Introduction
Hypertrophic scars are a malformation frequently
ecountered by plastic surgeons. Characteristic morphological and
ultrastructural changes in hypertrophic scars include the abnormal
and excessive deposition of extracellular matrix (ECM). Excessive
scar fibrosis results from the increased expression of several ECM
proteins, including collagen. The collagen genes pro-COL1A1 and
pro-COL1A2 are responsible for the synthesis of pro1 and pro2
polypeptides of type I collagen and they are synthesized at a ratio
of 2:1 to form a stable triple helical type I collagen molecule in
the extracellular space (1).
Fibrosis, the excessive accumulation of collagen, is induced by
persistent pro-fibrotic growth factor signaling, including
fibroblast growth factor (FGF), platelet-derived growth factor
(PDGF) and, most predominantly, transforming growth factor-β1
(TGF-β1). Sp1 is a transcription factor that regulates the
expression of a variety of ECM genes and is involved in numerous
growth factor-related signal transduction pathways, including
TGF-β1, FGF and PDGF signaling (2).
Sp1 belongs to a family of ubiquitous transcription
factors associated with GC-rich promoters that are inducible and
involved in inflammation (3). Sp1
exists in a variety of isoforms which bind with varying affinities
to consensus sequences that form a transcriptional network to
fine-tune gene expression. Sp1-dependent transcription is altered
during cell growth through increased gene expression.
Treating fibroblasts with TGF-β1 promotes the
synthesis of type I collagen (4).
Chronic exposure to TGF-β1 in lungs increases the deposition of
type I collagen, causing excessive fibrosis, which may increase
morbidity and mortality. Direct silencing of the COL1A1 gene is one
method to reduce collagen type I synthesis. A novel double-stranded
(ds) DNA decoy with phosphorothioate (PT) linkages containing the
TGF-β cis-element has been observed in the distal promoter region
of the COL1A1 gene (5). This decoy
blocks collagen type I synthesis in human hypertrophic scar
fibroblasts (HSFs) stimulated by TGF-β1 (6). These findings support the concept
that a decoy ODN may inhibit collagen type I synthesis at the
transcriptional and translational levels. Therefore, we performed
this ex vivo study to determine whether a Sp1 decoy ODN
could inhibit HSF proliferation, TGF-β signaling and collagen
production.
Materials and methods
Cell culture
Control human fibroblasts were isolated from normal
human skin (the individual biopsies ranged in size from 80 to 150
mm3; n=5). The size of hypertrophic scars used for
culture ranged from 500 to 1,300 mm3 (n=5). The samples
were collected at the Shanghai 6th People’s Hospital (Shanghai,
China) after approval by the ethics committee for human studies.
The patients provided informed consent and the patient clinical
characteristics are listed in Table
I. None of the patients had a systemic disease and none had
been previously treated for the scars. Primary human fibroblasts
were isolated from each hypertrophic scar sample before scar tissue
was fixed in formalin for routine histological examination. The
tissue sections were cut into 1–3 mm cubes and incubated with 200
U/ml type I collagenase for 4 h at 37°C. The cell cultures were
maintained in Dulbecco’s modified Eagle’s medium (DMEM)
supplemented with 10% fetal bovine serum (FBS), 2 mM glutamine, 100
U/ml penicillin and 100 mg/ml streptomycin at 37°C in a humidified
incubator with 5% CO2. HSF cultures were analyzed at
passages 3–8.
 | Table IPatient characteristics. |
Table I
Patient characteristics.
| Age (years) | Gender | Biopsy site | Duration (years) |
|---|
| 32 | Male | Chest | 3 |
| 32 | Male | Shoulder | 4 |
| 18 | Female | Earlobe | 1 |
| 21 | Female | Earlobe | 2 |
| 28 | Female | Chest | 3 |
Synthesis and modification of the Sp1
decoy ODNs
The sense ODN containing the binding sequence for
Sp1 was synthesized and PT-modified at the first and last three
bases. The antisense ODN was 5-carboxyfluorescein (5′-FAM) and
3′-biotin-labeled (Bio Basic Inc., Shanghai, China). The ds PT
decoy ODNs were prepared by annealing the sense and antisense
strands in 200 mM NaCl at 95°C for 7 min and slowly cooling to 4°C.
These were designated Sp1 decoy ODNs.
Mutated Sp1 decoy ODNs (designated mut-Sp1 decoy
ODNs) were mutated at two positions (shown as lowercase letters) in
the Sp1 binding sequence of the Sp1 decoy ODNs. Inspection of the
resulting nucleotide sequences showed no sequence homology to other
known transcription factors by searching databases on
transcriptional regulation for Sp1 decoy ODNs
(5′-gccccgatcttttgatcggggcggggcgagcttttgctcgcccc-3′) and
scrambled-Sp1 decoy ODNs
(5′-ctgactgactttttagtcagtcagtcagtcagtcttttcagtcagtca-3′).
Sp1 decoy ODN distribution test
Control fibroblasts and HSFs were plated on glass
coverslips and the coverslips were put into 35-mm dishes in DMEM
containing 10% FBS. When the cells reached 60–70% confluence, the
cells were serum-starved for 24 h in DMEM containing 0.5% FBS and
transfected with FAM-labeled-Sp1 decoy ODNs. Briefly, 1 μg of the
Sp1 decoy ODNs were mixed with 3–6 μl of lipofectamine plus reagent
(Invitrogen, Carlsbad, CA, USA) and transfected with modified
supplier instructions. The Sp1 decoy ODNs and liposome mixtures
were added into each well and incubated at 37°C for 3, 6, 12, 24
and 48 h. The HSFs were stained with 4′,6-diamidino-2-phenylindole
(DAPI) and observed by laser scanning confocal fluorescence
microscopy.
ODN transfection
We transfected Sp1 decoy ODNs into HSFs using the
cationic liposome method. Briefly, HSFs were seeded on a 6-well
plate and cultured in DMEM containing 10% FBS with antibiotics.
