Introduction
Neuropathic pain is a condition that is refractory
to analgesics and significantly decreases the quality of life.
Elucidation of the mechanisms underlying neuropathic pain
represents the first step in the discovery and selection of
effective therapies for this condition. In recent years, the
presence of non-coding RNA, termed microRNA (miRNA), consisting of
approximately 20 bases has become a major focus of research. It is
now known that miRNA binds to the 3′-untranslated region of its
target mRNA. Binding of miRNA to a target that has partially
complementary sequences inhibits mRNA translation, and binding to a
target that has perfectly complementary mRNA contributes to the
degradation of the mRNA (1–4).
It has been reported that a large number of these small RNAs exist
in the mammalian nervous system and that they play an important
role in nerve generation and degeneration (5,6).
The dorsal horn of the spinal cord is the location
of the secondary neurons that receive nociceptive stimuli in
neuropathic pain. The dorsal horn plays an important role in pain
control and treatment as well as in the generation of neuropathic
pain and is also responsible for central sensitization and
inhibition of the transmission of nociceptive stimuli via the
descending inhibitory system. It is therefore presumed that many
miRNAs play a role in the dorsal horn of the spinal cord in
neuropathic pain. Although it has been suggested that determination
of global changes in miRNA would help to clarify the mechanisms
underlying neuropathic pain, a comprehensive analysis of miRNA
expression in the dorsal horn of the spinal cord in chronic
constrictive injury (CCI) model rats has not been reported.
Previous reports have demonstrated that some miRNA expression is
associated with pain (7–10). Thus, we comprehensively analyzed
miRNA expression in the dorsal horn of the CCI rat model using the
TaqMan® Low Density Array (TLDA).
Materials and methods
Experimental animals
All experimental procedures were approved by the
Institutional Committee on Laboratory Animals of Nippon Medical
School (approval no. 22–162) and were performed under the
guidelines of the International Association for the Study of Pain
(11).
Male Sprague-Dawley rats (n=30; 6–7 weeks old;
weight, 200–250 g; Saitama Experimental Animals, Saitama, Japan)
were used for all experiments. Experimental neuropathy was produced
as described elsewhere (12–15). In the present study, a sciatic
nerve injury model was used. All surgical procedures were performed
on rats that were deeply anesthetized with sodium pentobarbital (50
mg/kg intraperitoneally).
To assess pain thresholds, we performed the plantar
test and the von Frey behavioral tests. The details of the test
methods have been discussed in previous reports (12,14–16). Briefly, the plantar test (Ugo
Basile, Comerio, Italy) was used to examine thermal hyperalgesia.
Mechanical allodynia was measured using a set of von Frey filaments
(Muromachi Kikai, Saitama, Japan) with bending forces ranging from
0.04 to 72.0 g.
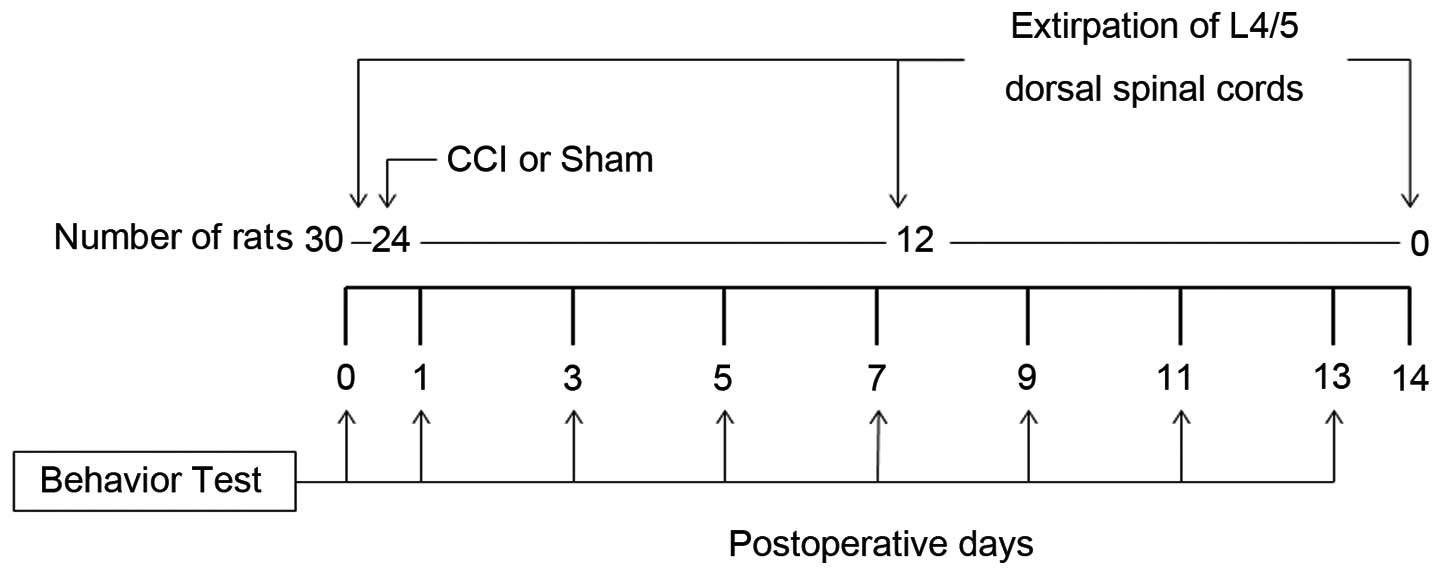
Following behavioral testing, the 30 rats were
divided into three groups. In the Day 0 group rats (n=6), that did
not undergo any operation, the L4/5 spinal cord was removed
immediately following behavioral testing. The Day 7 group rats
(n=12) underwent either sciatic nerve ligation (CCI model rats,
n=6) or sham operation (n=6). Seven days after the operation and
the behavioral testing, the L4/5 spinal cord was removed. Day 14
group rats (n=12) underwent either sciatic nerve ligation (n=6) or
sham operation (n=6). Fourteen days after the operation and the
behavioral testing, the L4/5 spinal cord was removed. The
behavioral test was practiced at some point after the operation
(nerve ligation or sham). The experimental protocol is shown in
