Introduction
The incidence of prostate cancer has increased over
several decades such that it is now the most commonly diagnosed
cancer among males in Europe and the USA (1,2). The
exact mechanisms for the carcinogenesis and progression of prostate
cancer are not yet clear. Several prostate cancer-associated genes
have been identified to have significant roles in these processes
(3). The inadequacies of current
diagnostic, prognostic, predictive and therapeutic markers have
created a requirement for novel prostate cancer-associated genes
(4,5), therefore it is vital to search for
these new genes.
In the present study, we performed comprehensive
expression analysis of expressed sequence tags (ESTs), microarray
data and serial analysis of gene expression (SAGE) to screen
candidate prostate cancer-associated genes in silico.
Prostate cancer-associated gene 1 (PCAG1) was identified. mRNA and
protein expression levels were evaluated in human common normal
tissues, common malignant tumors, prostate cancers and paired
adjacent normal prostate tissues. PCAG1 mRNA was highly expressed
in prostate cancer tissues, while few normal tissues showed a low
level of expression. tissues with low level of expression. PCAG1
protein was highly expressed in prostate cancer tissues, while few
common normal tissues, other common malignant tumors and paired
adjacent normal prostate tissues demonstrated even lower levels of
expression. Furthermore, PCAG1 protein was mainly shown to be
localized within the cytoplasm and mitochondria using
immunofluorescent staining.
Materials and methods
Tissue specimens
Paired prostate adenocarcinoma and adjacent normal
tissues were collected for RT-PCR from 14 patients who underwent
surgery between February 2009 and December 2010. The fresh tissues
were immediately immersed in RNAlater solution (Qiagen, Hilden,
Germany) following surgical resection, stored at 4°C overnight to
allow thorough penetration of the tissue and then frozen at −80°C
until RNA extraction was performed. In addition, 38 paired
paraffin-embedded samples of renal cell carcinoma and adjacent
normal renal tissue that had been collected between 2008 and 2010
were stored for immunohistochemical assay. The patients had not
received any therapy for prostate cancer prior to surgery. All
samples were pathologically confirmed. Collection and use of
patient samples were reviewed and approved by the Hospital Ethics
Committees and informed consent was obtained from all patients.
Multiple organ normal tissue microarrays (catalog
number, FDA999a) and high-density multiple organ tumor and normal
tissue microarrays (catalog number, MC5003) were purchased from
Biomax (Rockville, MD, USA). The FDA999a tissue microarray
contained 3 cases of 32 common types of normal human organs,
including tissue from the cerebrum, cerebellum, adrenal gland,
ovary, pancreas, parathyroid gland, hypophysis, testis, thyroid
gland, breast, spleen, tonsil, thymus, bone marrow, lung, cardiac
muscle, esophagus, stomach, small intestine, colon, liver, salivary
gland, kidney, prostate, endometrium, uterine cervix, skeletal
muscle, skin, peripheral nerve, mesothelium, retina and larynx.
Each sample type was obtained from 3 normal human individuals and
had a single core per case. The MC5003 tissue microarray contained
20 common types of human cancer tissues and had a single core per
case. These tissues were from cases of bladder urothelial
carcinoma, brain astrocytoma/cerebrum glioblastoma, breast invasive
ductal carcinoma, cervical squamous cell carcinoma, colon
adenocarcinoma, esophageal carcinoma, head and neck carcinoma,
kidney clear cell carcinoma, liver hepatocellular carcinoma, lung
carcinoma, lymph node diffuse B cell lymphoma and Hodgkin disease,
malignant melanoma, ovary carcinoma, pancreas adenocarcinoma,
prostate adenocarcinoma, soft tissue fibrosarcoma, stomach
adenocarcinoma, testicular cancer, thyroid papillary carcinoma and
uterus endometrioid adenocarcinoma.
