Introduction
Osteosarcoma (OS) is a common malignant bone cancer
with high aggressiveness and rapid systemic metastasis (1). OS is usually formed in the bones of
children and adolescents, with more male than female patients
(2). Furthermore, the incidence of
OS is high between the ages of 10 and 19 years, with an annual
incidence of ~10 patients with OS per million worldwide (3). With the development of surgery and
chemotherapy, the 5-year survival rate of patients with OS has
reached 60-70% (4). Thus,
identifying novel therapies for OS is essential.
Long non-coding RNAs (lncRNAs) are a class of
non-coding RNAs of ~200 nucleotides in length that play a critical
role in various physiological and pathological processes, including
growth, development and oncogenesis (5,6).
lncRNAs have been involved in the progression of multiple cancers,
including OS (7). A large number of
lncRNAs, such as HCG9, MELTF-AS1 and ZEB2-AS1 (8-10),
have been found to exert a regulatory effect on cell proliferation,
invasion, migration and apoptosis in OS (11). LncRNA MALAT1 is a novel lncRNA that
contributes to the occurrence and development of various tumors,
including OS (12). Recently, it
has been reported that MALAT1 expression is aberrantly regulated in
various cancers (13). For
instance, previous studies have shown that MALAT1 is upregulated in
OS tissues and cells, and promotes OS cell growth and metastasis by
binding to microRNA (miRNA/miR)-140-5p or regulating the
miR-34a/cyclin D1 axis in OS (14,15).
miRNAs consist of 18-25 nucleotides and are highly
conserved short non-coding RNAs. miRNAs have been identified as
post-transcriptional regulators in biological and pathological
processes (16), including cell
proliferation, apoptosis, migration and invasion (17). In addition, a large number of miRNAs
have been identified to be associated with cancer. miRNAs act as
oncogenes or tumor suppressors by modulating different target mRNAs
(18). Previous studies have shown
that miR-590-3p plays a role in a variety of tumors. For example,
miR-590-3p is a tumor suppressor that inhibits
epithelial-mesenchymal transition (EMT) and metastasis of
glioblastoma multiforme cells by targeting zinc finger
E-box-binding homeobox (ZEB)1 and ZEB2(19). In breast cancer cells, miR-590-3p
suppresses tumor progression by targeting sirtuin 1(20). Non-coding RNAs including MALAT1,
miR-590-3p and TUG1 have been proven to be key regulators of OS
(21). The expression level of
miR-590-3p in OS tissues and cells is significantly downregulated,
and miR-590-3p inhibits the progression of OS by targeting
SOX9(22). However, the
relationship between MALAT1 and miR-590-3p in OS progression
remains unknown.
The present study discovered that MALAT1 contained
some complementary pairing with the seed region of miR-590-3p in
143B and MG-63 cells using bioinformatics analysis. Hence, the
study aimed to illuminate whether the involvement of MALAT1 in OS
function was mediated by miR-590-3p.
Materials and methods
Tissue samples
A total of 45 pairs of OS tissues and matched
adjacent healthy tissues were obtained from patients (30 male
patients and 15 female patients; mean age, 22.7±6.18 years) who
underwent surgery at First Hospital of Lanzhou University (Lanzhou,
China) between June 2018 and March 2021. The inclusion criteria
were as follows: i) No other distant metastatic lesions; ii) none
of the patients received chemotherapy or radiotherapy prior to
surgery; iii) no history of other tumors; iv) patients had complete
clinical data. The exclusion criteria were as follows: i) Patients
were pregnant or lactating; ii) multiple lesions were present at
the first diagnosis; iii) patients had other diseases that would
affect the progression of treatment. Written informed consent was
obtained from all patients, and the research method was approved by
the Ethics Committee of the First Hospital of Lanzhou University
(Lanzhou, China). All tissue samples were immediately treated with
liquid nitrogen and then stored at -80˚C. A total of 100 mg of each
tissue sample was ground in liquid nitrogen for reverse
transcription-quantitative (RT-q)PCR.
Cell culture
Human fetal osteoblastic cells (hFOB1.19) and OS
cell lines (HOS, U2OS, 143B and MG-63) were obtained from American
Type Culture Collection. These cells were cultured in DMEM (Gibco;
Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Gibco;
Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100 mg/ml
streptomycin (Gibco; Thermo Fisher Scientific, Inc.) at 37˚C in a
humidified atmosphere with 5% CO2.
Plasmids and cell transfection
Small interfering RNA (siRNA) against MALAT1
(si-MALAT1: sense strand: 5'-AAGAAAAAUAAAAGCUUUCCU-3', antisense
strand: 5'-GAAAGCUUUUUAUUUUUCUUCC-3') and corresponding negative
control siRNA (Scramble: sense strand: 5'-UUCUCCGAACGUGUCACGUTT
-3', antisense strand: 5'-ACGUGACACGUUCGGAGAATT-3'), miR-590-3p
mimic (miR-590-3p: 5'-TAATTTTATGTATAAGCTAGT-3') and the negative
control (NC: 5'-UUCUCCGAACGUGUCACGUTT-3') mimic, miR-590-3p
inhibitor (anti-miR-590-3p: 5'-ACTAGCTTATACATAAAATTA-3') and the NC
inhibitor (anti-NC: 5'-CAGUACUUUUGUGUAGUACAA-3'), MALAT1
overexpression vector (MALAT1) and the empty pcDNA3.1 vector
(Vector; Invitrogen; Thermo Fisher Scientific, Inc.) were purchased
from Guangzhou RiboBio Co., Ltd. Cell transfection was performed
using Lipofectamine® 2000 reagent (Invitrogen; Thermo
Fisher Scientific, Inc.). When cell confluence reached ~90%, a
mixture of 0.8 µg plasmid or 20 nM oligonucleotides and 2 µl
Lipofectamine was added to each well of the 96-well plate and
incubated in a CO2 incubator at 37˚C, followed by
incubation for 48 h. Meanwhile, untreated cells acted as the
Control group.
