Introduction
Approximately 2 billion people worldwide are
infected with human hepatitis B virus (HBV) (1) and >350 million are chronically
infected and at high risk of progression to cirrhosis, liver
failure or hepatocellular carcinoma (2,3).
More than half of all liver cancers are attributable to HBV
infection; and HBV-related liver diseases remain a major public
health concern, leading to ~1 million deaths per year (3).
Advances in established cost-effective and reliable
experimental models have accelerated the development of therapeutic
modalities for these life-threatening viral infections (3). Although progress has been made, in
vitro and in vivo HBV assays are being developed, but
are impeded by a lack of understanding of HBV specificity for and
entry into human liver cells, as well as reliable cell and animal
infection models (4). In addition
to humans, only chimpanzees and a small mammal, the treeshrew
(families Tupaiidae and Ptilocercidae), are known to be susceptible
to human HBV infection (5–7). Previously, basic studies of HBV and
the development of antiviral therapeutics were hindered by the lack
of suitable in vitro infection systems and animal models
(8). Currently, only suboptimal
cell transfection- and murine hydrodynamic injection (HI)-based
assays are available for HBV research (9).
To analyze clinical HBV replication, isolates are
obtained by the transfection of replicative recombinant HBV DNA
into hepatoma cell lines or HI of replication-competent replicons
into the tail veins of C57BL/6 mice which are able to replicate and
secrete HBV virions (10,11). Previously, in order to obtain a HBV
murine model, 10 mg HBV plasmid DNA was injected into the tail
veins of mice in a volume of phosphate-buffered saline equivalent
to 8% of the mouse body weight, with the total volume being
delivered within 5–8 sec (12–14).
In a previous study, the plasmid, pAAV-HBV1.3, did not replicate
properly in mice, thus it was difficult to detect HBV DNA in the
liver and serum (14). The HBV
core antigen (HBcAg) was challenging to detect, particularly in HBV
mutants with weaker replication capacity compared with wild-type
(WT) HBV (14). In order to
enhance HBV replication in mice, 10 mg plasmid pAAV-HBV1.3 and 20
μl Lipofectamine 2000 (LP) transfection reagent (Invitrogen Life
Technologies, Carlsbad, CA, USA) were mixed and administered via HI
into the tail veins of C57BL/6 mice. Plasmid DNA is well
distributed and packaged with LP in liposomes, which facilitates
the transfer of DNA to the murine liver cells. The optimized murine
HBV-LP model was superior to the murine HBV model without LP based
on HBV challenge via HI in mice, at the DNA and protein levels.
HBV has a 3.2-kb, relaxed circular (RC), partially
double-stranded overlapped DNA genome containing four overlapping
open reading frames (ORFs): P, S, C and X (15,16).
HBV possesses the smallest known DNA viral genome, the latter of
which optimized genomic organization through an overlapping gene
strategy (17). The S ORF encodes
the surface proteins, L, M and S (the HBsAg), through alternative
translation initiation from three in-frame start codons (18). In our previous study, a methionine
(M) to threonine (T) substitution at residue 75 (sM75T) was found
to be associated with reduced HBV replication and HBsAg expression
in vitro. In the present study, the previously modified HBV
murine model was used to determine whether sM75T influenced HBV
replication and HBsAg expression in vitro similar to that
in vivo. The results obtained were as expected.
Materials and methods
Cell culture and treatment
Huh7 human hepatocarcinoma cells were cultured in
Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen, Carlsbad, CA,
USA) at 37°C in a 5% CO2 atmosphere supplemented with
10% fetal bovine serum (Gibco-BRL, Carlsbad, CA, USA), 2 mM/l
glutamine, 100 IU/ml penicillin, and 100 IU/ml streptomycin. Huh7
cells were seeded in 6-well plates and transfected with 2 μg
plasmids per well using LP (19).
Female C57BL/6 (H-2b) mice (age, 6–8 weeks) were raised under
specific pathogen-free conditions in the Central Animal Laboratory
of Shaoxing Centre for Disease Control and Prevention (Shaoxing,
China) and were handled following animal ethics guideline
standards, according to the principles of animal protection, animal
welfare and ethical inspection. The study was approved by the
Medical Laboratory Animal Management Committee of Zhejiang Province
(Department of Health of Zhejiang Province, Zhejiang, China).