Perl programming to screen candidate
genes in silico
To screen out new prostate cancer-associated genes
in silico, the following steps were applied. Firstly, a
secondary classification database for EST libraries was generated
based on the Cancer Genome Anatomy Project (CGAP) information on
EST libraries (6). The CGAP EST
libraries were classified into two classes: libraries from
non-fetal, non-germinal and non-placental normal tissues (NTs) and
libraries from prostate cancer. Secondly, Unigene clusters with
<20 ESTs from NT libraries and >2 ESTs from TCC libraries
were screened out. Thirdly, the frequency of the best SAGE tag in
NT for each candidate gene was counted based on CGAP SAGE data and
Unigene clusters, while <20 SAGE tags from NTs were retained for
further analysis. Lastly, the candidate genes were analyzed
manually with Affymetrix HG-U133A/B microarray data of normal
tissues downloaded from the University of California at Los Angeles
public core.
RT-PCR
Pooled cDNA from 16 normal tissues was obtained from
Clontech (Palo Alto, CA, USA). Total RNA from cancerous tissues was
extracted using the TRIzol solution (Invitrogen, Carlsbad, CA, USA)
according to the manufacturer’s instructions and with RNase-free
DNase I used to remove the DNA contamination. Total RNA (2 μg) was
treated with M-MLV Reverse Transcriptase (Fermentas, Pittsburgh,
PA, USA) to synthesize the first-strand cDNA according to the
manufacturer’s recommendations. The cDNA was then subjected to
RT-PCR for evaluation of the relative mRNA levels of PCAG1 and
GAPDH (as an internal control) with the corresponding primer pairs:
PCAG1 sense, 5′-CCGCAGAAGAACAGAAGACC-3′ and antisense,
5′-CAGAGAAACCATGGGCACTT-3′; GAPDH sense,
5′-CACCAGGGCTGCTTTTAACTC-3′ and antisense,
5′-GAAGATGGTGATGGGATTTC-3′. The amplification conditions were 50°C
(2 min) and 95° (2 min) for 1 cycle, then 95°C (15 sec), 55°C (30
sec) and 72°C (40 sec) for 35 cycles. Each PCR product was
separated by 7% polycrylamide gel electrophoresis and visualized
with ethidium bromide, excised from each gel and dissolved
overnight using 0.5 N quaternary ammonium hydroxide in toluene
(Soluenen 350; Packard Instruments Co., Meriden, CT, USA). After
the addition of scintillation fluid, the samples were measured by
scintillation spectroscopy. The mRNA results are expressed as a
ratio of counts per min (cpm) of PCAG1 divided by GAPDH and were
assigned to a semiquantitative score (+ to +++).
Generation of anti-PCAG1 antibody
Full-length cDNA of PCAG1 was inserted into pET23d
(Novagen, Darmstadt, Germany) and expressed in Escherichia
coli. The polypeptide was purified by applying it to a Ni-NTA
resin (Qiagen) and used to immunize rabbits. Rabbit antiserum was
then purified by applying it to the polypeptide column.
Immunohistochemistry assay
Immunohistochemistry procedures were performed with
classical protocols. In brief, paraffin-embedded specimens were cut
into 5-μm sections and baked at 65°C for 30 min. The sections were
deparaffinized with xylene and rehydrated. Sections were submerged
in 0.01 mol citrate antigenic retrieval buffer (pH 6.0) and
microwaved for antigenic retrieval. The sections were then treated
with 3% hydrogen peroxide in methanol to quench the endogenous
peroxidase activity, followed by incubation with 10% bovine serum
albumin to block the non-specific binding. The PCAG1 protein was
detected using a rabbit polyclonal antibody against PCAG1. The
specimens were incubated with anti-PCAG1 antibody (1:100) overnight
at 4°C. The negative immunohistochemical control procedure included
replacement of the primary antibodies by antibody diluent.
Subsequent to being washed in PBS, sections were
treated with MaxVision HRP-Polymer anti-Rabbit IHC kit (Maixin Bio,
Fujian, China) at 37°C for 15–20 min. The tissue sections were
immersed in 3-amino-9-ethylcarbazole, counterstained with Mayer’s
hematoxylin, dehydrated and mounted in a crystal mount.