Fig. 1. A two-tailed t-test was
used to compare the latencies or threshold values in behavioral
tests between CCI and sham rats. A one-way analysis of variance
(ANOVA) followed by Tukey's test was used to compare latencies or
threshold values obtained in behavioral tests performed on Day 0
(undergoing no operation; n=6) and at some point after the
operation.
Extirpation and preservation of
samples
The rats were deeply anesthetized with sodium
pentobarbital (50 mg/kg intraperitoneally) and the L4/5 spinal
cords were dissected. The L4/5 spinal cords were washed twice with
cold phosphate-buffered saline (PBS) and immediately divided into
contralateral (non-ligated side or right side) and ipsilateral
(ligated side or left side) samples, and stored in RNAlater
solution (Applied Biosystems, Foster City, CA, USA). Each
ipsilateral sample was further divided into ventral and dorsal
samples and the dorsal spinal cords were stored at −20°C in
RNAlater solution.
Total RNA isolation/miRNA isolation
The samples were defrosted and RNAlater solution was
rapidly separated from the samples. Total RNA was extracted using a
mirVana miRNA Isolation kit (Applied Biosystems). RNA quantity and
quality were assessed using a NanoDrop ND-1000 spectrophotometer
(Thermo Fisher Scientific, Waltham, MA, USA), and A260/280 nm ≥1.8
was determined for quantitative analysis using the procedures
described in previous studies (10,17,18).
TLDA
The miRNA expression profile of the L4/5 dorsal
spinal cord of CCI rats was analyzed using TLDA Rodent MicroRNA
Cards v.3 A and B (Applied Biosystems). We used the standard method
that was evaluated in a previous study (10,17–19). Each card contains 375 preloaded
rodent miRNA targets, all catalogued in the miRBase database, and
three endogenous controls: Mamm U6, U87 and Y1. In this study, we
used U87 as the endogenous normalizer. TLDAs were performed using a
four-step process. The first step was the Megaplex RT Reaction in
which 60 ng of total RNA per sample was reverse-transcribed using
Megaplex RT primer Pool A and B, with up to 381 stem-looped primers
per pool, and the TaqMan MicroRNA Reverse Transcription kit
(Applied Biosystems). The next step was the preamplification
reaction in which TaqMan PreAmp Master Mix, 2X and Megaplex PreAmp
Primers (Applied Biosystems) were added to each complementary DNA
(cDNA), and the reaction was performed. The preamplified product
was diluted with 0.1X TE at pH8.0 (Wako, Tokyo, Japan). In the
third step, each of the resulting cDNAs after the preamplification
reaction were diluted with the TaqMan Universal PCR master mix
(Applied Biosystems) and deionized distilled water (Wako), and were
loaded into one of the eight fill ports on the TLDA microfluidic
card. The card was centrifuged for 1 min at 331 × g to distribute
samples to multiple wells connected to the fill ports and was then
sealed to prevent well-to-well contamination. Finally, the cards
were processed and analyzed using a 7900 HT Real Time PCR System
(Applied Biosystems).
Real-time RT-PCR data analysis was performed using
DataAssist software v2.0 (Applied Biosystems). The resulting data
are shown as threshold cycle (Ct) values, where Ct represents a
unitless value defined as the fractional cycle number at which the
sample fluorescence signal passes a fixed threshold above baseline.
The expression level was calculated using the comparative Ct (ΔΔCt)
method and was further analyzed by comparing the fold change
relative to basal levels in the Day 0 sample. The results of the
TLDA analysis were converted into a graphic display as a heat map
based on hierarchical clustering using DataAssist version 2.0.
Distances between samples and assays were calculated for
hierarchical clustering based on ΔCt values using Euclidean
Distance.