C57BL/6 mice were challenged with HBV replication-competent
plasmids with or without LP administered by HI. The mouse assay was
repeated three times, six mice were used in each group and there
were seven groups in total: negative control, pBSK-HBV1.3,
pAAV-HBV1.3, pAAV-HBV1.3-LP, pBSK-HBV1.3-T378C, pAAV-HBV1.3 T378C
and pAAV-HBV1.3- T378C-LP.
Plasmid constructs
HBV mutants were constructed using fusion polymerase
chain reaction (PCR), with primers carrying aimed mutations and WT
pBSK-HBV1.3 as a template, which is a replication-competent plasmid
1.3-fold greater in length than the HBV genome. The sM75T was
included in the pBSK-HBV1.3 plasmid to construct pBSK-HBV1.3-sM75T
in our previous study (19). For
the in vivo assay, pAAV-HBV-1.3 and pAAV-HBV1.3-rtXs were
constructed based on pHBV1.3 and pHBV1.3-rtXs, respectively.
pHBV1.3-WT/MT and pAAV were digested with the SacI and
SacII restriction endonuclease (Takara Bio, Dalian, China),
then end-filled with T4 and Klenow DNA polymerase (Takara Bio),
respectively (14). The recovered
products were digested by HindIII (Takara Bio) and ligated
by T4 ligase (New England BioLabs, Beijing, China) to generate
pAAV-HBV1.3 and pAAV-sM75T. All primers used in our study were
manufactured by Sangon Biotech (Shanghai, China).
Enzyme-linked immunosorbent assay
(ELISA)
Huh7 cells were transfected with the indicated
HBV-bearing plasmids and the hepatitis B surface antigen (HBsAg)
and the hepatitis B e antigen (HBeAg) in the supernatant at 96 h
post-transfection (hpt) and were detected using an ELISA kit for
the detection of HBsAg/HBeAg (Shanghai Kehua Diagnostic Medical
Products Co., Ltd., Shanghai, China), according to the
manufacturer’s instructions (20).
qPCR
When the cell lysis solution had been treated with
DNase I (Roche Applied Science, Penzberg, Germany) at 37°C for 30
min to digest the plasmid DNA, total HBV DNA was purified from the
lysates of Huh7 cells 96 hpt and used as the template for qPCR
which was conducted using SYBR-Green I (Roche, Mannheim, Germany)
on a Light Cycler real-time PCR unit (Applied Biosystems, Inc.,
Foster City, CA, USA) according to the manufacturer’s instructions.
Primers (sense: 5′-GTTGCCCGTTTGTCCTCTAATTC-3′ and antisense:
5′-GGAGGGATACATAGAGGTTCCTT-3′ for RC) hybridized to the HBV surface
gene were designed to quantify the HBV DNA RC genomes (100-bp
fragments) by qPCR relative to an external plasmid DNA
standard.
Analysis of encapsidated HBV DNA
Replication-competent HBV WT and mutant-type (MT)
plasmids were transfected into Huh7 cells or injected into C57BL/6
mice, which were previously challenged via HI of
replication-competent pAAV-HBV1.3 or -T378C, respectively, with or
without LP, as described previously (12). HBV DNA replicative intermediates
from intracellular core particles were extracted and subjected to
agarose gel electrophoresis, followed by denaturation and southern
blotting with a 32P-labeled full-length HBV probe, as
described previously (14).
Hybridization signals were visualized and analyzed using a
phosphoimager (Cyclone® Plus Storage Phosphor System;
Parkard Instrument Company, Inc., Meriden, CT, USA). Data from the
densitometric analyses were quantified using OptiQuant software
(PerkinElmer, Inc., Waltham, MA, USA).
Immunohistochemical staining
Liver tissues were collected from mice sacrificed at
the indicated time points. Intrahepatic HBcAg of mice was
visualized by immunohistochemical staining of formalin-fixed
paraffin-embedded tissues by rabbit anti-HBc antibodies (DAKO) and
secondary goat antirabbit IgG HRP antibody (DAKO), using an and
Envision System (DAKO). The Envision system is also know as the
enhanced labeled polymer system. The liver sections were also
stained with hematoxylin and eosin (12,14).
Statistical analysis
The statistical analysis was conducted using
GraphPad (GraphPad software, San Diego, CA, USA). Differences in
multiple comparisons were determined for statistical significance
using the Student’s t-test. P<0.05 was considered to indicate a
statistically significant difference. Results are presented as the
mean ± SD.