When the cells reached 80–90% confluence, the cells were
transfected with 10 nM Sp1 and the scrambled Sp1 decoy-ODNs in
Lipofectamine Plus (0.7 μg DNA: 2 μl lipid; Invitrogen). For the
decoy ODN, lipid mixture was added to the cells according to the
manufacturer’s instructions. Following incubation at 37°C for 4 h,
the cells were cultured in fresh medium with 10% serum and
maintained until use.
Cytotoxicity assay of HSFs
HSFs were seeded in a 96-well plate
(5×103 cells/well) and cultured in DMEM containing 10%
FBS. After 24 h, the cells were transfected with Sp1 or scrambled
Sp1 decoy ODNs. Briefly, 10, 50, 100 or 200 nM Sp1 or scrambled Sp1
ODNs was mixed with Lipofectamine Plus reagent (Invitrogen) and
transfected with modified supplier instructions. After 48 h, a
WST-8 cell viability assay was performed with the Cell Counting
kit-8 (Dojindo, Kumamoto, Japan) according to the manufacturer’s
instructions. HSFs that were not transfected with cationic
liposomes were used as controls. The experiments were replicated in
triplicate.
Cell proliferation analysis
Cultured HSF proliferation was measured using the
Cell Counting kit-8 (Dojindo). HSFs were rendered quiescent by
incubation for 24 h in serum-free media. To evaluate the effect of
Sp1 decoy ODNs on HSF proliferation, Lipofectamine Plus and Sp1
decoy ODNs (100 nM) were added to the wells and the cells were
incubated at 37°C for an additional 4 h. The cell proliferation
index was determined after 48 h. Cells without transfection of
scrambled Sp1 decoy ODNs were used as controls. Each experiment was
performed in triplicate. Control fibroblast proliferation was
measured with the Cell Counting kit-8.
Electrophoretic mobility shift assays
(EMSAs)
EMSAs were performed on nuclear extracts prepared
from the control and HSFs using the ds PT decoy ODNs containing the
Sp1 binding site. The LightShift Chemiluminescent EMSA kit (Pierce,
Rockford, IL, USA) was used. In brief, 20 fmol decoy ODN probes,
with 3′-end labeled with biotin, were incubated with 1 μg nuclear
extract, 1 μg/μl poly(dI:dC), 50 mM NaCl, 200 mM EDTA, 1 M KCl, 100
mM MgCl2, 1% NP-40 and 50% glycerol. After incubation,
the samples were separated on 6% native polyacrylamide gels with a
1X Tris-borate-EDTA (TBE) running buffer (450 mM Tris, 450 mM boric
acid, 10 mM EDTA, pH 8.3) and biotin-labeled DNA was detected by
chemiluminescence. For competition experiments, 100-fold molar
excess of the unlabeled probe was added to the reaction mixture
before the addition of the labeled probe.
RNA isolation and reverse
transcription-polymerase chain reaction (RT-PCR)
Total RNA was isolated from confluent fibroblasts
using TRIzol reagent (Invitrogen) and the integrity of the RNA was
determined by agarose gel electrophoresis. For the RT-PCR, 2 μg
total RNA was reverse transcribed at 37°C for 1 h in a 25-μl
reaction volume containing 250 mM Tris-HCl, 375 mM KCl, 15 mM
MgCl2, 50 mM dithiothreitol, 10 mM dNTPs, 0.5 μg oligo
(DT) 20 primer, 100 units reverse transcriptase Moloney Murine
Leukemia Virus (reverse transcriptase M-MLV) and 25 units
ribonuclease inhibitor (Takara Bio, Inc., Shiga, Japan) and
subjected to PCR amplification with the primers listed in Table II. Glyceraldehyde-3-phosphate
dehydrogenase (GAPDH) was amplified as the internal control. RT
products (0.5-l μg) were amplified with 1 unit Taq DNA
polymerase (Takara Bio, Inc.) and 1 mM of each primer in 50 μl of
reaction mix containing 50 mM KCl, 10 mM Tris-HCl, 1.5 mM
MgCl2 and 0.02 mM each of dNTPs as follows: initial
denaturation for 3 min at 94°C, 30 cycles of amplification, 1 min
of denaturation at 94°C, different annealing temperatures for each
pair of primers, 1 min of extension at 72°C and a final elongation
for 5 min at 72°C. Parallel RT-PCR assays without reverse
transcriptase were performed for each sample to confirm that the
PCR products resulted from cDNA rather than from genomic DNA. The
PCR products (10 μl) were analyzed by 2% agarose gel
electrophoresis. Relative mRNA abundance was calculated by
densitometric analysis using computer software (Kodak Digital
Science 1D Image Analysis software; Eastman Kodak Co., Rochester,
NY, USA).
 | Table IIPrimer design and product lengths for
RT-PCR products. |
Table II
Primer design and product lengths for
RT-PCR products.
| Gene | Primer | Length (bp) | Tm |
|---|
| COL1A1 |
5-AAAGACGGGAGGGCGAGTG-3 | | |
|
5-GCCATAGGACATCTGGGAAGCAA-3 | 242 | 62 |
| COL1A2 |
5-CTCCAAGGAAATGGCAACTCA-3 | | |
|
5-AGGAACGGCAGGCGAGAT-3 | 288 | 58 |
| COL3A1 |
5-CCCACAGCCTTCTTCTACACCT-3 | | |
|
5-ACCCATTCCTCCCACTCC-3 | 241 | 60 |
| FN |
5-GCCGAATGTAGATGAGGAG-3 | | |
|
5-GTCGAGTCGCACTGGTAGA-3 | 249 | 54 |
| GAPDH |
5-GTCGTGGAGTCTACTGGCGTCTT-3 | | |
|
5-CAGTCTTCTGAGTGGCAGTGATGG-3 | 280 | 58 |
Statistical analysis
Statistical Package for Social Sciences (SPSS, Inc.,
Chicago, IL, USA), version 13.0, was used for statistical analyses
and the Wilcoxon signed rank test was used to determine if there
were differences between the groups. P<0.05 was considered to
indicate a statistically significant result.