The degree of immunostaining of formalin-fixed,
paraffin-embedded sections was reviewed and scored by two
independent observers. The intensity of the staining varied from
weak to strong. The staining intensity was graded according to the
mean optical density method (7–9): 0,
no staining; 1, weak staining (light yellow); 2, moderate staining
(yellow brown); and 3, strong staining (brown).
Immunofluorescence assay
The PC3 cells were fixed in 4% paraformaldehyde,
permeabilized with 0.2% Triton X-100 and then incubated with
anti-PCAG1 antibody at a concentration of 20 mg/ml for 1 h at room
temperature, followed by an incubation with anti-rabbit IgG-FITC
(Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA). Intracellular
localization of the PCAG1 protein was examined under fluorescent
microscopy.
Statistical analysis
All quantitative data were analyzed using Student’s
t-test. Statistical analysis was performed using the SPSS 17.0
package and P<0.05 was considered to indicate a statistically
significant difference.
Results
Results of in silico screening and RT-PCR
evaluation
A total of 45 candidate clusters were first screened
out by perl programming based on EST data; after secondary analysis
using SAGE and microarray data, the result was narrowed to twelve
clusters with reconfirmed low expression in normal tissues. The
twelve clusters were ranked according to the number of ESTs from
prostate cancer and RT-PCR was performed to evaluate the prostate
cancer specificity. In the first five genes evaluated, C12orf62
(chromosome 12 open reading frame 62, Gene ID: 84987) showed high