RT-qPCR
Total RNA was extracted from tissues and cell lines
(hFOB1.19, and OS cell lines HOS, U2OS, 143B and MG-63) using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). RNA was reversely transcribed into cDNA using the
High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher
Scientific, Inc.) or MicroRNA Reverse Transcription Kit (Thermo
Fisher Scientific, Inc.), according to the manufacturer's
protocols. The expression levels were detected using
SYBR® Premix Ex Taq™ (Takara Biotechnology Co., Ltd.)
and quantified using 2-ΔΔCq method. GAPDH and U6 were
used as internal references. Primer sequences were as follows:
MALAT1-forward (F), 5'-AAAGCAAGGTCTCCCCACAAG-3' and MALAT1-reverse
(R), 5'-GGTCTGTGCTAGATCAAAAGGCA-3'; miR-590-3p-F,
5'-GCGTAATTTTATGTATAAGC-3' and miR-590-3p-R,
5'-GTATCCAGTGCGTGTCGTGGAGT-3'; GAPDH-F,
5'-GACTCCACTCACGGCAAATTCA-3' and GAPDH-R,
5'-TCGCTCCTGGAAGATGGTGAT-3'); U6-F, 5'-CTCGCTTCGGCAGCACA-3' and
U6-R, 5'-AACGCTTCACGAATTTGCGT-3'.
Cell Counting Kit-8 (CCK-8) assay
A total of 2x103 143B and MG-63 cells
were seeded into 96-well plates (Corning, Inc.) after transfection.
A final 10% concentration of the CCK-8 solution (Dojindo
Laboratories, Inc.) was added to each well after incubation for 24,
48 and 72 h. After 2 h incubation at 37˚C, the absorbance was
measured at 450 nm using a microplate reader (Bio-Rad Laboratories,
Inc.).
Cell apoptotic assay
Briefly, 143B and MG-63 cells (1x106)
were digested with trypsin and washed with PBS. After being
resuspended with binding buffer, the cells were stained with an
Annexin V-FITC/PI kit (BD Pharmingen; BD Biosciences) for 15 min at
room temperature in the dark. Then, the apoptotic rate was
determined by the FACScan flow cytometry (BD Biosciences) and
analyzed using FlowJo v.10.1 (FlowJo, LLC).
Transwell assay
For cell migration assay, the transfected 143B and
MG-63 cells (5x104 cell/well) were seeded in serum-free
DMEM and plated into the upper chamber of a 24-well Transwell with
8-µm polycarbonate membrane filters (Corning, Inc.). DMEM containing
10% FBS was added to the lower chamber. Cells were incubated for 24
h in a humidified atmosphere of 5% CO2 at 37˚C. The cells
adhering to the lower surface were fixed using 4% paraformaldehyde
at 37˚C for 20 min, stained with 1% crystal violet at 37˚C for 10
min and counted under a light microscope (200x magnification;
Olympus Corporation) in three random fields. For the invasion
assay, the upper layer of the polycarbonate membrane was coated
with 100 µl of diluted (1:1) Matrigel (BD Biosciences) and
incubated at 37˚C for 4 h to dry into a gel. The invasive cells
were analyzed using the same methods as aforementioned.
Western blot assay
Total protein was extracted from 143B and MG-63
cells using RIPA buffer (Thermo Fisher Scientific, Inc.). Proteins
were quantified using BCA Protein Assay Kit (Pierce; Thermo Fisher
Scientific, Inc.). A total of 50 µg protein was separated by
SDS-PAGE on 10% gels and transferred to PVDF membranes
(MilliporeSigma). The membranes were blocked with 5% skim milk for
2 h at room temperature and then incubated overnight at 4˚C with
primary antibodies against E-cadherin (cat. no. ab40772), Vimentin
(cat. no. ab92547), N-cadherin (cat. no. ab76011), Snail (cat. no.
ab216347) and GAPDH (cat. no. ab9485) (all 1:2,000; all from
Abcam). The membranes were washed twice in TBS-1% Tween 20 and
incubated with horseradish peroxidase-conjugated goat-anti-rabbit
secondary antibody (cat. no. ab97051; 1:100,000; Abcam) for 2 h at
room temperature. Finally, the protein bands were detected using an
ECL system (Pierce; Thermo Fisher Scientific, Inc.), and the
intensity of the target protein was determined by ImageJ software
v1.8.2 (National Institutes of Health). GAPDH was used as an
internal reference.
Luciferase reporter assay
The binding sequences of MALAT1 and miR-590-3p were
predicted using online software starBase v2.0 (https://starbase.sysu.edu.cn/). The sequences of
wild-type (wt) MALAT1 (MALAT1-wt) and mutant (mut) MALAT1
(MALAT1-mut1 and MALAT1-mut2) containing miR-590-3p binding sites
were cloned into pGL3 luciferase reporter vector (Promega
Corporation). Then, pGL3 luciferase reporter vectors were
co-transfected with miR-590-3p or NC into 143B and MG-63 cells
using Lipofectamine 2000. Luciferase activity was examined using
Dual-Luciferase Reporter Assay System (Promega Corporation) at 48 h
after transfection according to the manufacturer's instructions.
The activity of Renilla luciferase was used for
normalization.
RNA immunoprecipitation (RIP)
assay
RIP was performed with EZ-Magna RNA-Binding Protein
Immunoprecipitation Kit (car. no. 17-701; MilliporeSigma) in
accordance with the manufacturer's protocol. Briefly, 143B and
MG-63 cells were transfected as aforementioned with miR-590-3p or
NC, lysed at 4˚C using RIP lysis buffer for 5 min, and then
incubated overnight at 4˚C with magnetic beads conjugated with
argonaute 2 (AGO2; cat. no. ab186733; 1:50; Abcam) antibody. After
purification from AGO2 immunoprecipitation complex, the relative
MALAT1 expression was analyzed by RT-qPCR.
RNA pull-down assay
Biotin-labeled miR-590-3p probe (Bio-miR-590-3p) and
the corresponding control probe (Bio-NC) were purchased from
Guangzhou RiboBio Co., Ltd. In brief, the biotinylated probe at a
final concentration of 100 nM was incubated with 100 µl M-280
Streptavidin Dynabeads (Invitrogen; Thermo Fisher Scientific, Inc.)
at room temperature for 2 h to form probe-coated beads. Then, 143B
and MG-63 cells were transfected with Bio-miR-590-3p or Bio-NC,
according to the aforementioned protocol. Subsequently, the cells
were lysed with Pierce IP lysis buffer (Thermo Fisher Scientific,
Inc.) at 4˚C for 5 min and incubated with probe-coated beads at 4˚C
for 3 h. After RNA isolation, the enrichment of MALAT1 was examined
by RT-qPCR.