Results
Design and identification of Sp1 decoy
ODNs
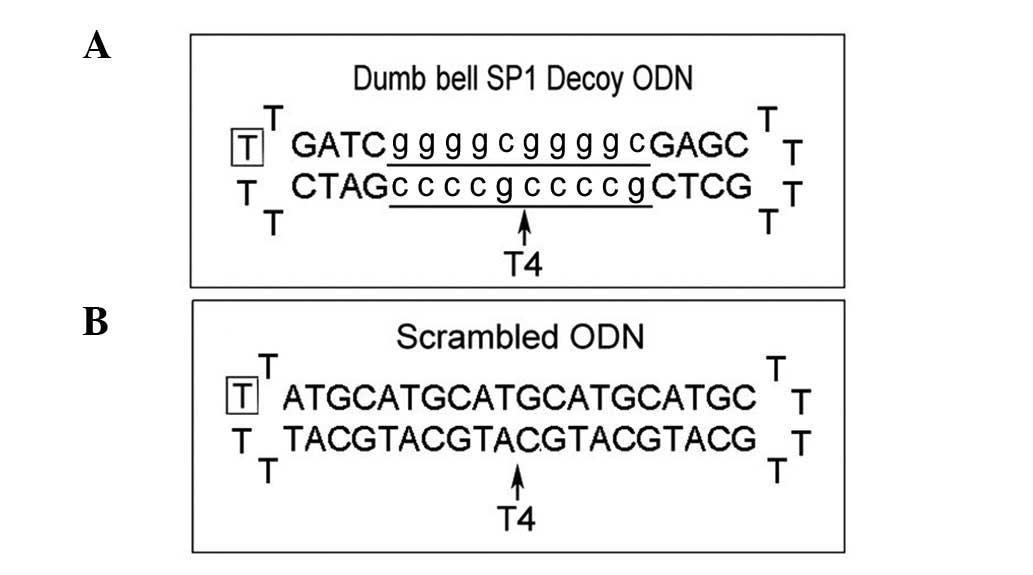
Dumbbell Sp1 decoy ODNs and scrambled Sp1 ODNs were
designed and the structure and sequence of the ODNs are shown in
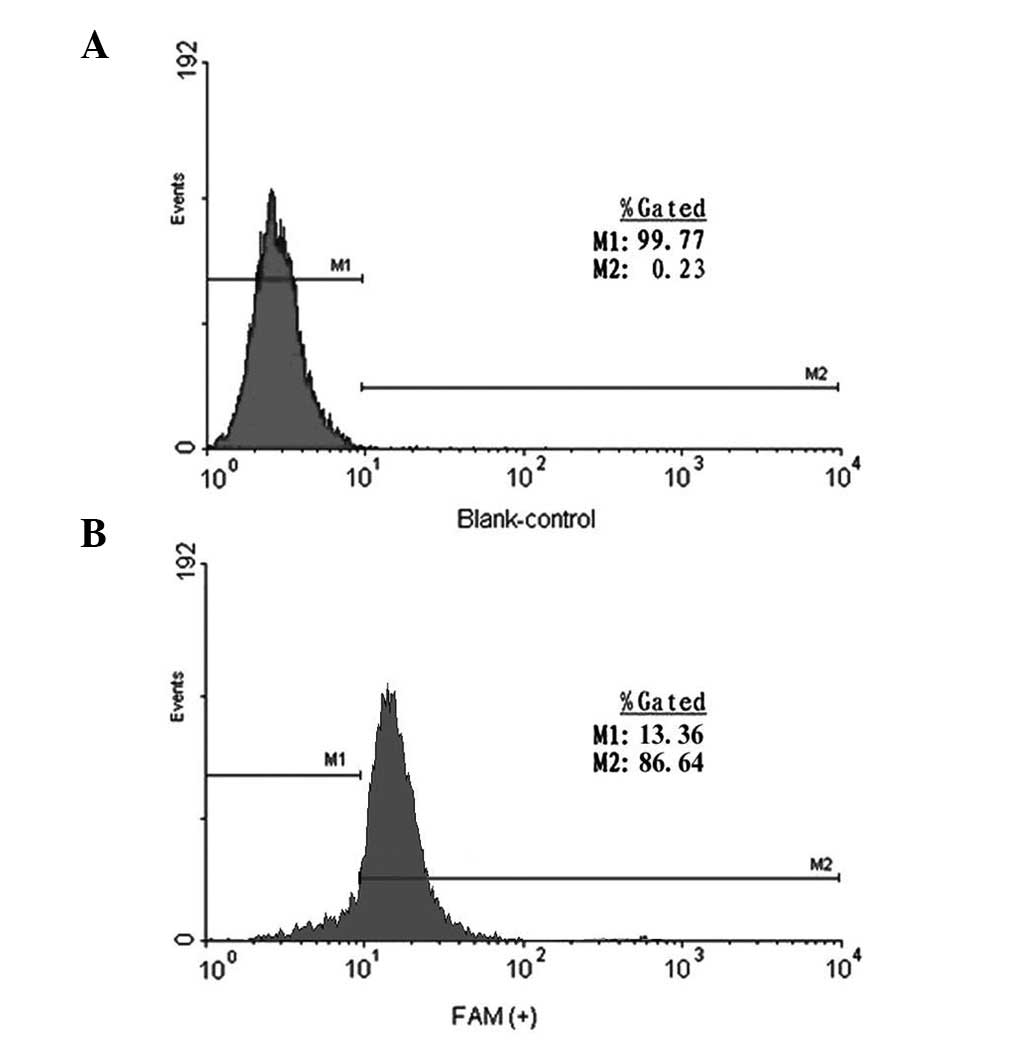
Fig. 1. Cells transfected with 100
nM FAM-labeled ODNs showed positive results by flow cytometry, with
the positive efficiency being >99%. No fluorescence was detected
in cells transfected with the blank control (Fig. 2).