Results
Replication and protein expression of WT
and MT plasmids
The plasmid pBSK-HBV1.3 bearing a
replication-competent WT HBV, which was 1.3-fold greater in length
than the genotype A genome (GenBank accession no. U95551, ayw), was
used as a backbone to construct mutant pBSK-HBV1.3. The sM75T in
the s gene was introduced into pBSK-HBV1.3 to obtain
pBSK-HBV1.3-sM75T. For the in vivo assay, pAAV-HBV-1.3 and
pAAV-HBV1.3-sM75T were constructed based on plasmids pBSK-HBV1.3
and pBSK-HBV1.3-sM75T, respectively, as described above (Fig. 1A). Plasmids pBSK-HBV1.3,
pBSK-HBV1.3-sM75T, pAAV-HBV1.3 and pAAV-HBV1.3-sM75T were
transfected into Huh7 cells and the HBsAg and HBeAg in the culture
medium were detected by ELISA. Replicative HBV intermediates from
intracellular core particles were extracted from Huh7 cells and
detected by southern blot analysis, as previously described
(14). The data indicated that
pBSK-HBV1.3 and pAAV-HBV1.3 had similar replication levels of
HBs/eAg, while pBSK-HBV1.3-sM75T and pAAV-HBV1.3-sM75T had similar
replication levels to HBs/eAg. In addition, the pBSK-HBV1.3-sM75T
and pAAV-HBV1.3-sM75T exhibited moderately decreased levels of HBV
DNA and HBeAg compared with the WT and a markedly decreased
expression level of HBsAg (Fig. 1B and
C).
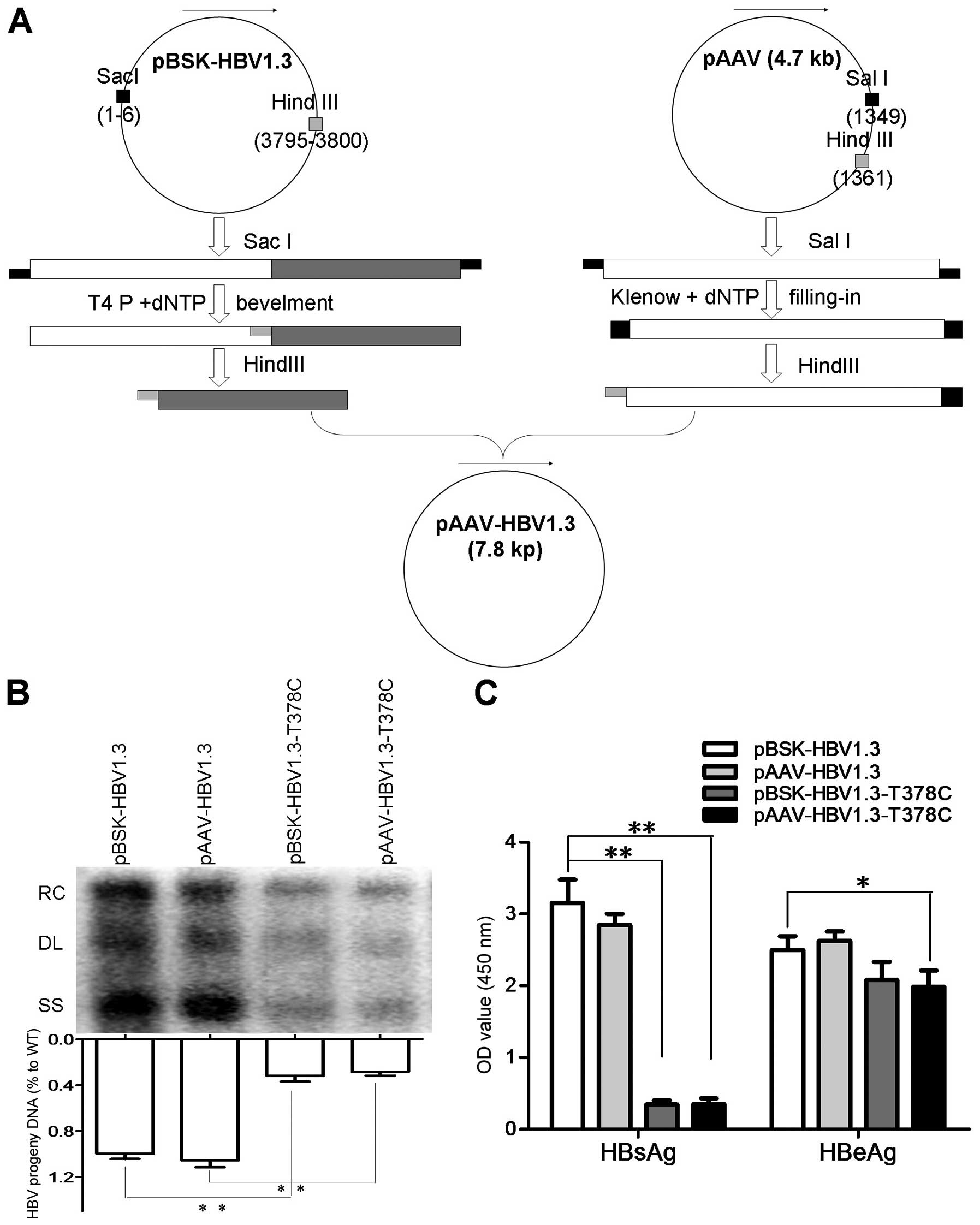 | Figure 1Construction strategy of pAAV-HBV1.3,
replication and protein expression of WT and MT plasmids. (A) For
the in vivo assay, pAAV-HBV-1.3 and pAAV-HBV1.3-T378C were
constructed based on pHBV1.3 and pHBV1.3-T378C plasmids,
respectively. The pHBV1.3/pHBV1.3-T378C and pAAV plasmids were
digested with SacI, then end-filled with T4 and Klenow DNA
polymerase, respectively. The recovered products were digested with
HindIII and ligated by T4 ligase to generate pAAV-HBV-1.3 or
pAAV-HBV1.3-T378C. (B) Detection of HBV replication intermediates
by southern blot analysis. HBV replication RC, DL, and SS HBV DNAs
are indicated (upper panel). Intracellular encapsidated HBV DNA