Statistical analysis
All data are presented as the mean ± standard
deviation of three independent experiments. Pearson correlation
analysis was used to analyze the association between the expression
levels of MALAT1 and miR-590-3p in OS tissues. Statistical analysis
was performed using unpaired or paired Student's t-test for two
groups and one-way ANOVA followed by Turkey's post hoc test for
multiple groups. Notably, the paired Student's t-test was used to
compare tumor tissues and adjacent non-tumor tissues of the same
patients. Statistical analysis was implemented using GraphPad Prism
7 software (GraphPad Software, Inc.). P<0.05 was considered to
indicate a statistically significant difference.
Results
MALAT1 is upregulated in OS tissues
and cells
To investigate the expression of MALAT1 in OS
tissues and cells, RT-qPCR was performed in OS tissues and cell
lines. The results demonstrated that MALAT1 was significantly
increased in OS tissues compared with adjacent non-tumor tissues
(Fig. 1A). Next, the expression of
MALAT1 was significantly upregulated in OS cells (HOS, U2OS, 143B
and MG-63) compared with human fetal osteoblastic cells (hFOB1.19)
(Fig. 1B). These data suggested
that dysregulation of MALAT1 may be associated with OS.
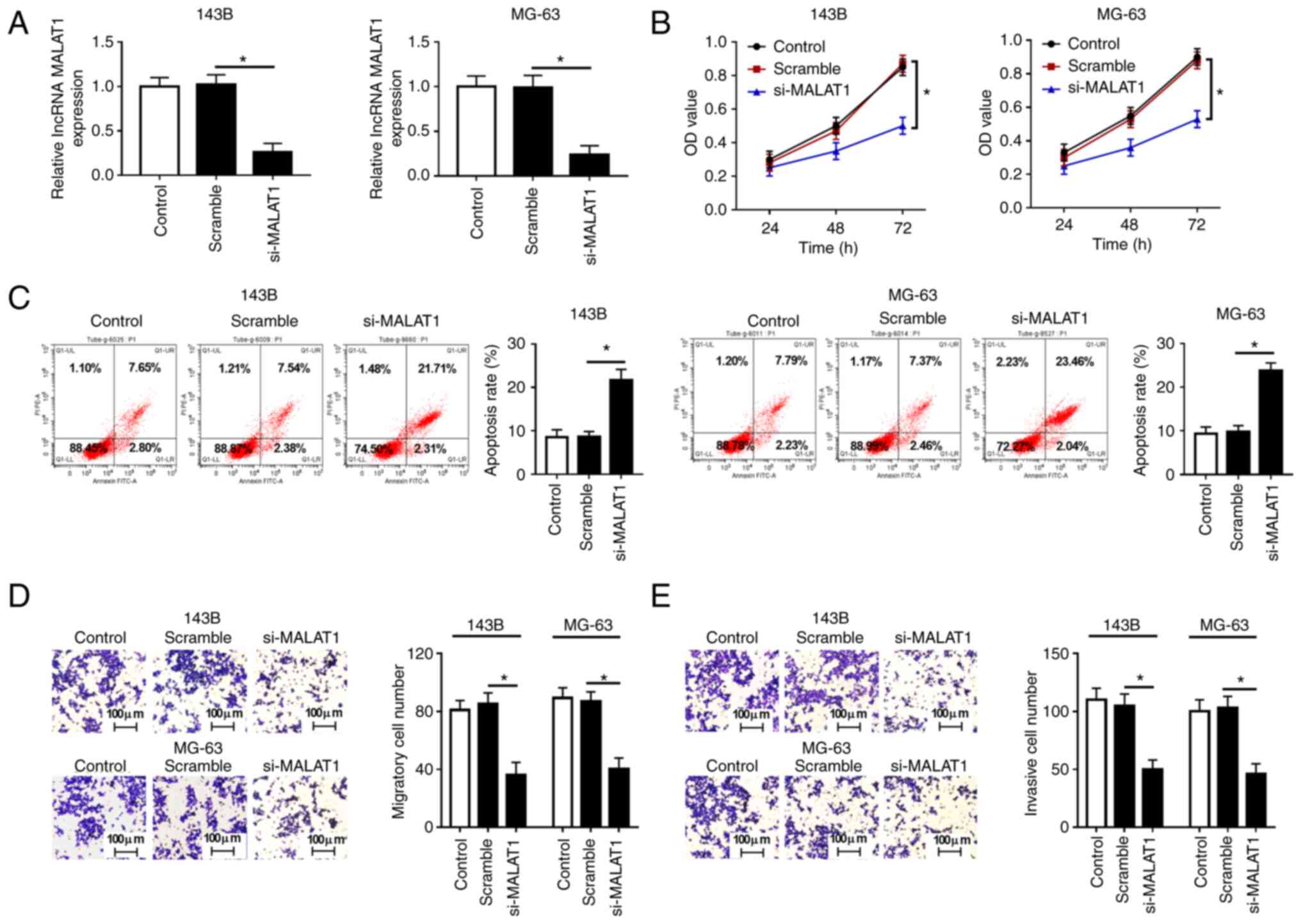
MALAT1 knockdown suppresses
proliferation, migration, invasion and promotes apoptosis in OS
cells
Loss-of-function experiments were performed to
examine the function of MALAT1 in OS cell proliferation and
apoptosis. Since the expression levels of MALAT1 were higher in the
143B and MG-63 cell lines than those in the other OS cell lines
(HOS and U2OS), 143B and MG-63 cells were selected for subsequent
experiments. Firstly, the expression of MALAT1 was detected in 143B
and MG-63 cells transfected with Scramble or si-MALAT1, and the
results demonstrated that transfection with si-MALAT1 significantly
decreased MALAT1 expression compared with Scramble (Fig. 2A). CCK-8 assay showed that MALAT1
knockdown significantly inhibited the proliferation of 143B and
MG-63 cells (Fig. 2B). Flow
cytometry assay revealed that the decrease of MALAT1 increased the
apoptosis rate of 143B and MG-63 cells (Fig. 2C). Furthermore, Transwell assay
demonstrated that MALAT1 knockdown markedly suppressed the
migratory and invasive rate of 143B and MG-63 cells compared with
cells transfected with Scramble (Fig.