Inhibition of collagen type I and III
expression by Sp1 decoy ODNs
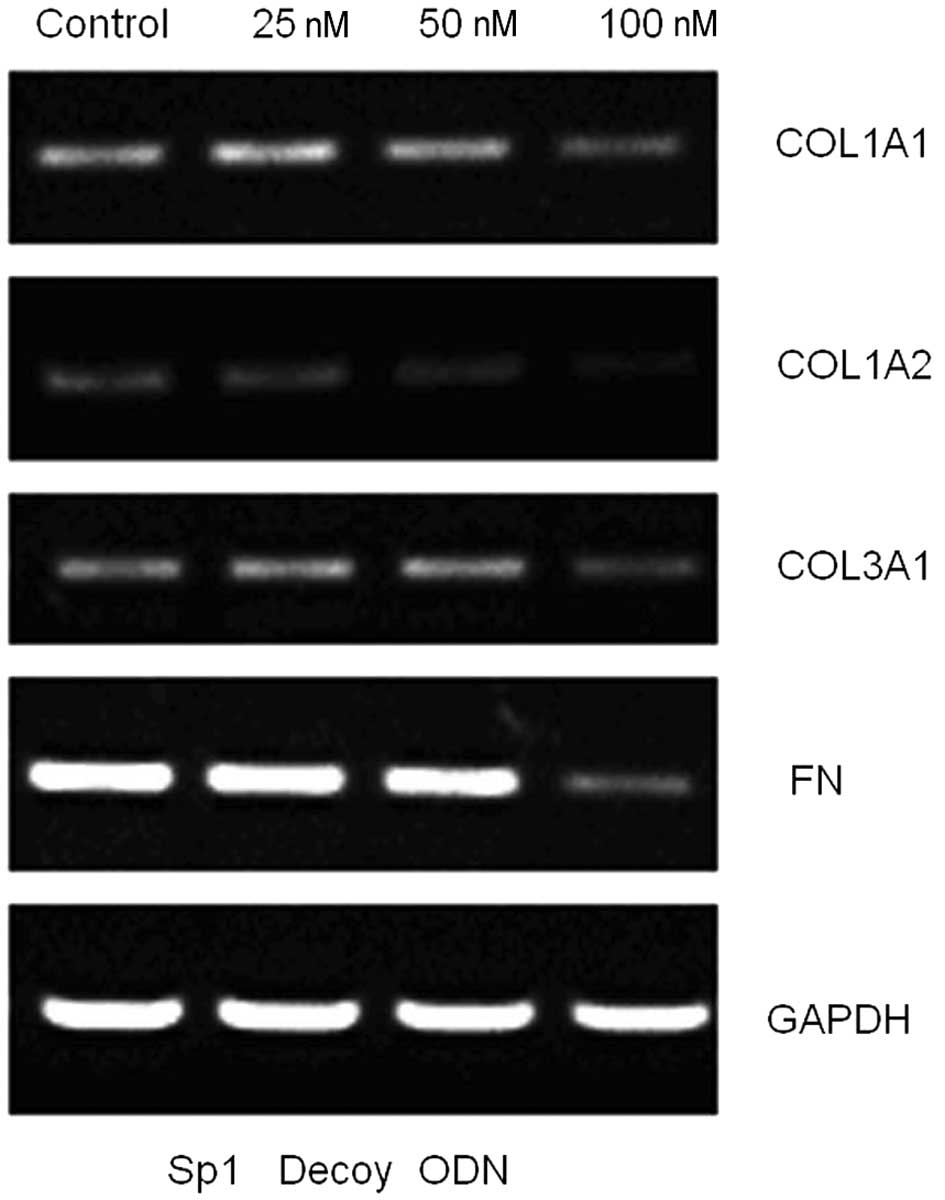
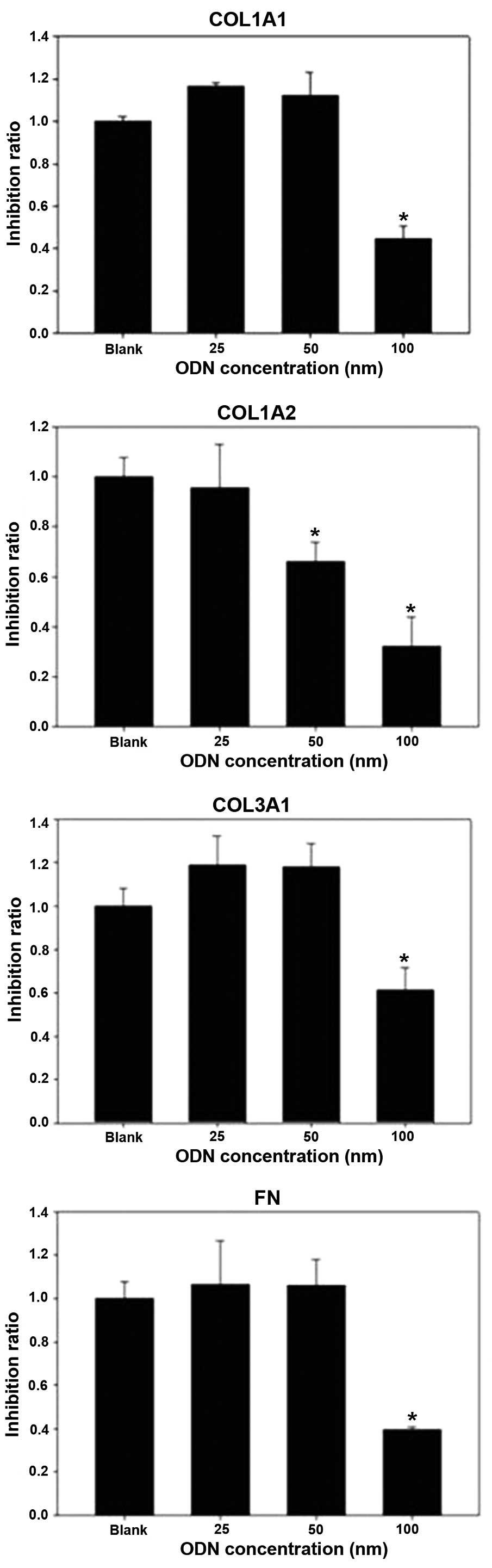
At 48 h after transfection with Sp1 decoy ODNs,
COL1A1 and COL3A1 mRNA levels were significantly reduced (both
P<0.01; Fig. 3). However, the
control ODNs had no inhibitory effect on the expression of collagen
type I or III (Fig. 4;
P>0.05).