Statistical analysis
For statistical analyses of behavioral data, we used
a two-tailed t-test and an ANOVA followed by Tukey's test. To
compare the gene expression levels of sham-operated rats (Day 7,
n=6; Day 14, n=6) with TLDA dates in the L4/5 dorsal spinal cords
of CCI rats (Day 7, n=6; Day 14, n=6) within the same group, ANOVA
was used followed by Tukey's test. All statistical procedures were
performed using KyPlot 5.0 (KyensLab, Inc., Tokyo, Japan). All
values are expressed as the means ± standard deviation. A P-value
<0.01 was considered to indicate statistically significant
differences.
Results
Behavioral tests
Compared with the values from sham-operated rats,
the latencies of paw withdrawal thresholds in response to
mechanical stimulation (Fig 2A)
and paw withdrawal from thermal stimulation (Fig 2B) on the ligated ipsilateral side
in CCI rats were significantly decreased at Day 1 after surgery.
The hypersensitivity peaked at 7 days, and was then maintained at
the same level until Day 13 after surgery (P<0.01) (Fig. 2).
TLDA analysis
Using the TLDA card, 375 out of 750 miRNAs were
peculiarly expressed in the rat tissue. In CCI rats (Day 7 and/or
Day 14), 111 of 375 miRNAs were significantly regulated compared
with sham-operated rats (Day 7 and/or Day 14). The expression of 21
miRNAs (up, 8 miRNAs; down, 13 miRNAs) was significantly altered
only in Day 7 CCI rats compared to Day 7 sham-operated rats. The
expression of 65 miRNAs (up, 20 miRNAs; down, 45 miRNAs) was
significantly altered only in Day 14 CCI rats compared to Day 14
sham rats. In the Day 7 and Day 14 groups, 25 miRNAs significantly
changed expression (up, 3 miRNAs; down, 22 miRNAs). Details of the
results are shown in Table I. We
illustrated a clustergram of the samples and the significant
differentially expressed miRNAs in the dorsal horn of the spinal
cord as a heat map (Fig. 3). Heat
maps are commonly used for visualization of high-dimensional data
on a two-dimensional image with colors representing the intensity
values. Heat maps are typically used in gene expression analysis to
represent the level of expression of many genes across a number of
comparable samples. After individually clustering columns (samples)
and rows (miRNAs), the heat map simultaneously displays the
separate samples and miRNA clustering in one graphic. Red and blue
colors indicate relatively high and low expression, respectively.
The dendrogram at the top of the figure indicates the relatedness
of the samples based on overall miRNA expression values. The
dendrogram on the left side of the figure orders miRNAs into groups
based on the divergence of miRNA expression values among the
samples. In the top dendrogram of the heat map, these 111 miRNAs
separate three branches, which are the sham group (Day 7 and Day
14), the Day 7 CCI group and the Day 14 CCI group. Sham samples
from Day 7 and Day 14 rats are not separated from each other. These
results demonstrated no difference in the expression pattern
between Day 7 and Day 14 sham-operated rats, and the role of miRNAs
in the dorsal horn of the spinal cord of CCI rats is changed with
time.
 | Table IThe varied expression of microRNAs
(miRNAs) on chronic constrictive injury (CCI) rats compared to the
sham rats without ligated sciatic nerve. |
Table I
The varied expression of microRNAs
(miRNAs) on chronic constrictive injury (CCI) rats compared to the