2D and E). These results
indicated that MALAT1 knockdown inhibited cell proliferation,
migration and invasion, and induced apoptosis in OS cells.
MALAT1 knockdown inhibits EMT in OS
cells
The effects of MALAT1 on EMT-related markers were
determined using western blot assay. The results revealed that
knockdown of MALAT1 increased the protein level of E-cadherin, and
decreased the protein levels of N-cadherin, Vimentin and Snail in
143B and MG-63 cells transfected with si-MALAT1 compared with the
Scramble group (Fig. 3A and
B), which suggested that low levels
of MALAT1 may inhibit EMT of OS cells.
Overexpression of miR-590-3p inhibits
cell proliferation, migration, invasion and induces apoptosis in OS
cells
Firstly, it was demonstrated that the expression of
miR-590-3p was decreased in OS tissues compared with adjacent
healthy tissues (Fig. 4A), and
miR-590-3p was downregulated in HOS, U2OS, 143B and MG-63 cells
compared with hFOB1.19 cells (Fig.
4B). After transfection of NC or miR-590-3p into 143B and MG-63
cells, the overexpression efficiency of miR-590-3p was determined
by RT-qPCR (Fig. 4C). CCK-8 assay
indicated that the proliferation of 143B and MG-63 cells was
significantly inhibited in the miR-590-3p group compared with the
NC group (Fig. 4D). Flow cytometry
assay demonstrated that the apoptosis rate of both 143B and MG-63
cells transfected with miR-590-3p was increased (Fig. 4E and S1A). Transwell assay revealed that
miR-590-3p overexpression markedly suppressed the migration and
invasion of 143B and MG-63 cells (Fig.
4F and G; Fig. S1C and E). In addition, the protein level of
E-cadherin was increased, and the protein levels of N-cadherin,
Vimentin and Snail were decreased in 143B and MG-63 cells
transfected with miR-590-3p compared with the NC group (Fig. 4H). These data suggested that
miR-590-3p overexpression inhibited cell proliferation, migration,
invasion and EMT, and induced apoptosis in OS cells.
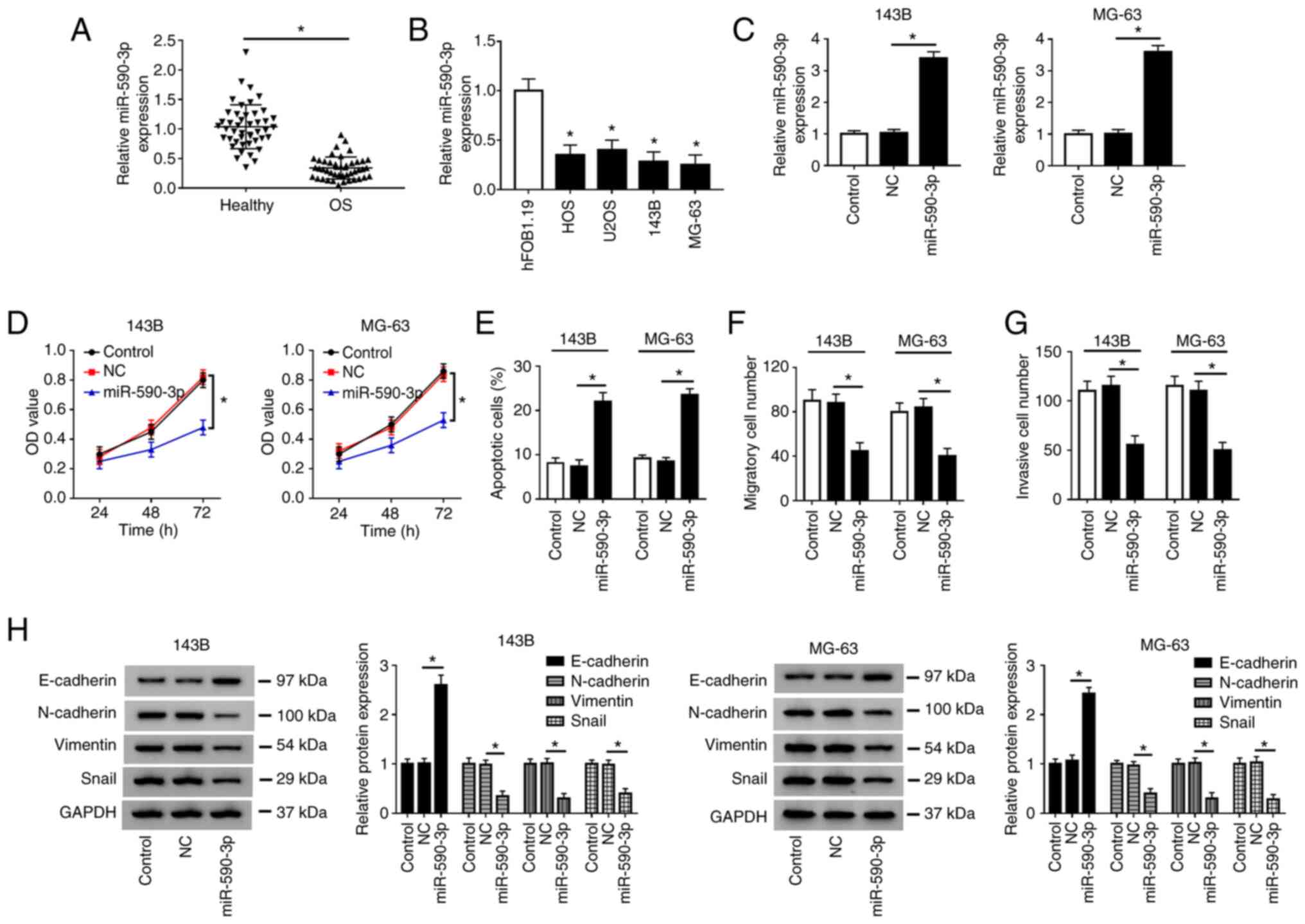 | Figure 4Overexpression of miR-590-3p inhibits
cell proliferation, migration, invasion and induces apoptosis in OS
cells. (A) Expression of miR-590-3p in OS tissues and adjacent
healthy tissues was measured by RT-qPCR. (B) miR-590-3p expression
was detected in OS cells (HOS, U2OS, 143B and MG-63) and human
fetal osteoblastic cells (hFOB1.19). (C) miR-590-3p level was
measured by RT-qPCR in 143B and MG-63 cells transfected with NC or
miR-590-3p. (D) Cell proliferation was evaluated by Cell Counting
Kit-8 assay at 24, 48 and 72 h after transfection. (E) Cell
apoptotic rate was detected by flow cytometry. (F) Cell migration
and (G) invasion were evaluated by Transwell assay. (H) Expression
of epithelial-mesenchymal transition-related proteins (E-cadherin,
N-Cadherin, Vimentin and Snail) was detected by western blotting.