Suppression of cell proliferation by Sp1
decoy ODNs
Cultured HSFs were transfected with FAM-labeled ODNs
targeting Sp1 transcriptional factor, with the negative control
containing a random sequence. Transfection with 100 nM Sp1 decoy
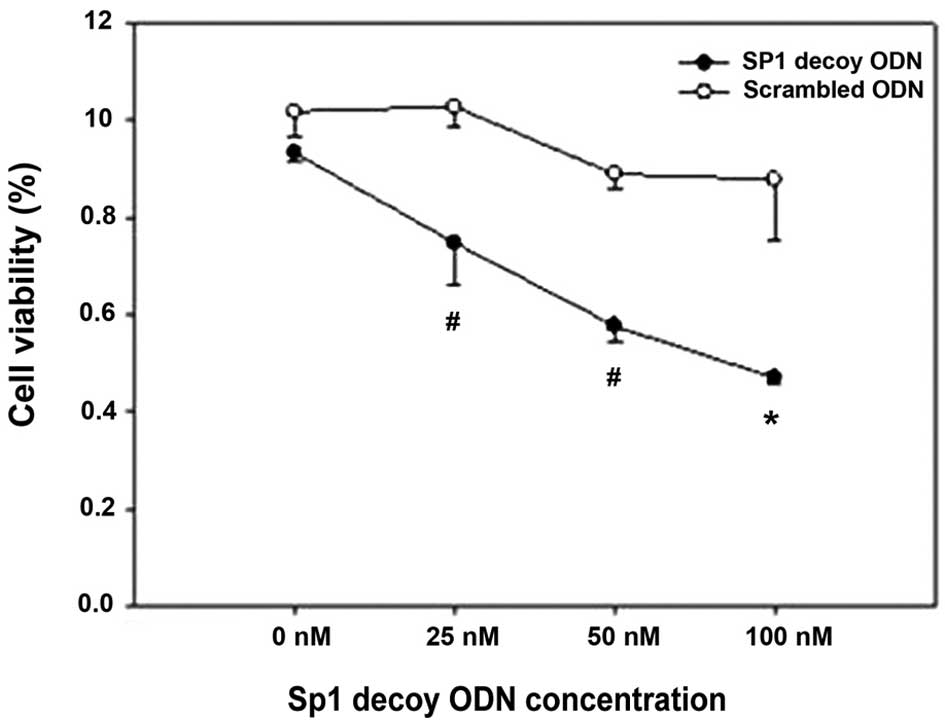
ODN resulted in minimal cell viability (Fig. 5). There were significant
differences between cells transfected with the Sp1 decoy ODN and
scrambled ODN at the 25, 50 and 100 nM doses. No significant
difference was observed between cells transfected with the Sp1
decoy ODN and scrambled Sp1 ODN effect at 0 nM (P=0.21). HSFs
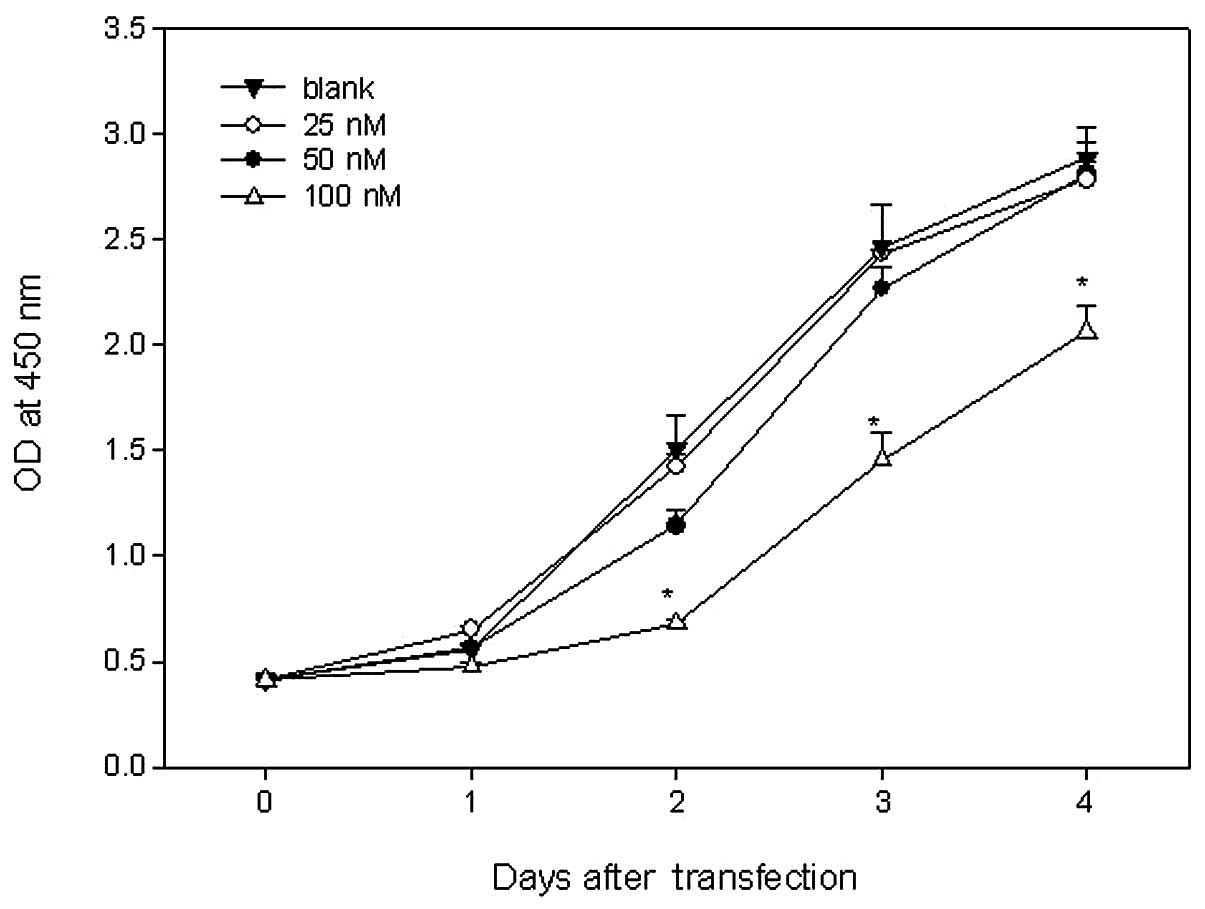
treated with 100 nM Sp1 ODN had minimal growth potential. There
were significant differences between cells treated with 100 nM Sp1
ODN and the other concentrations at 2 days after transfection.