specificity for prostate cancer and we temporarily named it
prostate cancer-associated gene 1 (PCAG1).
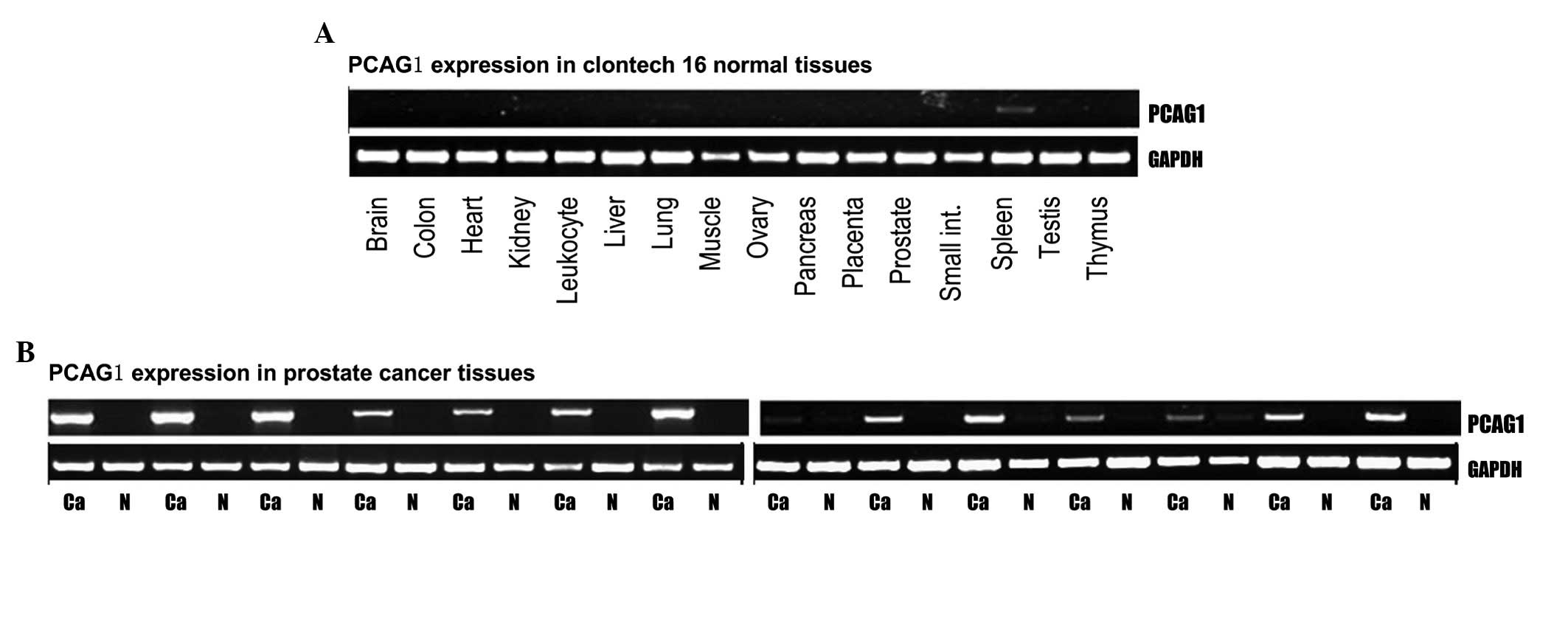
PCAG1 mRNA expression in normal tissues
and prostate cancer tissues analyzed by RT-PCR
The transcriptional level of PCAG1 was determined by
RT-PCR assays in all 16 normal human tissues (Clontech). Our
results showed that PCAG1 mRNA was absent in the 15 pooled normal
tissues (including normal prostate tissue) but registered at a low
level in spleen tissue (+; Fig.
1A). By contrast, expression of PCAG1 mRNA in each of the 14
cases of prostate cancer was significantly higher than in the
paired adjacent normal prostate tissues with approximately half of
the cases scoring a high expression level (+++; Fig. 1B).
Immunohistochemical observations of PCAG1
protein expression in 32 types of normal tissue and 20 types of
human common malignant tumor
In order to determine the expression of the PCAG1
protein, rabbit anti-PCAG1 antibody was used as described in
Materials and methods. The expression of PCAG1 protein was
determined by immunohistochemistry in microarrays of 32 types of
normal human tissues (Biomax; Fig.
2A). Of these tissues, 29/32 types (including normal prostate
tissue) showed negative staining of PCAG1 while low levels were
observed in the adrenal gland, parathyroid gland and liver tissues
(Fig. 2A).
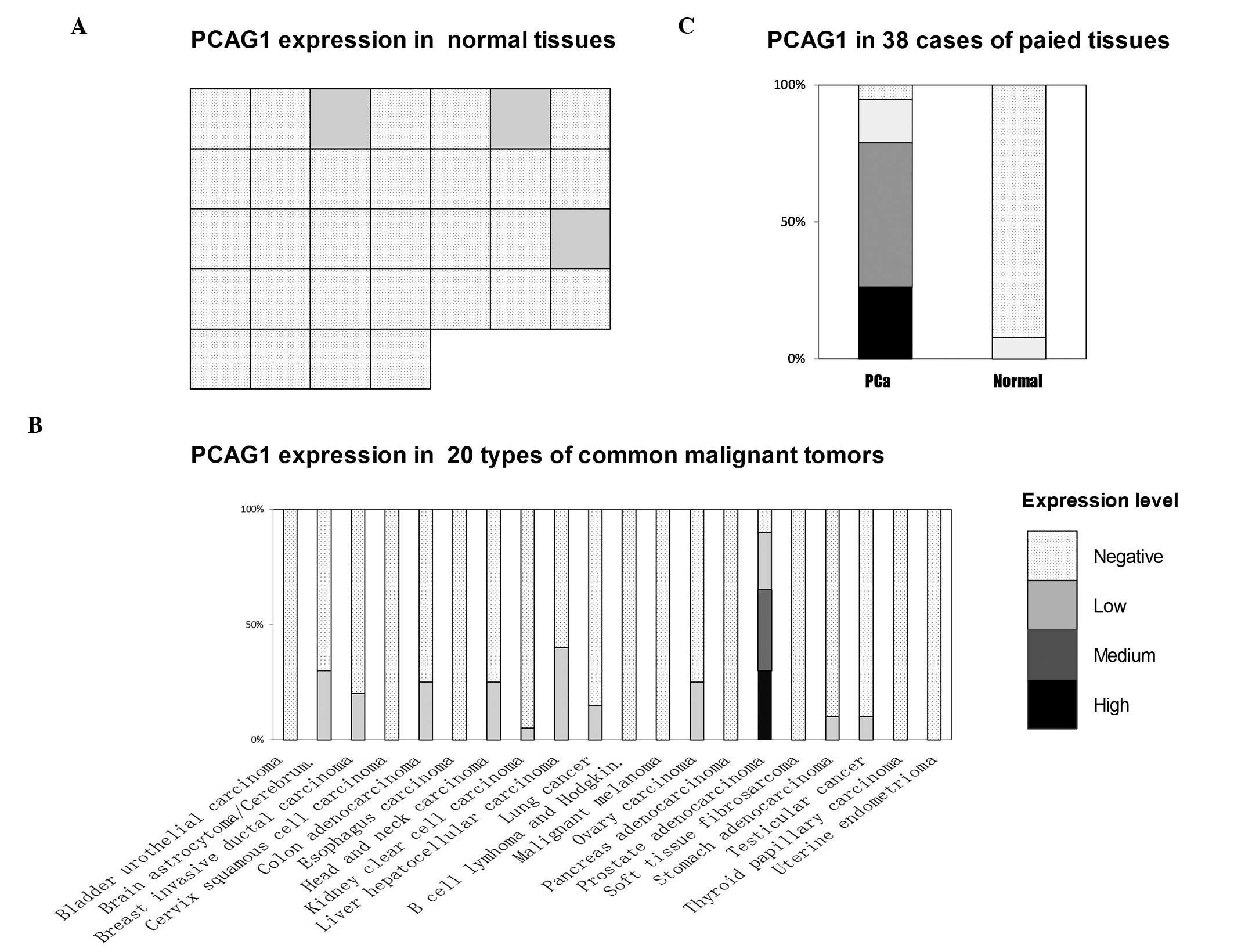 | Figure 2Expression of PCAG1 protein analyzed
with immunohistochemistry (IHC). (A) Expression of PCAG1 protein in
32 types of normal human tissues. Each IHC dot (square) represents