sham rats without ligated sciatic nerve.
| Fold change ±
standard deviation (SD) | |
|---|
|
| |
|---|
| Assay | CCI7
CCI14 | Sham7
Sham14 | P-value |
|---|
| miRNAs downregulated
in both the Day 7 and Day 14 groups | | | |
| hsa-miR-22# | 1.134±0.098 | 0.387±0.062 |
1.74×10−6 |
| 0.757±0.224 | 0.440±0.068 | 0.00172 |
| mmu-miR-496 | 1.231±0.259 | 0.227±0.119 |
1.74×10−6 |
| 1.312±0.204 | 0.078±0.042 |
1.74×10−6 |
| mmu-miR-151-3p | 1.156±0.262 | 0.310±0.095 |
2.32×10−6 |
| 0.899±0.236 | 0.468±0.158 | 0.00581 |
| mmu-miR-24-2# | 0.883±0.099 | 0.500±0.075 |
9.80×10−6 |
| 0.803±0.102 | 0.384±0.072 |
3.52×10−6 |
| mmu-miR-324-5p | 0.994±0.195 | 0.398±0.096 |
8.50×10−5 |
| 0.957±0.300 | 0.467±0.056 | 0.00102 |
| rno-miR-345-3p | 0.956±0.159 | 0.289±0.216 | 0.000157 |
| 1.261±0.313 | 0.181±0.039 |
1.80×10−6 |
| mmu-miR-127 | 1.269±0.174 | 0.676±0.116 | 0.000162 |
| 1.313±0.329 | 0.632±0.124 |
2.37×10−5 |
| mmu-miR-125b-5p | 1.057±0.202 | 0.454±0.075 | 0.000262 |
| 1.034±0.345 | 0.494±0.128 | 0.00100 |
| mmu-miR-221 | 1.360±0.224 | 0.519±0.205 | 0.000334 |
| 1.334±0.519 | 0.391±0.109 |
7.29×10−5 |
| mmu-miR-296-5p | 1.531±0.388 | 0.710±0.085 | 0.000337 |
| 1.250±0.404 | 0.618±0.212 | 0.00603 |
| rno-miR-377 | 0.967±0.165 | 0.158±0.087 | 0.000416 |
| 1.425±0.570 | 0.090±0.052 |
1.91×10−6 |
| mmu-miR-365 | 1.492±0.387 | 0.276±0.238 | 0.000918 |
| 1.681±0.906 | 0.028±0.035 |
1.59×10−5 |
| mmu-miR-598 | 1.083±0.085 | 0.577±0.147 | 0.00104 |
| 1.259±0.318 | 0.664±0.144 | 0.000139 |
| mmu-miR-7a | 0.996±0.215 | 0.485±0.152 | 0.00206 |
| 1.093±0.231 | 0.623±0.202 | 0.00486 |
| mmu-miR-101b | 1.002±0.119 | 0.690±0.135 | 0.00319 |
| 1.070±0.149 | 0.452±0.107 |
1.90×10−6 |
| mmu-miR-29b | 1.004±0.202 | 0.481±0.187 | 0.00324 |
| 1.009±0.278 | 0.396±0.143 | 0.000553 |
| rno-miR-336 | 0.976±0.203 | 0.099±0.088 | 0.00362 |
| 1.500±0.761 | 0.057±0.030 |
6.75×10−6 |
| hsa-miR-493-3p | 0.794±0.156 | 0.373±0.153 | 0.00384 |
| 1.204±0.244 | 0.420±0.083 |
2.44×10−6 |
| mmu-miR-322 | 1.068±0.196 | 0.641±0.146 | 0.00469 |
| 1.016±0.256 | 0.531±0.112 | 0.00123 |
| mmu-miR-21 | 1.400±0.084 | 0.612±0.119 | 0.00677 |
| 1.887±0.747 | 0.718±0.179 |
6.76×10−5 |
| mmu-miR-27b | 1.018±0.111 | 0.545±0.126 | 0.00687 |
| 1.220±0.391 | 0.620±0.137 | 0.000534 |
| rno-miR-632 | 1.078±0.177 | 0.571±0.260 | 0.00901 |
| 1.163±0.360 | 0.324±0.087 |
2.30×10−5 |
|
| Assay | CCI7 | Sham7 | P-value |
|
| miRNAs
downregulated only in the Day 7 group | | | |
| mmu-miR-193# | 1.014±0.058 | 0.368±0.062 |
1.74×10−6 |
| hsa-miR-29a# | 0.984±0.193 | 0.448±0.159 | 0.000410 |
| mmu-miR-19b | 1.279±0.195 | 0.657±0.167 | 0.000139 |
| rno-miR-339-3p | 1.046±0.178 | 0.464±0.246 | 0.000662 |
| mmu-miR-106b | 0.985±0.187 | 0.408±0.124 | 0.000213 |
| mmu-miR-500 | 1.194±0.208 | 0.534±0.130 | 0.000216 |
| mmu-miR-375 | 1.667±0.438 | 0.800±0.314 | 0.00113 |
| hsa-miR-378 | 0.914±0.052 | 0.487±0.235 | 0.00142 |
| mmu-miR-28 | 0.934±0.170 | 0.478±0.079 | 0.00172 |
| mmu-miR-30b | 1.201±0.180 | 0.775±0.125 | 0.00278 |
| mmu-miR-381 | 1.157±0.380 | 0.397±0.349 | 0.00390 |
| mmu-miR-337 | 0.974±0.106 | 0.572±0.114 | 0.00581 |
| mmu-miR-203 | 0.999±0.223 | 0.491±0.169 | 0.00906 |
|
| Assay | CCI7
CCI14 | Sham7
Sham14 | P-value |
|
| miRNAs upregulated
in both the Day 7 and Day 14 groups | | | |
| mmu-miR-539 | 1.077±0.279 | 1044±240.5 |
1.74×10−6 |
| 0.840±0.222 | 949.9±201.6 |
