and α1,3-fucosylated glycans. Oncogenesis.
6(e358)2017.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Wu J, Tao Y, Shang A, Wang W, Zhang Y, Hu
L, Wang J, Wang Y and Guo N: Effect of the interaction between
MiR-200b-3p and DNMT3A on cartilage cells of osteoarthritis
patients. J Cell Mol Med. 21:2308–2316. 2017.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Keeley TS, Yang S and Lau E: The diverse
contributions of fucose linkages in cancer. Cancers (Basel).
11(1241)2019.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Cheng L, Luo S, Jin C, Ma H, Zhou H and
Jia L: FUT family mediates the multidrug resistance of human
hepatocellular carcinoma via the PI3K/Akt signaling pathway. Cell
Death Dis. 4(e923)2013.PubMed/NCBI View Article : Google Scholar
|
|
19
|
De Benedetti F, Pignatti P, Biffi M, Bono
E, Wahid S, Ingegnoli F, Chang SY, Alexander H, Massa M, Pistorio
A, et al: Increased expression of alpha(1,3)-fucosyltransferase-VII
and P-selectin binding of synovial fluid T cells in juvenile
idiopathic arthritis. J Rheumatol. 30:1611–1615. 2003.PubMed/NCBI
|
|
20
|
Isozaki T, Ruth JH, Amin MA, Campbell PL,
Tsou PS, Ha CM, Haines GK, Edhayan G and Koch AE:
Fucosyltransferase 1 mediates angiogenesis, cell adhesion and
rheumatoid arthritis synovial tissue fibroblast proliferation.
Arthritis Res Ther. 16(R28)2014.PubMed/NCBI View
Article : Google Scholar
|
|
21
|
Hu J, Wang Z, Pan Y, Ma J, Miao X, Qi X,
Zhou H and Jia L: MiR-26a and miR-26b mediate osteoarthritis
progression by targeting FUT4 via NF-κB signaling pathway. Int J
Biochem Cell Biol. 94:79–88. 2018.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Chaby R: Lipopolysaccharide-binding
molecules: Transporters, blockers and sensors. Cell Mol Life Sci.
61:1697–1713. 2004.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Huang ZY, Stabler T, Pei FX and Kraus VB:
Both systemic and local lipopolysaccharide (LPS) burden are
associated with knee OA severity and inflammation. Osteoarthritis
Cartilage. 24:1769–1775. 2016.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Sun T, Li X, Song H, Gao F, Zhou G, Li X,
Chen Z and Chen L: MiR-146a aggravates LPS-induced inflammatory
injury by targeting CXCR4 in the articular chondrocytes. Cell
Physiol Biochem. 44:1282–1294. 2017.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Wang Y and Kong D: MicroRNA-136 promotes
lipopolysaccharide-induced ATDC5 cell injury and inflammatory
cytokine expression by targeting myeloid cell leukemia 1. J Cell
Biochem. 119:9316–9326. 2018.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Hubertsson J, Petersson IF, Thorstensson
CA and Englund M: Risk of sick leave and disability pension in
working-age women and men with knee osteoarthritis. Ann Rheum Dis.
72:401–405. 2013.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
28
|
van Middelkoop M, Arden NK, Atchia I,
Birrell F, Chao J, Rezende MU, Lambert RG, Ravaud P, Bijlsma JW,
Doherty M, et al: The OA Trial Bank: meta-analysis of individual
patient data from knee and hip osteoarthritis trials show that
patients with severe pain exhibit greater benefit from
intra-articular glucocorticoids. Osteoarthritis Cartilage.
24:1143–1152. 2016.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Hwang HS and Kim HA: Chondrocyte apoptosis
in the pathogenesis of osteoarthritis. Int J Mol Sci.
16:26035–2654. 2015.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Feldmann M: Pathogenesis of arthritis:
Recent research progress. Nat Immunol. 2:771–773. 2001.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Li F, Sun J, Huang S, Su G and Pi G:
LncRNA GAS5 overexpression reverses LPS-induced inflammatory injury
and apoptosis through up-regulating KLF2 expression in ATDC5
chondrocytes. Cell Physiol Biochem. 45:1241–1251. 2018.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Pan L, Liu D, Zhao L, Wang L, Xin M and Li
X: Long noncoding RNA MALAT1 alleviates lipopolysaccharide-induced
inflammatory injury by upregulating microRNA-19b in murine
chondrogenic ATDC5 cells. J Cell Biochem. 119:10165–10175.
2018.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Wu J, Cui H, Zhu Z and Wang L:
MicroRNA-200b-3p suppresses epithelial-mesenchymal transition and
inhibits tumor growth of glioma through down-regulation of ERK5.
Biochem Biophys Res Commun. 478:1158–1164. 2016.PubMed/NCBI View Article : Google Scholar
|
|
34
|
He M, Liu Y, Deng X, Qi S, Sun X, Liu G,
Liu Y, Liu Y and Zhao M: Down-regulation of miR-200b-3p by low p73
contributes to the androgen-independence of prostate cancer cells.
Prostate. 73:1048–1056. 2013.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Gwam CU, Etcheson JI, George NE, Mistry
JB, Mohamed N, Patel A, Gwam PN, Piuzzi NS and Delanois RE:
Presentation of knee osteoarthritis in the emergency department: A
problem worth mentioning? Surg Technol Int. 31:277–284.
2017.PubMed/NCBI
|