the expression level of PCAG1 in one tissue. From left to right and
top to bottom, these dots represent cerebrum, cerebellum, adrenal
gland, ovary, pancreas, parathyroid gland, hypophysis, testis,
thyroid gland, breast, spleen, tonsil, thymus, bone marrow, lung,
cardiac muscle, esophagus, stomach, small intestine, colon, liver,
salivary gland, kidney, prostate, endometrium, uterine cervix,
skeletal muscle, skin, peripheral nerve, mesothelium, retina and
larynx. The expression levels of PCAG1 in the adrenal gland,
parathyroid gland and liver were low while the other 29 tissues
revealed negative results, including the normal prostate tissue.
(B) Expression of PCAG1 protein in 20 types of common human
malignant tumors. (C) Expression of PCAG1 protein in 38 cases of
paired prostate adenocarcinoma (PCa) and paired adjacent normal
prostate tissues. The expression in prostate adenocarcinoma
revealed 10 cases showing high expression levels, 20 showing medium
levels, 6 showing low levels and 2 with a negative result. Only
3/38 cases of paired adjacent normal prostate tissues showed
positive (low) levels and 35/38 cases gave negative results. PCAG1,
prostate cancer-associated gene 1. |
Expression of PCAG1 protein was also determined by
immunohistochemistry in microarrays of 20 types of human common
malignant tumors (Biomax; Fig. 2B
and Table I). While 18/20 cases of
prostate adenocarcinoma showed positive results, PCAG1 protein
expression in other types of cancer was scarce when present at all;
only 41/380 other cancer cases demonstrated positive results at
even a low level (Table I).