There were no significant differences between the cells treated
with the 25 and 50 nM dose (P=0.23; Fig. 6).
Location of Sp1 decoy ODNs in cells after
transfection
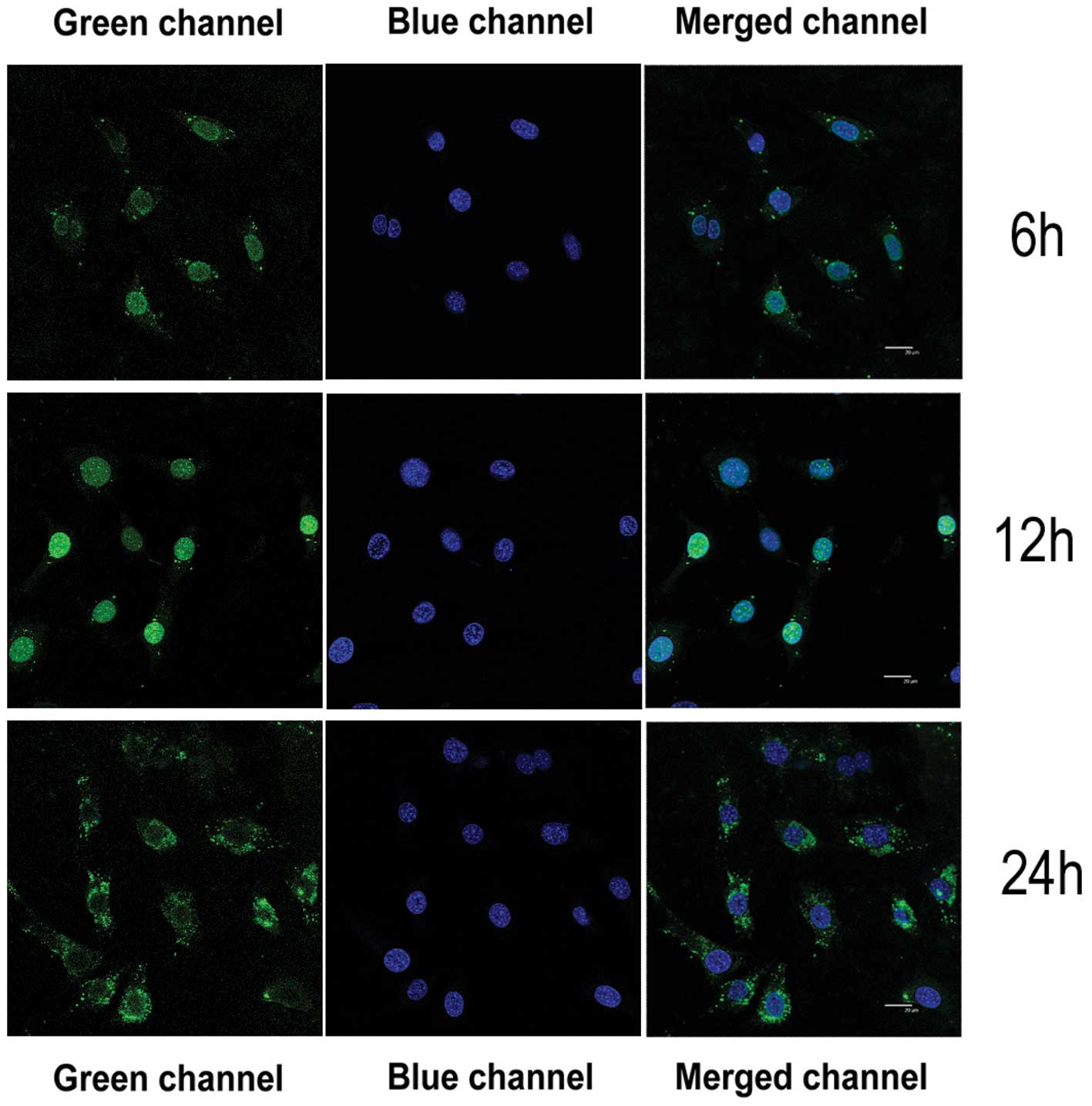
Yellow-green fluorescence appeared in the cell
nucleus after HSFs were transfected with the 100 nM Sp1 decoy ODN
for 6–12 h. By 24 h post-transfection, the yellow-green
fluorescence was observed in the cytoplasm of the infected cells
(Fig. 7).