1.74×10−6 |
| rno-miR-381 | 1.192±0.157 | 7.123±1.091 |
1.74×10−6 |
| 1.733±0.458 | 6.369±1.251 |
1.74×10−6 |
| mmu-miR-323-3p | 1.024±0.190 | 2.541±0.682 |
3.29×10−5 |
| 0.948±0.207 | 2.591±0.594 |
1.09×10−5 |
|
| Assay | CCI7 | Sham7 | P-value |
|
| miRNAs upregulated
only in the Day 7 group | | | |
| mmu-miR-193 | 0.913±0.147 | 53.40±13.73 |
1.74×10−6 |
| rno-miR-489 | 0.850±0.230 | 16.59±1.874 | 0.000107 |
| rno-miR-346 | 1.075±0.423 | 7.038±4.246 | 0.000256 |
| rno-miR-409-5p | 0.365±0.059 | 16.68±10.57 | 0.000315 |
| hsa-miR-99b# | 1.204±0.287 | 3.063±1.397 | 0.00117 |
| hsa-miR-136 | 1.010±0.201 | 1.855±0.269 | 0.00228 |
| mmu-miR-770-5p | 1.160±0.159 | 2.202±0.319 | 0.00252 |
| mmu-miR-376a | 1.241±0.241 | 2.307±0.677 | 0.000343 |
|
| Assay | CCI14 | Sham14 | P-value |
|
| miRNAs upregulated
only in the Day14 group | | | |
| mmu-let-7d# | 0.058±0.009 | 21.43±9.389 |
1.97×10−6 |
| mmu-miR-139-3p | 0.335±0.271 | 3.056±0.900 |
3.12×10−6 |
| mmu-miR-296-3p | 0.997±0.186 | 24.75±14.28 |
1.70×10−5 |
| mmu-miR-134 | 0.915±0.253 | 2.687±1.000 |
2.88×10−5 |
|
rno-miR-219-1-3p | 0.837±0.196 | 2.625±0.777 |
3.76×10−5 |
| mmu-miR-342-3p | 1.284±0.237 | 5.354±2.611 |
9.37×10−5 |
| mmu-miR-126-3p | 0.915±0.142 | 2.171±0.801 | 0.000226 |
|
| Assay | CCI14 | Sham14 | P-value |
|
| miRNAs upregulated
only in the Day14 group | | | |
| mmu-miR-186 | 1.443±0.502 | 5.076±2.501 | 0.000289 |
|
mmu-miR-125a-3p | 0.886±0.227 | 2.700±1.261 | 0.000328 |
| mmu-miR-449a | 1.874±0.577 | 6.181±3.405 | 0.00113 |
| mmu-miR-191 | 1.092±0.312 | 2.671±1.134 | 0.00120 |
| mmu-miR-764-5p | 0.827±0.762 | 3.243±1.237 | 0.00123 |
| mmu-miR-139-5p | 0.951±0.195 | 2.003±0.781 | 0.00175 |
| mmu-miR-146a | 1.303±0.409 | 2.670±1.048 | 0.00179 |
| mmu-miR-672 | 0.755±0.124 | 3.127±1.685 | 0.00211 |
| mmu-miR-150 | 0.885±0.263 | 1.848±0.731 | 0.00246 |
| mmu-miR-511 | 0.073±0.014 | 1.710±1.402 | 0.00329 |
| mmu-miR-380-5p | 1.066±0.347 | 1.833±0.412 | 0.00441 |
| mmu-miR-187 | 0.861±0.281 | 1.657±0.507 | 0.00449 |
| hsa-miR-124# | 2.265±3.467 | 12.67±9.371 | 0.00688 |
|
| Assay | CCI14 | Sham14 | P-value |
|
| miRNAs
downregulated only in the Day 14 group | | | |
| mmu-miR-374-5p | 1.570±0.363 | 0.291±0.191 |
1.76×10−6 |
| mmu-miR-137 | 1.487±0.231 | 0.688±0.153 |
2.04×10−6 |
| mmu-miR-27a | 1.743±0.367 | 0.847±0.148 |
2.94×10−6 |
| rno-miR-505 | 1.720±0.443 | 0.743±0.072 |
3.13×10−6 |
| rno-miR-421 | 3.157±1.519 | 0.122±0.206 |
4.06×10−6 |
| rno-miR-29c# | 1.317±0.283 | 0.674±0.105 |
7.19×10−6 |
| mmu-miR-369-5p | 1.092±0.276 | 0.331±0.099 |
1.06×10−5 |
| mmu-miR-879 | 2.539±0.761 | 1.043±0.309 |
1.17×10−5 |
| mmu-miR-190 | 2.182±0.699 | 0.964±0.168 |
2.58×10−5 |
| mmu-miR-674# | 0.948±0.187 | 0.526±0.098 |
4.36×10−5 |
| mmu-miR-487b | 1.225±0.264 | 0.703±0.148 |
4.33×10−5 |
| mmu-miR-325 | 1.885±0.388 | 1.044±0.200 |
4.43×10−5 |
| mmu-miR-542-3p | 1.455±0.388 | 0.649±0.232 |
4.66×10−5 |
| mmu-miR-96 | 1.170±0.310 | 0.526±0.145 |
7.87×10−5 |
| rno-miR-99a# | 1.038±0.222 | 0.574±0.124 |
8.45×10−5 |
| mmu-miR-26b | 1.635±0.333 | 0.995±0.149 | 0.000101 |
| hsa-miR-27a# | 1.159±0.182 | 0.423±0.236 | 0.000127 |
| mmu-miR-32 | 1.708±0.243 | 0.871±0.358 | 0.000145 |
| rno-miR-207 | 2.362±0.314 | 0.858±0.934 | 0.000188 |
| hsa-miR-423-3P | 1.093±0.156 | 0.709±0.182 | 0.000258 |
| mmu-miR-196a# | 1.600±0.582 | 0.520±0.233 | 0.000332 |