 | Table IExpression of PCAG1 in various
malignant tumor tissues evaluated with IHC. |
Table I
Expression of PCAG1 in various
malignant tumor tissues evaluated with IHC.
| Expression |
|---|
|
|
|---|
| Malignant tumors | Positive
(rate,%) | High | Medium | Low | Negative | Total |
|---|
| Bladder urothelial
carcinoma | 0/20 (0) | 0 | 0 | 0 | 20 | 20 |
| Brain
astrocytoma/cerebrum glioblastoma | 6/20 (30) | 0 | 0 | 6 | 14 | 20 |
| Breast invasive
ductal carcinoma | 4/20 (20) | 0 | 0 | 4 | 16 | 20 |
| Cervix squamous cell
carcinoma | 0/20 (0) | 0 | 0 | 0 | 20 | 20 |
| Colon
adenocarcinoma | 5/20 (25) | 0 | 0 | 5 | 15 | 20 |
| Esophageal
carcinoma | 0/20 (0) | 0 | 0 | 0 | 20 | 20 |
| Head and neck
carcinoma | 5/20 (25) | 0 | 0 | 5 | 15 | 20 |
| Kidney clear cell
carcinoma | 1/20 (5) | 0 | 0 | 1 | 19 | 20 |
| Liver hepatocellular
carcinoma | 8/20 (40) | 0 | 0 | 8 | 12 | 20 |
| Lung cancer | 3/20 (15) | 0 | 0 | 3 | 17 | 20 |
| B cell lymphoma and
Hodgkin disease | 0/20 (0) | 0 | 0 | 0 | 20 | 20 |
| Malignant
melanoma | 0/20 (0) | 0 | 0 | 0 | 20 | 20 |
| Ovary carcinoma | 5/20 (25) | 0 | 0 | 5 | 15 | 20 |
| Pancreas
adenocarcinoma | 0/20 (0) | 0 | 0 | 0 | 20 | 20 |
| Prostate
adenocarcinoma | 18/20 (90) | 6 | 7 | 5 | 2 | 20 |
| Soft tissue
fibrosarcoma | 0/20 (0) | 0 | 0 | 0 | 20 | 20 |
| Stomach
adenocarcinoma | 2/20 (10) | 0 | 0 | 2 | 18 | 20 |
| Testicular
cancer | 2/20 (10) | 0 | 0 | 2 | 18 | 20 |
| Thyroid papillary
carcinoma | 0/20 (0) | 0 | 0 | 0 | 20 | 20 |
| Uterus endometrioid
adenocarcinoma | 0/20 (0) | 0 | 0 | 0 | 20 | 20 |
Immunohistochemical observations of PCAG1
protein expression in 38 cases of paired prostate adenocarcinoma
and paired adjacent normal prostate tissues
In order to further determine the expression level
of PCAG1 protein in prostate adenocarcinoma, immunohistochemistry
assays were conducted in 38 cases of paired prostate adenocarcinoma
and paired adjacent normal prostate tissues. As demonstrated in
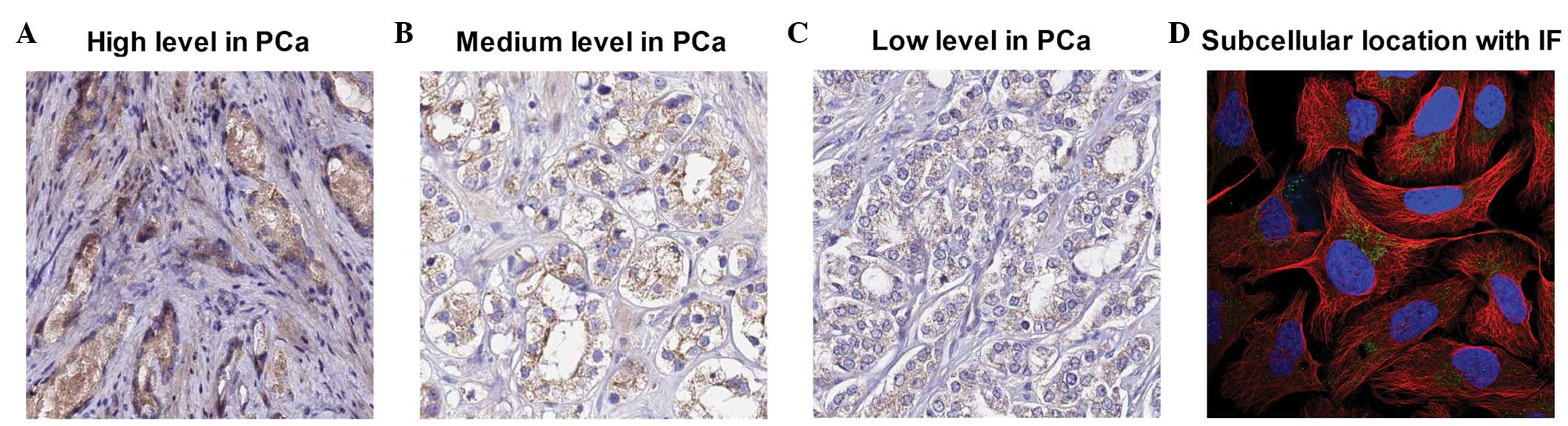
Fig. 2C, the most substantial
PCAG1-positive expression results were identified by cytoplasmic
staining in 36/38 prostate adenocarcinoma cases (Fig. 3A-C), with 10 cases showing high
expression levels, 20 showing medium levels and 6 showing low