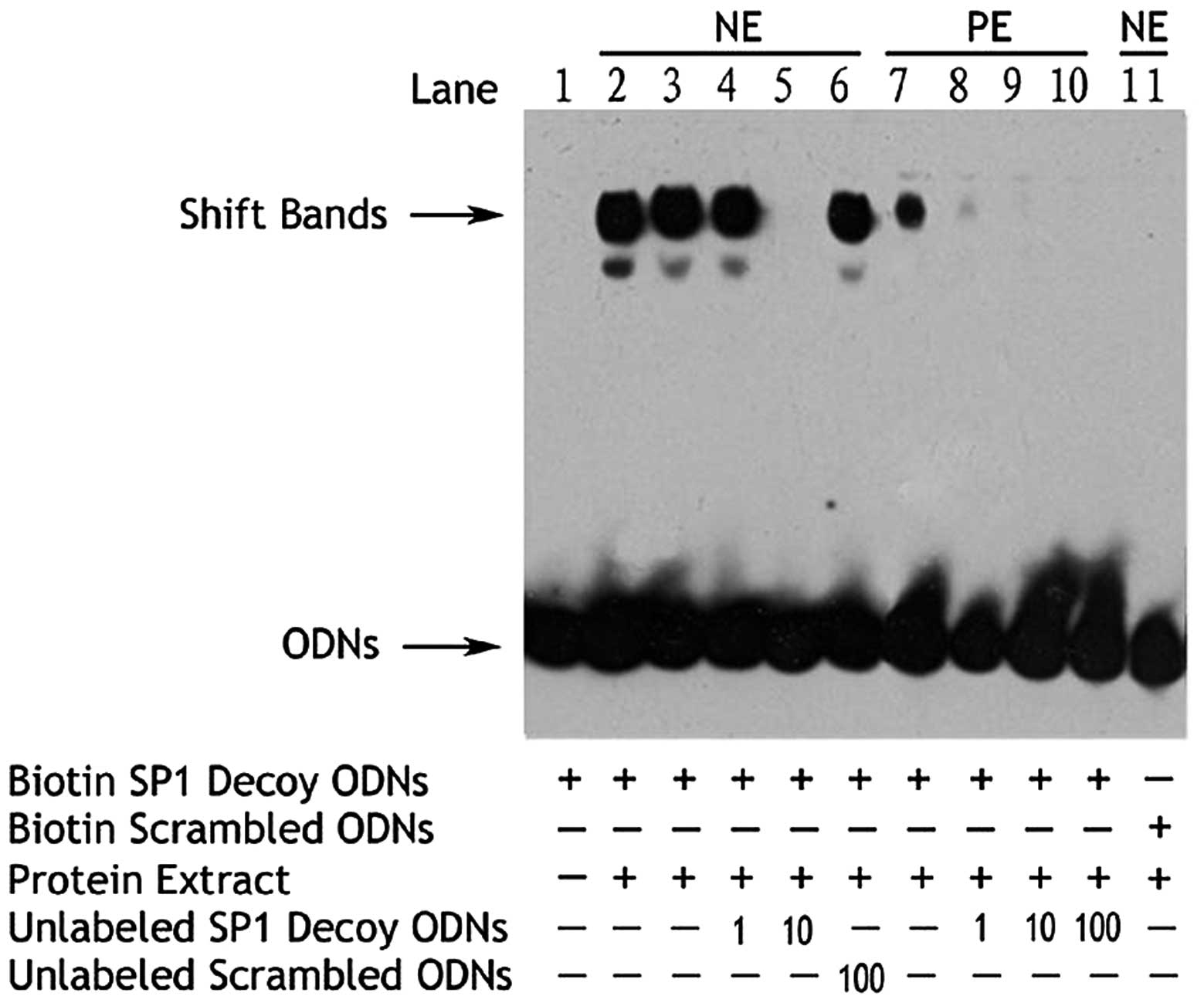
The EMSA assay revealed that a 10-fold increase in
the unlabeled Sp1 decoy ODN inhibited binding of the nuclear
protein and biotin-labeled Sp1 decoy ODN (Fig. 8). A 100-fold increase in the
unlabeled scrambled Sp1 ODN showed no inhibitory effect, while the
biotin-labeled scrambled Sp1 ODN was unable to bind nuclear
protein.
Discussion
Enhanced HSF proliferation and increased collagen
production are principal contributors to hypertrophic scar
formation (7). This process is
governed by signal transduction cascades that are finely regulated
by growth factors. The transcription factor Sp1 is ubiquitously
expressed and plays a significant role in signal transduction
(8). The most significant findings
of the current study were that the Sp1 decoy ODN inhibited cell
proliferation and collagen expression. These results demonstrate
that Sp1 decoy ODNs may provide a promising therapeutic option for
the treatment of hypertrophic scars.
Complementary approaches have also demonstrated that
Sp1 targeting may be a potentially powerful therapeutic approach
for reducing ECM accumulation in hypertrophic scars (9). Recently, a number of new technologies
have been developed to inhibit target gene expression in a
sequence-specific manner and these technologies have been
investigated as treatment modalities for a variety of diseases
(10,11). While there are numerous studies on
strategies targeting mRNA via antisense ODNs, ribozymes and RNA
interference, targeting proteins that regulate expression of a
specific gene is a relatively novel approach (12). Decoy technology has recently been
developed in an attempt to reduce the activity of a specific
transcription factor through the use of cis-element ds ODNs
containing the consensus binding sequence of the transcription
factor (13).
The greatest restriction to using ODNs is that they
are easily degraded by nucleases or by readily nonspecific
reactions with the control strand. To circumvent these problems,
various modified DNA analogs, particularly ODNs with PT-modified
ODNs, are often employed. These modified DNA analogs replace the
oxygen atoms in the phosphodiester bond of the terminal nucleotides
with sulfur atoms. This structural modification increases
resistance to nuclease attack (14). In this study, we used dumbbell Sp1
decoy ODNs to enhance the stability of the decoy. Using this
method, Sp1 regulated the expression of collagen. Similar
approaches have been used to deliver gene-regulating molecules,
such as synthetic ds ODNs that mimic cis-acting promoter elements
(15,16).
In the present study, Sp1 decoy ODNs inhibited HSF
proliferation and downregulated collagen synthesis. Sp1 decoy ODNs
at concentrations above 10 nM significantly inhibited HSF
proliferation. In addition, the Sp1 decoy ODN suppressed TGF-β1 and
fibronectin mRNA levels, as well as cell proliferation induced by
PDGF (12). EMSA evaluation showed
that the DNA-protein complex induced by PDGF induction was reduced
significantly in HSFs transfected with Sp1 decoy ODNs, whereas the
scramble decoy ODNs had no effect on DNA binding activity. Using
reporter plasmids containing the Sp1 binding motif in the TGF-β1
and fibronectin promoters, the enhancement in Sp1-dependent
transcription activity in response to serum stimulation was reduced
significantly by the Sp1 decoy ODN (17).