| mmu-miR-130b | 1.406±0.410 | 0.562±0.185 | 0.000342 |
| hsa-miR-9# | 1.104±0.252 | 0.619±0.129 | 0.000354 |
| mmu-miR-335-5p | 1.455±0.413 | 0.801±0.132 | 0.000374 |
| mmu-miR-26a | 1.663±0.443 | 0.965±0.161 | 0.000398 |
| hsa-miR-136# | 1.505±0.268 | 0.959±0.241 | 0.000656 |
| hsa-miR-340 | 1.085±0.204 | 0.632±0.055 | 0.000769 |
| rno-miR-504 | 0.969±0.166 | 0.586±0.170 | 0.000993 |
| mmu-miR-130a | 1.242±0.337 | 0.468±0.151 | 0.00153 |
| hsa-miR-455 | 0.989±0.161 | 0.643±0.131 | 0.00157 |
| mmu-miR-142-3p | 1.974±0.970 | 0.730±0.300 | 0.00188 |
| hsa-miR-26b# | 1.233±0.226 | 0.781±0.201 | 0.00194 |
|
| Assay | CCI14 | Sham14 | P-value |
|
| miRNAs
downregulated only in the Day 14 group | | | |
| rno-miR-450a | 1.131±0.305 | 0.536±0.134 | 0.00257 |
| mmu-let-7i | 1.313±0.331 | 0.750±0.234 | 0.00292 |
| rno-miR-532-5p | 1.238±0.355 | 0.731±0.074 | 0.00297 |
| mmu-miR-34c | 1.439±0.615 | 0.691±0.181 | 0.00306 |
| mmu-miR-129-5p | 2.072±0.537 | 1.345±0.284 | 0.00342 |
| mmu-miR-497 | 1.086±0.462 | 0.486±0.092 | 0.00360 |
| mmu-miR-135a | 1.936±0.384 | 1.294±0.340 | 0.00398 |
| rno-miR-351 | 1.517±0.374 | 0.843±0.286 | 0.00468 |
| mmu-miR-369-3
p | 1.837±0.561 | 1.021±0.477 | 0.00516 |
| mmu-miR-148b | 1.526±0.398 | 0.996±0.195 | 0.00585 |
| rno-miR-409-3P | 1.442±0.260 | 0.930±0.378 | 0.00667 |
| hsa-miR-140-3p | 1.698±0.502 | 0.948±0.425 | 0.00691 |
| mmu-miR-29c | 1.748±1.390 | 0.399±0.110 | 0.00910 |
Text mining of miRNA associations with
gene function
Next, we sought to identify the role of miRNA
changes in this study by text mining using PubMed. In 111 miRNAs,
there were 76 (68.5%) miRNAs analyzed in previous reports and 36
miRNAs (32.4%) played a role related to nerve development and
maintenance (14 miRNAs), tumors of the nervous system (18 miRNAs),
and neurodegenerative diseases (8 miRNAs). Four miRNAs overlapped.
There were three miRNAs related to neuropathic pain; miR-500, −221
and −21 (20,21).
Discussion
In the present study, we comprehensively analyzed
the dorsal horn of the spinal cord, which is an important organ for
synaptic plasticity, pain control and treatment of neuropathic
pain, in the CCI rat model using TLDA with the onset and
maintenance of hyperalgesia. Comparison between sham-operated and
CCI rats in the same group revealed expression changes in several
miRNAs, as hypothesized. This was particularly true for regulated
miRNAs, anticipated to be associated with the mechanisms underlying
neuropathic pain. In the pain field, many reports on miRNA
expression have been published since Bai et al (22) reported miRNA expression following
muscle pain (7–10,23). However, the available data do not
clarify the role of miRNA expression in the dorsal horn of the
spinal cord in neuropathic pain. In this study, it is suggested,
for the first time, that the expression of many miRNAs is
regulated, and that the number of changed miRNAs increases with the
passage of time after ligation of the sciatic nerve in the dorsal
horn of the spinal cord in the neuropathic pain model, although
hypersensitivity has been stable. In a previous study using mice,
the expression of miRNA in the dorsal horn of the spinal cord was
changed 14 days after sciatic nerve injury, later than the change
in the dorsal root ganglion (7).
We speculated that part of the role of the dorsal horn of the
spinal cord may be to maintain the symptoms of neuropathic pain.