*P<0.05 as indicated or vs. hFOB1.19. RT-qPCR,
reverse transcription-quantitative PCR; OD, optical density; miR,
microRNA; NC, negative control; OS, osteosarcoma. |
MALAT1 directly interacts with
miR-590-3p
Bioinformatics software starBase v2.0 predicted the
binding sites between MALAT1 and miR-590-3p (Fig. 5A). Luciferase reporter assay showed
that miR-590-3p overexpression decreased the luciferase activity of
MALAT1-wt in 143B and MG-63 cells, but failed to inhibit the
luciferase activity of MALAT1-mut1+2 (Fig. 5B). Luciferase activity was also
reduced in the MALAT1-mut1 and MALAT1-mut 2 groups, because both
binding sites are active. Therefore, a single mutation in one of
the binding sites allows miRNAs to bind to the unmutated site.
Then, the specific binding between MALAT1 and miR-590-3p was
validated using RIP and pull-down assays. RIP analysis showed that
MALAT1 was notably enriched in the miR-590-3p group after
precipitation with AGO2 antibody compared with the NC group
(Fig. 5C). In addition, pull-down
analysis revealed that MALAT1 was significantly enriched by binding
to Bio-miR-590-3p, but not Bio-NC (Fig.
5D). Taken together, these results demonstrated that MALAT1
directly bound to miR-590-3p.
 | Figure 5MALAT1 directly interacts with
miR-590-3p. (A) Bioinformatics software predicted the putative
binding sites between MALAT1 and miR-590-3p. (B) 143B and MG-63
cells were co-transfected with MALAT1-wt, MALAT1-mut1, MALAT1-mut2
or MALAT1-mut1+2 and miR-590-3p or NC, and the luciferase activity
was examined at 48 h after transfection. (C) 143B and MG-63 cells
were transfected with miR-590-3p or NC. RNA immunoprecipitation and
RT-qPCR assays were performed to determine MALAT1 enrichment in
AGO2 immunoprecipitation complex. (D) 143B and MG-63 cells were
transfected with Bio-miR-590-3p or Bio-NC. RNA pull-down assay was
carried out to detect MALAT1 enrichment. (E) 143B and MG-63 cells
were transfected with Scramble, si-MALAT1, Vector or MALAT1. The
levels of miR-590-3p were detected by RT-qPCR. (F) The correlation
between MALAT1 and miR-590-3p in OS tissues was examined.
*P<0.05. RT-qPCR, reverse transcription-quantitative
PCR; miR, microRNA; NC, negative control; wt, wild-type; mut,
mutant; si, small interfering; Bio, biotinylated; lncRNA, long
non-coding RNA. |
Next, the overexpression efficiency of MALAT1 was
determined in 143B and MG-63 cells transfected with Vector or
MALAT1 (Fig. S2A). The results
showed that knockdown of MALAT1 markedly increased the expression
of miR-590-3p in 143B and MG-63 cells compared with those in the
Scramble group, whereas overexpression of MALAT1 notably reduced
the expression of miR-590-3p in OS cells compared with in the
control group (Vector) (Fig. 5E).
Furthermore, the expression levels of MALAT1 and miR-590-3p were
negatively correlated in OS tissues (Fig. 5F). These results indicated that
MALAT1 negatively regulated miR-590-3p.
Knockdown of miR-590-3p reverses the
effects of MALAT1 knockdown on the proliferation, apoptosis,
migration, invasion and EMT of OS cells
To further explore the relationship between MALAT1
and miR-590-3p in OS, 143B and MG-63 cells were transfected with
Scramble, si-MALAT1, si-MALAT1 + anti-NC and si-MALAT1 +
anti-miR-590-3p, respectively. Firstly, the knockdown efficiency of
anti-miR-590-3p was detected by RT-qPCR in 143B and MG-63 cells
transfected with anti-NC or anti-miR-590-3p (Fig. S2B). Then, the results indicated
that miR-590-3p was upregulated after knockdown of MALAT1, whereas
miR-590-3p was markedly reduced after transfection with si-MALAT1
and anti-miR-590-3p (Fig. 6A).
Knockdown of MALAT1 significantly suppressed cell proliferation
(Fig. 6B), promoted apoptosis
(Fig. 6C and S1B), and inhibited migration and invasion
(Fig. 6D and E; Fig.
S1D and F) in 143B and MG-63
cells. In addition, 143B and MG-63 cells transfected with si-MALAT1
had higher E-cadherin protein level and lower N-cadherin, Vimentin
and Snail protein levels compared with the Scramble group (Fig. 6F). However, these results caused by
MALAT1 knockdown could be reversed by inhibiting miR-590-3p.