levels. In paired adjacent normal prostate tissues, only 3/38 cases
demonstrated low level positive staining while 35/38 cases were
negative.
Immunofluorescent staining for
subcellular localization of PCAG1 protein
To further determine the subcellular localization of
the PCAG1 protein, we conducted an immunofluorescence assay in the
human prostate cancer cell line PC3. As demonstrated in Fig. 3D, immunofluorescent staining of
human prostate cancer PC3 cells for PCAG1 revealed a positive
result in the mitochondria.
Discussion
In the present study, microarray data, ESTs and SAGE
were used to increase the efficacy of expression analysis, as each
technique had varying advantages and disadvantages. To screen out
the prostate cancer-associated genes based on the databases of
these three techniques, a specialized streamlined program was used
prior to the expression patterns of the candidate genes being
evaluated by immunohistochemistry and RT-PCR. Consequently, PCAG1
was identified. The results showed that screening in silico
with this comprehensive strategy made the procedure to explore new
prostate cancer-associated genes more efficient and accurate.
PCAG1 was temporarily named as PCAG1 in our
screening procedure for prostate cancer-associated genes but is
also designated as C12orf62 (chromosome 12 open reading frame 62)
(10–12).
The full-length cDNA of PCAG1 was mapped to the
12q13.12 positive strand with two exons. Using Uniprot (13) and Nextprot (14), PCAG1 was predicted to be a
transmembrane protein with a transmembrane domain of 22 amino
acids. The present study showed that the PCAG1 protein was mainly
localized within the cytoplasm and mitochondria.
The present study also demonstrated that PCAG1 mRNA
and protein were expressed highly in prostate cancer but were
almost absent in all normal tissues and other common malignant
tumors. This indicates that PCAG1 has the features of prostate
cancer-associated genes. Therefore, the functional significance of
PCAG1 in prostate cancer deserves further investigation. The
prostate cancer-specific expression of PCAG1 suggests a unique
transcriptional regulation. Clarification of the function and
transcriptional mechanism of PCAG1 may aid the elucidation of the
mechanisms of carcinogenesis and progression of prostate cancer.