Previous studies have identified a number of miRNAs expressed in
the spinal cord (24–27). The miR-29a/b/c, −26a, −142–3p and
−193 are highly expressed in the spinal cord. The miR-129, −146a
and −21 have also been reported to play a role in the
reorganization or recovery of the nervous system after injury and
in the nervous system development, apoptosis and synaptic
plasticity (28–31). Above all, miR-26a/b and miR-29 are
strongly expressed in astrocytes, and regulate the role of
astrocytes in the structure of the brain and spinal cord. They may
also be involved in the expression of neurotransmitters such as
glutamate transporters to modulate synaptic transmission and to
repair the nervous system (24,25). It is of note that miR-28 was
associated with μ-opioid receptors and/or expression of cAMP
response element-binding 1 (CREB-1), which has a role in neuronal
plasticity (32,33). In addition, miR-203 targets
γ-aminobutyric acid (GABA)-A receptors, which are a class of
receptors that respond to the neurotransmitter GABA, the chief
inhibitory neurotransmitter in the vertebrate central nervous
system (34). Although several
miRNAs changed expression, their roles in neuropathic pain have yet
to be analyzed. We speculate that numerous miRNAs play key roles in
the molecular mechanisms responsible for nerve regeneration,
synaptic plasticity or analgesia, similar to miR-26, miR-29, miR-28
and miR-203.
The comprehensive TLDA analysis performed in this
study demonstrated changes in a large number of miRNAs in addition
to those previously reported as being related to neuropathic pain.
These findings are expected to contribute to our understanding of
the role of miRNA in the spinal cord of patients with neuropathic
pain and of the mechanisms underlying neuropathic pain and may be
the first step towards the selection of effective therapeutic
methods.
In this TLDA study, we report that the expression of
many miRNAs is altered in the dorsal horn of the spinal cord in CCI
rats, and we suggest the possibility that these changes play a role
in the maintenance and development of, and therapy for, neuropathic
pain. However, these data are not sufficient to make strong
conclusions on the role of miRNA changes in neuropathic pain. In
the study of miRNA biology, it is critical to predict changes in
the target mRNA using available online target prediction software
such as miRecords, TargetScan and PicTar, and to prove that a miRNA
can bind to a target mRNA using a luciferase reporter assay. For
example, it would be of benefit to clarify the role of mRNA coding
mediators in the activation of microglia or in the control of
analgesia, serotonin, adrenaline and acetylcholine. Identification
of the target gene of these miRNAs should help to elucidate
mechanisms underlying neuropathic pain and identify promising
targets for future research.
References
|
1
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
He L and Hannon GJ: MicroRNAs: small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Grishok A, Pasquinelli AE, Conte D, Li N,
Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G and Mello CC: Genes
and mechanisms related to RNA interference regulate expression of
the small temporal RNAs that control C. elegans developmental
timing. Cell. 106:23–34. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hutvagner G and Zamore PD: A microRNA in a
multiple-turnover RNAi enzyme complex. Science. 297:2056–2060.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mehler MF and Mattick JS: Non-coding RNAs
in the nervous system. J Physiol. 575:333–341. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kosik KS: The neuronal microRNA system.
Nat Rev Neurosci. 7:911–920. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kusuda R, Cadetti F, Ravanelli MI, Sousa
TA, Zanon S, De Lucca FL and Lucas G: Differential expression of
microRNAs in mouse pain models. Mol Pain. 7:172011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Aldrich BT, Frakes EP, Kasuya J, Hammond
DL and Kitamoto T: Changes in expression of sensory organ-specific
microRNAs in rat dorsal root ganglia in association with mechanical
hypersensitivity induced by spinal nerve ligation. Neuroscience.
164:711–723. 2009. View Article : Google Scholar
|
|
9
|
Imai S, Saeki M, Yanase M, Horiuchi H, Abe
M and Narita M, Kuzumaki N, Suzuki T and Narita M: Change in
microRNAs associated with neuronal adaptive responses in the
nucleus accumbens under neuropathic pain. J Neurosci.
31:15294–15299. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
von Schack D, Agostino MJ, Murray BS, Li
Y, Reddy PS, Chen J, Choe SE, Strassle BW, Li C, Bates B, Zhang L,
Hu H, Kotnis S, Bingham B, Liu W, Whiteside GT, Samad TA, Kennedy
JD and Ajit SK: Dynamic changes in the microRNA expression profile
reveal multiple regulatory mechanisms in the spinal nerve ligation
model of neuropathic pain. PLoS One. 6:e176702011.PubMed/NCBI
|
|
11
|
Covino BG, Dubner R and Gybels J: Ethical
standards for investigations of experimental pain in animals. Pain.
9:141–143. 1980. View Article : Google Scholar
|
|
12
|
Jarvis MF and Boyce-Rustay JM: Neuropathic
pain: models and mechanisms. Curr Pharm Des. 15:1711–1716. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bennett GJ and Xie YK: A peripheral
mononeuropathy in rat that produces disorders of pain sensation
like those seen in man. Pain. 720:111–119. 1988.
|
|
14
|
Sato C, Sakai A, Ikeda Y, Suzuki H and
Sakamoto A: The prolonged analgesic effect of epidural ropivacine
in a rat model of neuropathic pain. Anesth Analg. 106:313–320.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Okabe T, Sato C and Sakamoto A: Changes in
neuropeptide Y gene expression in the spinal cord of chronic
constrictive injury model rats after electroconvulsive stimulation.