We also demonstrated that the in vitro
addition of the Sp1 decoy ODN effectively attenuated ECM mRNA
induction and the resulting ECM deposition (18). Therefore, these results suggest
that the Sp1 decoy ODN represents a potentially effective gene
therapy strategy to ameliorate dermal ECM accumulation during
hypertrophic scar formation (19).
In addition, Sp1 decoy ODNs markedly suppressed collagen IV mRNA
levels and fibronectin mRNA and protein in the hypertrophic scar
(20).
One limitation of this study is that Sp1 regulates
numerous genes, many of which are not involved in the etiology of
hypertrophic scars. The present study did not analyze any of these
genes. While this limitation precludes the decoy ODN approach from
being useful as a systemic therapeutic strategy, this approach
could be viable as a topical ointment applied directly to the skin.
Whether decoy ODNs would be useful for other diseases that involve
Sp1 upregulation remains to be investigated.
In conclusion, our data demonstrate that Sp1 is a
key transcription factor mediating HSF proliferation and ECM gene
expression that may prevent the pathogenesis of the hypertrophic
scar. Although additional in vitro and in vivo
studies need to be conducted to fully understand the potential of
decoy ODNs in preventing excessive collagen contraction in patients
with hypertrophic scars, this study provides a platform for these
experiments.
Acknowledgements
This study was supported by the National Scientific
Fund (30571929, 81000837), through the Experimental Study Center,
for the 6th People’s Hospital at Shanghai Jiaotong University
(Shanghai, China). We thank Medjaden Bioscience Limited for
assisting in the preparation of this manuscript.
References
|
1
|
Wang R, Ghahary A, Shen Q, Scott PG, Roy K
and Tredget EE: Hypertrophic scar tissues and fibroblasts produce
more transforming growth factor-beta1 mRNA and protein than normal
skin and cells. Wound Repair Regen. 8:128–137. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jagadeesan J and Bayat A: Transforming
growth factor beta (TGFbeta) and keloid disease. Int J Surg.
5:278–285. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chin GS, Liu W, Peled Z, et al:
Differential expression of transforming growth factor-beta
receptors I and II and activation of Smad 3 in keloid fibroblasts.
Plast Reconstr Surg. 108:423–429. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen SJ, Artlett CM, Jimenez SA and Varga
J: Modulation of human alpha1(I) procollagen gene activity by
interaction with Sp1 and Sp3 transcription factors in vitro. Gene.
215:101–110. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Boros DL, Singh KP, Gerard HC, Hudson AP,
White SL and Cutroneo KR: A novel nonsteroidal antifibrotic oligo
decoy containing the TGF-beta element found in the COL1A1 gene
which regulates murine schistosomiasis liver fibrosis. J Cell
Physiol. 204:370–374. 2005. View Article : Google Scholar
|
|
6
|
Chen SJ, Yuan W, Lo S, Trojanowska M and
Varga J: Interaction of smad3 with a proximal smad-binding element
of the human alpha2(I) procollagen gene promoter required for
transcriptional activation by TGF-beta. J Cell Physiol.
183:381–392. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Naim R, Naumann A, Barnes J, et al:
Transforming growth factor-beta1-antisense modulates the expression
of hepatocyte growth factor/scatter factor in keloid fibroblast
cell culture. Aesthetic Plast Surg. 32:346–352. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Meisler NT, Chiu JF and Cutroneo KR:
Promoter competitors as novel antifibrotics that inhibit
transforming growth factor-beta induction of collagen and
noncollagen protein synthesis in fibroblasts. J Cell Biochem.
75:196–205. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bowley E, O’Gorman DB and Gan BS:
Beta-catenin signaling in fibroproliferative disease. J Surg Res.
138:141–150. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Smith RA, Miller TM, Yamanaka K, et al:
Antisense oligonucleotide therapy for neurodegenerative disease. J
Clin Invest. 116:2290–2296. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lindow M and Kauppinen S: Discovering the
first microRNA-targeted drug. J Cell Biol. 199:407–412.
2012.PubMed/NCBI
|
|
12
|
Sato M: Upregulation of the
Wnt/beta-catenin pathway induced by transforming growth factor-beta
in hypertrophic scars and keloids. Acta Derm Venereol. 86:300–307.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Takamizawa J, Konishi H, Yanagisawa K, et
al: Reduced expression of the let-7 microRNAs in human lung cancers
in association with shortened postoperative survival. Cancer Res.
64:3753–3756. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Calin GA, Dumitru CD, Shimizu M, et al:
Frequent deletions and down-regulation of micro- RNA genes miR15
and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad
Sci USA. 99:15524–15529. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ahn JD, Kim CH, Magae J, et al: E2F decoy
oligodeoxynucleotides effectively inhibit growth of human tumor
cells. Biochem Biophys Res Commun. 310:1048–1053. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ahn JD, Morishita R, Kaneda Y, et al:
Inhibitory effects of novel AP-1 decoy oligodeoxynucleotides on
vascular smooth muscle cell proliferation in vitro and neointimal
formation in vivo. Circ Res. 90:1325–1332. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nusse R: Wnt signaling in disease and in
development. Cell Res. 15:28–32. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kato M, Zhang J, Wang M, et al:
MicroRNA-192 in diabetic kidney glomeruli and its function in
TGF-beta-induced collagen expression via inhibition of E-box
repressors. Proc Natl Acad Sci USA. 104:3432–3437. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cheon SS, Wei Q, Gurung A, et al:
Beta-catenin regulates wound size and mediates the effect of
TGF-beta in cutaneous healing. FASEB J. 20:692–701. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Berezikov E, Guryev V, van de Belt J,
Wienholds E, Plasterk RH and Cuppen E: Phylogenetic shadowing and
computational identification of human microRNA genes. Cell.
120:21–24. 2005. View Article : Google Scholar : PubMed/NCBI
|