The mechanisms behind prostate cancer carcinogenisis and
progression may be resolved by a further elucidation of the
functions and transcriptional mechanisms of PCAG1. Moreover, the
unique expression pattern of PCAG1 suggests its potential as a
marker of prostate cancer in early detection, forecasting disease
severity, choosing treatments and monitoring responses to
therapies.
In summary, the present study demonstrated that
PCAG1 mRNA was highly expressed in prostate cancer tissues while
being almost absent in all tested common normal tissues and paired
adjacent normal prostate tissues. Furthermore, PCAG1 protein was
also highly expressed in prostate cancer tissues while few common
normal tissues, other common malignant tumors and paired adjacent
normal prostate tissues showed even low levels of expression.
Further analysis of the functions and transcriptional mechanisms of
PCAG1 may aid in the understanding of the mechanisms for the
carcinogenesis and progression of prostate cancer. The unique
expression pattern of PCAG1 suggests its potential in certain
clinical applications.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (No. 81101922).
References
|
1
|
Bray F, Lortet-Tieulent J, Ferlay J,
Forman D and Auvinen A: Prostate cancer incidence and mortality
trends in 37 European countries: an overview. Eur J Cancer.
46:3040–3052. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel R, Ward E, Brawley O and Jemal A:
Cancer statistics, 2011: the impact of eliminating socioeconomic
and racial disparities on premature cancer deaths. CA Cancer J
Clin. 61:212–236. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Shen MM and Abate-Shen C: Molecular
genetics of prostate cancer: new prospects for old challenges.
Genes Dev. 24:1967–2000. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sardana G, Dowell B and Diamandis EP:
Emerging biomarkers for the diagnosis and prognosis of prostate
cancer. Clin Chem. 54:1951–1960. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shariat SF, Scherr DS, Gupta A, et al:
Emerging biomarkers for prostate cancer diagnosis, staging, and
prognosis. Arch Esp Urol. 64:681–694. 2011.(In English and
Spanish).
|
|
6
|
Krizman DB, Wagner L, Lash A, Strausberg
RL and Emmert-Buck MR: The Cancer Genome Anatomy Project: EST
sequencing and the genetics of cancer progression. Neoplasia.
1:101–106. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bao S, Ouyang G, Bai X, et al: Periostin
potently promotes metastatic growth of colon cancer by augmenting
cell survival via the Akt/PKB pathway. Cancer Cell. 5:329–339.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chow WH, Dong LM and Devesa SS:
Epidemiology and risk factors for kidney cancer. Nat Rev Urol.
7:245–257. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tsuchiya A, Sakamoto M, Yasuda J, et al:
Expression profiling in ovarian clear cell carcinoma:
identification of hepatocyte nuclear factor-1 beta as a molecular
marker and a possible molecular target for therapy of ovarian clear
cell carcinoma. Am J Pathol. 163:2503–2512. 2003. View Article : Google Scholar
|
|
10
|
Gerhard DS, Wagner L, Feingold EA, et al;
MGC Project Team. The status, quality, and expansion of the NIH
full-length cDNA project: the Mammalian Gene Collection (MGC).
Genome Res. 14:2121–2127. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lamesch P, Li N, Milstein S, et al:
hORFeome v3.1: a resource of human open reading frames representing
over 10,000 human genes. Genomics. 89:307–315. 2007.PubMed/NCBI
|
|
12
|
Strausberg RL, Feingold EA, Grouse LH, et
al; Mammalian Gene Collection Program Team. Generation and initial
analysis of more than 15,000 full-length human and mouse cDNA
sequences. Proc Natl Acad Sci USA. 99:16899–16903. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
UniProt Consortium. Reorganizing the
protein space at the Universal Protein Resource (UniProt). Nucleic
Acids Res. 40:D71–D75. 2012.PubMed/NCBI
|
|
14
|
Lane L, Argoud-Puy G, Britan A, et al:
neXtProt: a knowledge platform for human proteins. Nucleic Acids
Res. 40:D76–D83. 2012. View Article : Google Scholar : PubMed/NCBI
|