Biomed Res. 31:287–292. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shibata M, Wakisaka S, Inoue T and Yoshiya
I, Shimizu T and Yoshiya I: The effect of electroconvulsive
treatment on thermal hyperalgesia and mechanical allodynia in a rat
model of peripheral neuropathy. Anesth Analg. 86:584–587.
1998.PubMed/NCBI
|
|
17
|
Kodani M, Yang G, Conklin LM, Travis TC,
Whitney CG, Anderson LJ, Schrag SJ, Taylor TH Jr, Beall BW, Breiman
RF, Feikin DR, Njenga MK, Mayer LW, Oberste MS, Tondella ML,
Winchell JM, Lindstrom SL, Erdman DD and Fields BS: Application of
TaqMan low-density arrays for simultaneous detection of multiple
respiratory pathogens. J Clin Microbiol. 49:2175–2182. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Stanton MC, Chen SC, Jackson JV,
Rojas-Triana A, Kinsley D, Cui L, Fine JS, Greenfeder S, Bober LA
and Jenh CH: Inflammatory signals shift from adipose to liver
during high fat feeding and influence the development of
steatohepatitis in mice. J Inflamm (Lond). 8:82011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang B, Howel P, Bruheim S, Ju J, Owen LB,
Oystein F and Xi Y: Systematic evaluation of three microRNA
profiling platforms: microarray, beads array, and quantitative
real-time PCR array. PLoS One. 6:e171672011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yu B, Zhou S, Qian T, Wang Y, Ding F and
Gu X: Altered microRNA expression following sciatic nerve resection
in dorsal root ganglia of rats. Acta Biochim Biophys Sin
(Shanghai). 43:909–915. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yu B, Zhou S, Wang Y, Qian T, Ding G, Ding
F and Gu X: miR-221/222 promote Schwann cell proliferation and
migration by targeting LASS2 following sciatic nerve injury. J Cell
Sci. 125:2675–2683. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bai G, Ambalavanar R, Wei D and Dessem D:
Downregulation of selective microRNAs in trigeminal ganglion
neurons following inflammatory muscle pain. Mol Pain. 3:152007.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhao J, Lee MC, Momin A, Cendan CM,
Shepherd ST, Baker MD, Asante C, Bee L, Bethry A, Perkins JR,
Nassar MA, Abrahamsen B, Dickenson A, Cobb BS, Merkenschlager M and
Wood JN: Small RNAs control sodium channel expression, nociceptor
excitability, and pain thresholds. J Neurosci. 30:10860–10871.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Smirnova L, Gräfe A, Seiler A, Schumacher
S, Nitsch R and Wulczyn FG: Regulation of miRNA expression during
neural cell specification. Eur J Neurosci. 21:1469–1477. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hohjoh H and Fukushima T: Expression
profile analysis of microRNA (miRNA) in mouse central nervous
system using a new miRNA detection system that examines
hybridization signals at every step of washing. Gene. 391:39–44.
2007. View Article : Google Scholar
|
|
26
|
Bak M, Silahtaroglu A, Møller M,
Christensen M, Rath MF, Skryabin B, Tommerup N and Kauppinen S:
MicroRNA expression in the adult mouse central nervous system. RNA.
14:432–444. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tang X, Gal J, Zhuang X, Wang W, Zhu H and
Tang G: A simple array platform for microRNA analysis and its
application in mouse tissues. RNA. 13:1803–1822. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Strickland ER, Hook MA, Balaraman S, Huie
JR, Grau JW and Miranda RC: MicroRNA dysregulation following spinal
cord contusion: implications for neural plasticity and repair.
Neuroscience. 186:146–160. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liu NK, Wang XF, Lu QB and Xu XM: Altered
microRNA expression following traumatic spinal cord injury. Exp
Neurol. 219:424–429. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Madathil SK, Nelson PT, Saatman KE and
Wilfred BR: MicroRNAs in CNS injury: potential roles and
therapeutic implications. Bioessays. 33:21–26. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu G, Detloff MR, Miller KN, Santi L and
Houlé JD: Exercise modulates microRNAs that affect the PTEN/mTOR
pathway in rats after spinal cord injury. Exp Neurol. 233:447–456.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Purohit V, Rapaka RS, Rutter J and
Shurtleff D: Do opioids activate latent HIV-1 by down-regulating
anti-HIV microRNAs? J Neuroimmune Pharmacol. 7:519–523. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Leone V, D'Angelo D, Ferraro A, Pallante
P, Rubio I, Santoro M, Croce CM and Fusco A: A TSH-CREB1-microRNA
loop is required for thyroid cell growth. Mol Endocrinol.
25:1819–1830. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhao C, Huang C, Weng T, Xiao X, Ma H and
Liu L: Computational prediction of MicroRNAs targeting GABA
receptors and experimental verification of miR-181, miR-216 and
miR-203 targets in GABA-A receptor. BMC Res Notes. 5:912012.
View Article : Google Scholar : PubMed/NCBI
|