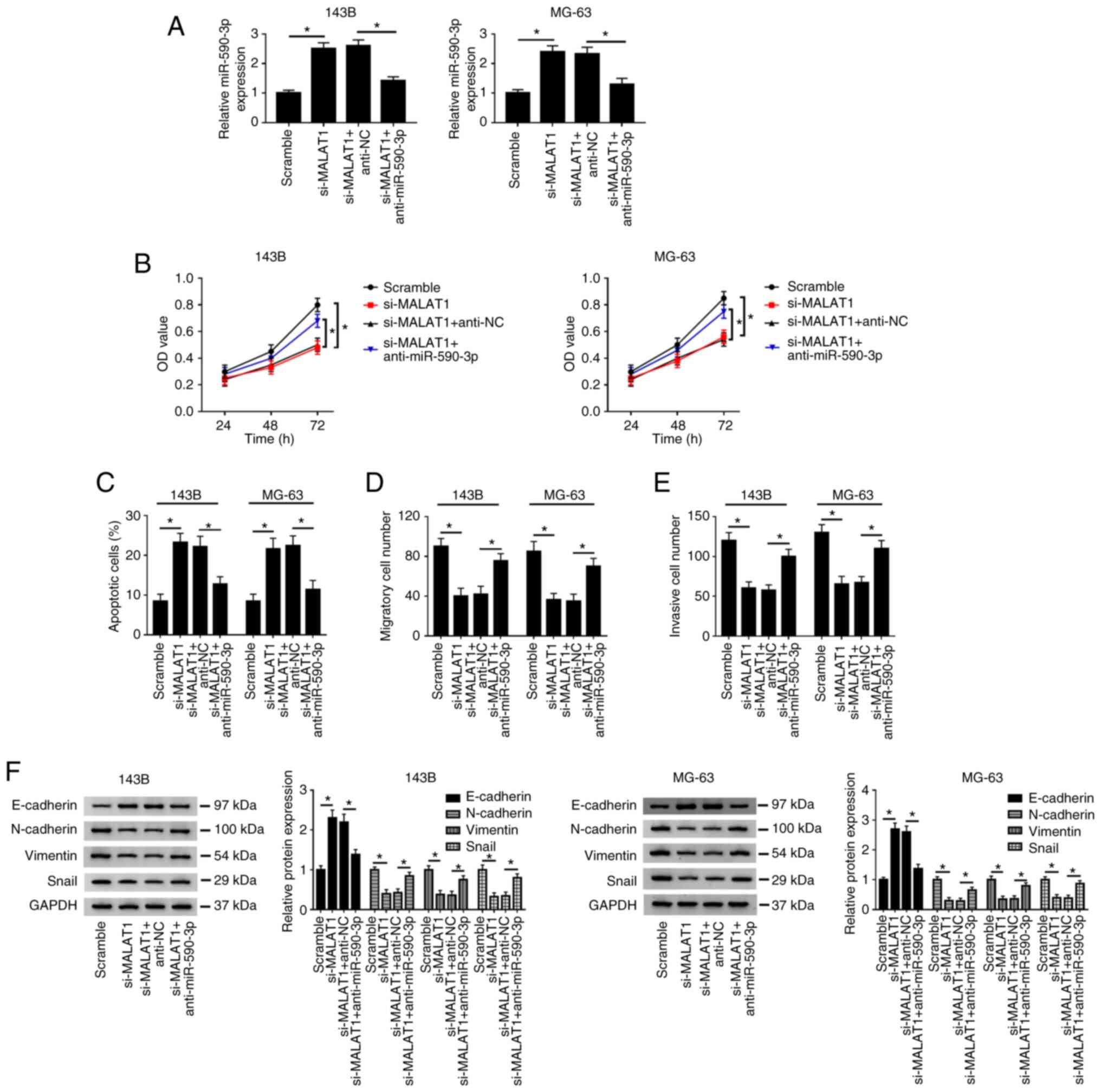 | Figure 6Inhibition of miR-590-3p reverses the
effects of MALAT1 knockdown on the proliferation, apoptosis,
migration, invasion and EMT of osteosarcoma cells. 143B and MG-63
cells were transfected with Scramble, si-MALAT1, si-MALAT1 +
anti-NC and si-MALAT1 + anti-miR-590-3p. (A) miR-590-3p expression
was measured by reverse transcription-quantitative PCR. (B) Cell
proliferative capacity was evaluated by Cell Counting Kit-8 assay
at 24, 48 and 72 h after transfection. (C) Cell apoptotic rate was
detected by flow cytometry. (D) Cell migration and (E) invasion
were evaluated by Transwell assay. (F) EMT-related protein
(E-cadherin, N-Cadherin, Vimentin and Snail) expression was
detected by western blotting. *P<0.05. EMT;
epithelial-mesenchymal transition; miR, microRNA; NC, negative
control; wt, wild-type; mut, mutant; si, small interfering; OD,
optical density. |
Discussion
It has been shown that numerous lncRNAs function as
competitive endogenous RNAs (ceRNAs) in human tumors (23), including OS. For example, Wang et
al confirmed that lncRNA HCG9 could contribute to OS growth
through RAD51 by acting as a ceRNA of miR-34b-3p (8). In addition, Ding et al reported
that lncRNA MELETF-AS1 may serve as a ceRNA of miR-485 to promote
OS cell migration and invasion in vitro (10). Non-coding RNAs, including lncRNAs
and miRNAs, could serve as potential tumor markers or therapeutic
targets in OS (1). lncRNA MALAT1
has been reported to be an oncogenic factor in OS. For example, Ren
et al (24) showed that
MALAT1 knockdown suppressed cell proliferation by regulating CDK9
and competitively binding to miR-206 in OS. MALAT1 positively
regulated cell proliferation, migration and invasion by sponging
miR-34a/c-5p and miR-449a/b in OS (25). Zhang et al (26) demonstrated that MALAT1 induced OS
cell growth and metastasis, and inhibited cell apoptosis by
modulating miR-509 to activate the Rac1/JNK pathway. Chen et
al (27) found that MALAT1
increased stem cell-like properties and activated the PI3K/Akt
signaling pathway by regulating Ret proto-oncogene and
competitively binding to miR-129-5p. However, the molecular
mechanisms of MALAT1 in OS cell proliferation, migration and
invasion require further elucidation. Consistent with previous
reports (24-27),
in the present study MALAT1 was highly expressed in OS tissues and
cell lines, and MALAT1 knockdown inhibited cell proliferation and
triggered cell apoptosis in OS.