levels of each construct were compared with that of the WT genome
(set as 100%, lower panel). (C) The expression of HBsAg and HBeAg
were measured using commercial enzyme-linked immunosorbent assay
kits. Each value is presented as the mean of three independent
experiments. The error bars represent the standard deviation.
*P<0.05 and **P<0.01. HBV, hepatitis B
virus; WT, wild-type; MT, mutant-type; RC, relaxed circular; DL,
double-stranded linear; SS, single-stranded; HBsAg, hepatitis B
surface antigen; HBeAG, hepatitis B e antigen. |
LP enhances HBV replication in mice
The LP transfection reagent is a proprietary
formulation for transfecting nucleic acids (DNA, RNA and mRNA) into
a wide range of eukaryotic cells and was used in the present study
to package HBV plasmid DNA allowing entry into the Huh7 cells.
However, it is not clear whether LP promotes HBV replication when
mixed and co-injected with pAAV-HBV1.3 into mice tail veins.
C57BL/6 mice were respectively challenged with pBSK-HBV1.3,
-HBV1.3-sM75T, pAAV-HBV1.3 and -HBV1.3-sM75T with or without LP by
HI. HBV DNA in the mouse sera and liver samples were measured by
qPCR targeted to RC and southern blot analysis at the indicated
time points, respectively. Based on the in vitro results, LP
significantly increased HBV replication in the sera (Fig. 2A and B) and liver (Fig. 2D) samples. The mouse model was
analyzed following HI to confirm the results of our previous study.
In line with our previous observations at the cellular level, the
sM75T substitution also significantly depressed HBV replication
in vivo at all time points (Fig. 2C), particularly in the pAAV and LP
pAAV groups. Serum HBV DNA reached the highest level at 12 days
postinfection (dpi) in all groups. By contrast, the pBSK group
demonstrated a maximum viral DNA level at 12 dpi, which rapidly
decreased thereafter. Moreover, on the background of the sM75T
substitution, LP enhanced HBV replication.