Accumulating evidence has demonstrated that ectopic
expression of miRNAs plays an important role in metastasis,
invasion and chemoresistance of human OS cells (28). miR-590-3p has been widely reported
as a tumor suppressor in multiple malignant tumors, such as breast
cancer (29), gastric cancer
(30) and glioblastoma (31). However, miR-590-3p has been
indicated to promote ovarian cancer cell proliferation, invasion
and spheroid formation by targeting cyclin G2 and FOXO3(32). In colon cancer cells, miR-590-3p
positively regulated cell proliferation, spheroid formation and
cell cycle by binding to Wnt inhibitory factor 1 and
Dickkopf-related protein 1 (DKK1) (33). Wang et al (22) demonstrated that miR-590-3p may be a
potential therapeutic target for OS by inhibiting SOX9 expression.
Hu et al (34) found that
RNA binding protein pumilio homolog 2 could inhibit OS cell
progression by competitively binding to STARD13 with miR-590-3p and
miR-9. The present study further demonstrated that miR-590-3p
expression was significantly downregulated in OS tissues and cells
compared with in adjacent healthy tissues and hFOB1.19 cells.
Furthermore, it was demonstrated that MALAT1 negatively regulated
miR-590-3p in OS tissues, and knockdown of miR-590-3p attenuated
the inhibitory effect of MALAT1 on proliferation, migration and
invasion and its promotive effect on apoptosis in OS cells. In
addition, the current study revealed that MALAT1 directly bound to
miR-590-3p. Bioinformatics analysis indicated that miR-590-3p and
MALAT1 have two complementary sites. Moreover, the interaction
between MALAT1 and miR-590-3p was verified by luciferase reporter,
RIP and RNA pull-down assays.
In conclusion, the results of the present study
indicated that MALAT1 promoted cell proliferation, migration,
invasion and EMT and inhibited cell apoptosis by suppressing
miR-590-3p in OS, which provided a promising therapeutic target or
diagnostic marker for OS therapy. Due to the lack of in-depth
analysis on tissue samples, the present study has certain
limitations. Therefore, the effects of MALAT1 and miR-590-3p on OS
progression require further investigation.
Supplementary Material
Images of flow cytometry and Transwell
assay. (A) Fow cytometry images of 143B and MG-63 cells transfected
with NC or miR-590-3p. (B) Flow cytometry image of 143B and MG-63
cells transfected with Scramble, si-MALAT1, si-MALAT1 + anti-NC and
si-MALAT1 + anti-miR-590-3p. (C) Migration images of 143B and MG-63
cells transfected with NC or miR-590-3p. (D) Migration images of
143B and MG-63 cells transfected with Scramble, si-MALAT1,
si-MALAT1 + anti-NC and si-MALAT1 + anti-miR-590-3p. (E) Invasion
images of 143B and MG-63 cells transfected with NC or miR-590-3p.
(F) Invasion images of 143B and MG-63 cells transfected with
Scramble, si-MALAT1, si-MALAT1 + anti-NC and si-MALAT1 +
anti-miR-590-3p (200x magnification). miR, microRNA; NC, negative
control; si, small interfering.
Transfection efficiency of MALAT1
overexpression and miR-590-3p knockdown. (A) MALAT1 level was
measured in 143B and MG-63 cells transfected with Vector or MALAT1.
(B) Expression of miR-590-3p was detected in 143B and MG-63 cells
transfected with anti-NC or anti-miR-590-3p. *P<0.05.
miR, microRNA; NC, negative control; lncRNA, long non-coding
RNA.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
HZ conceived and designed the study, and wrote the
first draft of the manuscript. HZ, YW, WH, XD and WW performed all
of the experiments. YW, WH, XD and WW analyzed and collated the
results. HZ and YW confirm the authenticity of all the raw data.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Written informed consent was obtained from all
patients, and the research method was approved by the Ethics
Committee of the First Hospital of Lanzhou University (Lanzhou,
China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Luo Z, Xiao L, Li J, Dong B and Wang C:
Integrative analysis reveals driver long non-coding RNAs in
osteosarcoma. Medicine (Baltimore). 98(e14302)2019.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Mirabello L, Troisi RJ and Savage SA:
International osteosarcoma incidence patterns in children and
adolescents, middle ages and elderly persons. Int J Cancer.
125:229–234. 2009.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Nie Z and Peng H: Osteosarcoma in patients
below 25 years of age: An observational study of incidence,
metastasis, treatment and outcomes. Oncol Lett. 16:6502–6514.
2018.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Luetke A, Meyers PA, Lewis I and Juergens
H: Osteosarcoma treatment-where do we stand? A state of the art
review. Cancer Treat Rev. 40:523–532. 2014.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009.PubMed/NCBI View
Article : Google Scholar
|
|
6
|
Zhang J, Hao X, Yin M, Xu T and Guo F:
Long non-coding RNA in osteogenesis: A new world to be explored.
Bone Joint Res. 8:73–80. 2019.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Li JP, Liu LH, Li J, Chen Y, Jiang XW,
Ouyang YR, Liu YQ, Zhong H, Li H and Xiao T: Microarray expression
profile of long noncoding RNAs in human osteosarcoma. Biochem
Biophys Res Commun. 433:200–206. 2013.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Wang L, Li S, Qi L and Ling L: Long
noncoding RNA HCG9 promotes osteosarcoma progression through RAD51
by acting as a ceRNA of miR-34b-3p. Mediators Inflamm: Aug 18, 2021
(Epub ahead of print).
|
|
9
|
Zhon Y, Feng D, Gu X, Gao A and Liu Y: The
role and clinical significance of long noncoding RNA zinc finger
E-box-binding homeobox two antisense RNA 1 in promoting
osteosarcoma cancer cell proliferation, inhibiting apoptosis and
increasing migration by regulating miR-145. Anticancer Drugs.
32:168–177. 2021.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Ding L, Liu T, Qu Y, Kang Z, Guo L, Zhang
H, Jiang J, Qu F, Ge W and Zhang S: lncRNA MELTF-AS1 facilitates
osteosarcoma metastasis by modulating MMP14 expression. Mol Ther
Nucleic Acids. 26:787–797. 2021.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Chen R, Wang G, Zheng Y, Hua Y and Cai Z:
Long non-coding RNAs in osteosarcoma. Oncotarget. 8:20462–20475.
2017.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Li ZX, Zhu QN, Zhang HB, Hu Y, Wang G and
Zhu YS: MALAT1: A potential biomarker in cancer. Cancer Manag Res.
10:6757–6768. 2018.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Li Z, Yu X and Shen J: Long non-coding
RNAs: Emerging players in osteosarcoma. Tumour Biol. 37:2811–2816.
2016.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Sun Y and Qin B: Long noncoding RNA MALAT1
regulates HDAC4-mediated proliferation and apoptosis via decoying
of miR-140-5p in osteosarcoma cells. Cancer Med. 7:4584–4597.
2018.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Duan G, Zhang C, Xu C, Xu C, Zhang L and
Zhang Y: Knockdown of MALAT1 inhibits osteosarcoma progression via
regulating the miR34a/cyclin D1 axis. Int J Oncol. 54:17–28.
2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Lynam-Lennon N, Maher SG and Reynolds JV:
The roles of microRNA in cancer and apoptosis. Biol Rev Camb Philos
Soc. 84:55–71. 2009.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Di Leva G, Garofalo M and Croce CM:
MicroRNAs in cancer. Annu Rev Pathol. 9:287–314. 2014.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Chen B, Xia Z, Deng YN, Yang Y, Zhang P,
Zhu H, Xu N and Liang S: Emerging microRNA biomarkers for
colorectal cancer diagnosis and prognosis. Open Biol.
9(180212)2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Pang H, Zheng Y, Zhao Y, Xiu X and Wang J:
miR-590-3p suppresses cancer cell migration, invasion and
epithelial-mesenchymal transition in glioblastoma multiforme by
targeting ZEB1 and ZEB2. Biochem Biophys Res Commun. 468:739–745.
2015.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Abdolvahabi Z, Nourbakhsh M, Hosseinkhani
S, Hesari Z, Alipour M, Jafarzadeh M, Ghorbanhosseini SS, Seiri P,
Yousefi Z, Yarahmadi S and Golpour P: MicroRNA-590-3P suppresses
cell survival and triggers breast cancer cell apoptosis via
targeting sirtuin-1 and deacetylation of p53. J Cell Biochem.
120:9356–9368. 2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Dou Y, Zhu K, Sun Z, Geng X and Fang Q:
Screening of disorders associated with osteosarcoma by integrated
network analysis. Biosci Rep: May 21, 2019 (Epub ahead of
print).
|
|
22
|
Wang WT, Qi Q, Zhao P, Li CY, Yin XY and
Yan RB: miR-590-3p is a novel microRNA which suppresses
osteosarcoma progression by targeting SOX9. Biomed Pharmacother.
107:1763–1769. 2018.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Long J, Bai Y, Yang X, Lin J, Yang X, Wang
D, He L, Zheng Y and Zhao H: Construction and comprehensive
analysis of a ceRNA network to reveal potential prognostic
biomarkers for hepatocellular carcinoma. Cancer Cell Int.
19(90)2019.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Ren D, Zheng H, Fei S and Zhao JL: MALAT1
induces osteosarcoma progression by targeting miR-206/CDK9 axis. J
Cell Physiol. 234:950–957. 2018.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Sun Z, Zhang T and Chen B: Long non-coding
RNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT1)
promotes proliferation and metastasis of osteosarcoma cells by
targeting c-Met and SOX4 via miR-34a/c-5p and miR-449a/b. Med Sci
Monit. 25:1410–1422. 2019.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Zhang Y, Dai Q, Zeng F and Liu H: MALAT1
Promotes the proliferation and metastasis of osteosarcoma cells by
activating the Rac1/JNK pathway via targeting MiR-509. Oncol Res:
Apr 27, 2018 (Epub ahead of print).
|
|
27
|
Chen Y, Huang W, Sun W, Zheng B, Wang C,
Luo Z, Wang J and Yan W: LncRNA MALAT1 promotes cancer metastasis
in osteosarcoma via activation of the PI3K-Akt signaling pathway.
Cell Physiol Biochem. 51:1313–1326. 2018.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Maximov VV, Akkawi R, Khawaled S, Salah Z,
Jaber L, Barhoum A, Or O, Galasso M, Kurek KC, Yavin E and Aqeilan
RI: MiR-16-1-3p and miR-16-2-3p possess strong tumor suppressive
and antimetastatic properties in osteosarcoma. Int J Cancer.
145:3052–3063. 2019.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Rohini M, Gokulnath M, Miranda PJ and
Selvamurugan N: miR-590-3p inhibits proliferation and promotes
apoptosis by targeting activating transcription factor 3 in human
breast cancer cells. Biochimie. 154:10–18. 2018.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Gu L, Lu LS, Zhou DL and Liu ZC: UCA1
promotes cell proliferation and invasion of gastric cancer by
targeting CREB1 sponging to miR-590-3p. Cancer Med. 7:1253–1263.
2018.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Chen L, Wang W, Zhu S, Jin X, Wang J, Zhu
J and Zhou Y: MicroRNA-590-3p enhances the radioresistance in
glioblastoma cells by targeting LRIG1. Exp Ther Med. 14:1818–1824.
2017.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Salem M, Shan Y, Bernaudo S and Peng C:
miR-590-3p targets cyclin G2 and FOXO3 to promote ovarian cancer
cell proliferation, invasion, and spheroid formation. Int J Mol
Sci. 20(1810)2019.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Feng ZY, Xu XH, Cen DZ, Luo CY and Wu SB:
miR-590-3p promotes colon cancer cell proliferation via
Wnt/beta-catenin signaling pathway by inhibiting WIF1 and DKK1. Eur
Rev Med Pharmacol Sci. 21:4844–4852. 2017.PubMed/NCBI
|
|
34
|
Hu R, Zhu X, Chen C, Xu R, Li Y and Xu W:
RNA-binding protein PUM2 suppresses osteosarcoma progression via
partly and competitively binding to STARD13 3'UTR with miRNAs. Cell
Prolif. 51(e12508)2018.PubMed/NCBI View Article : Google Scholar
|