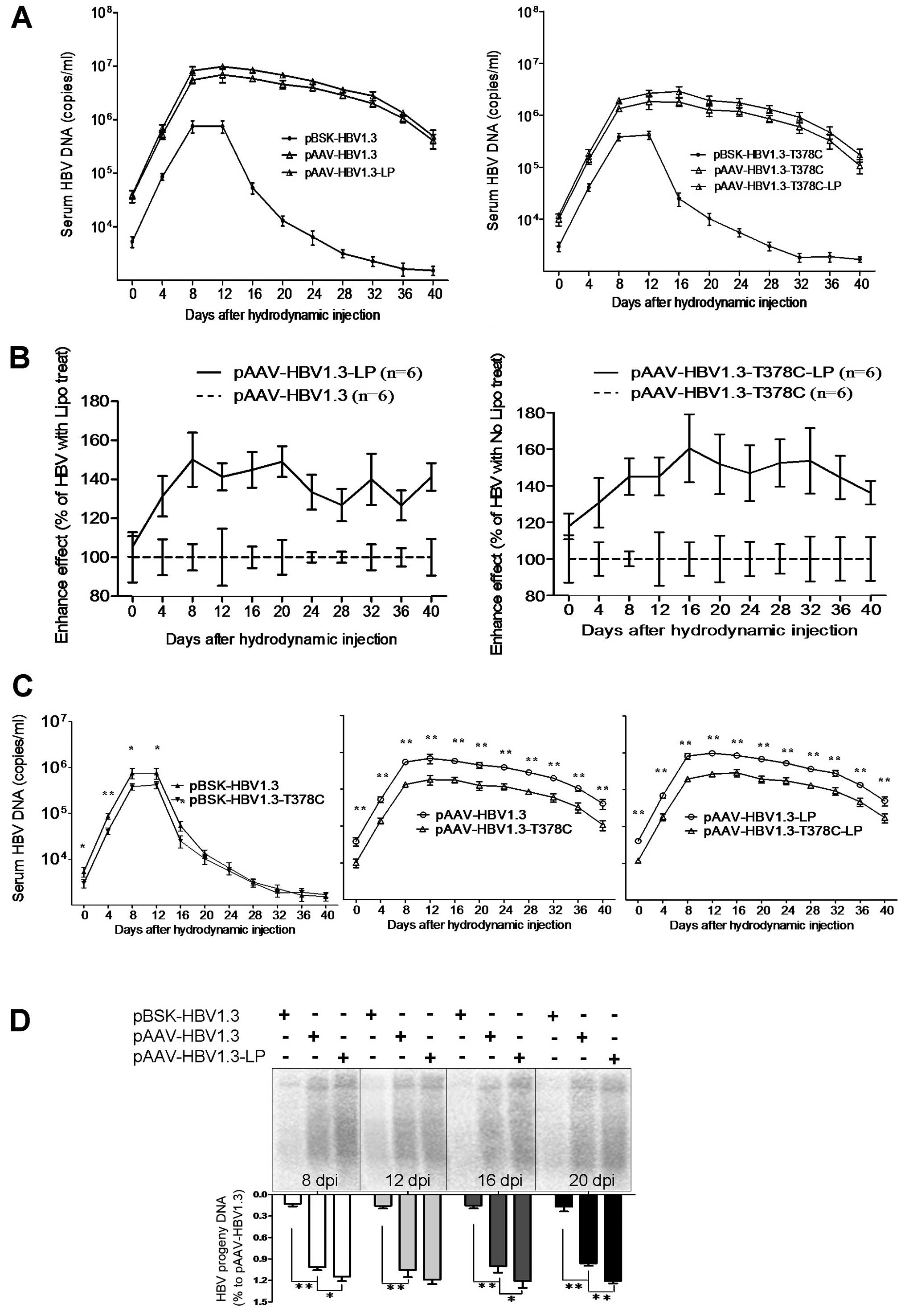 | Figure 2Replication of pBSK/pAAV-HBV1.3 and
-T378C in the murine model. C57BL/6 (H-2b) mice were challenged
with pBSK/pAAV-HBV1.3 and -T378C by tail vein administration of
hydrodynamic injection. At the indicated time points, HBV DNA and
proteins in the sera and liver samples were measured by qPCR
targeted to HBV RC DNA, southern blot analysis, enzyme-linked
immunosorbent assay and immunohistochemical analysis, respectively.
(A–C) HBV DNA serum analysis of mice from the pBSK/pAAV-HBV1.3 and
-T378C groups by qPCR. (A) Mice were divided into the wild-type and
T378C groups. The kinetics of HBV replication expression of each
group, which contained pBSK, pAAV and pAAV-Lipo subgroups. (B) The
enhanced HBV replication effect of LP in vivo is shown as
the upregulation of serum HBV DNA in LP-mixed mice compared with
that of mice without LP. The average HBV DNA copy number of the
unmixed control group at each of the indicated time points was set
as 100%. (C) The effect of T378C in vivo on the pBSK, pAAV
and pAAV-Lipo subgroups is shown. (D) To detect HBV DNA in the
liver, mice from each group were sacrificed at the indicated time
points. Total DNA was isolated from the liver tissue and subjected
to southern blot analysis. HBV, hepatitis B virus; LP,
Lipofectamine 2000. |
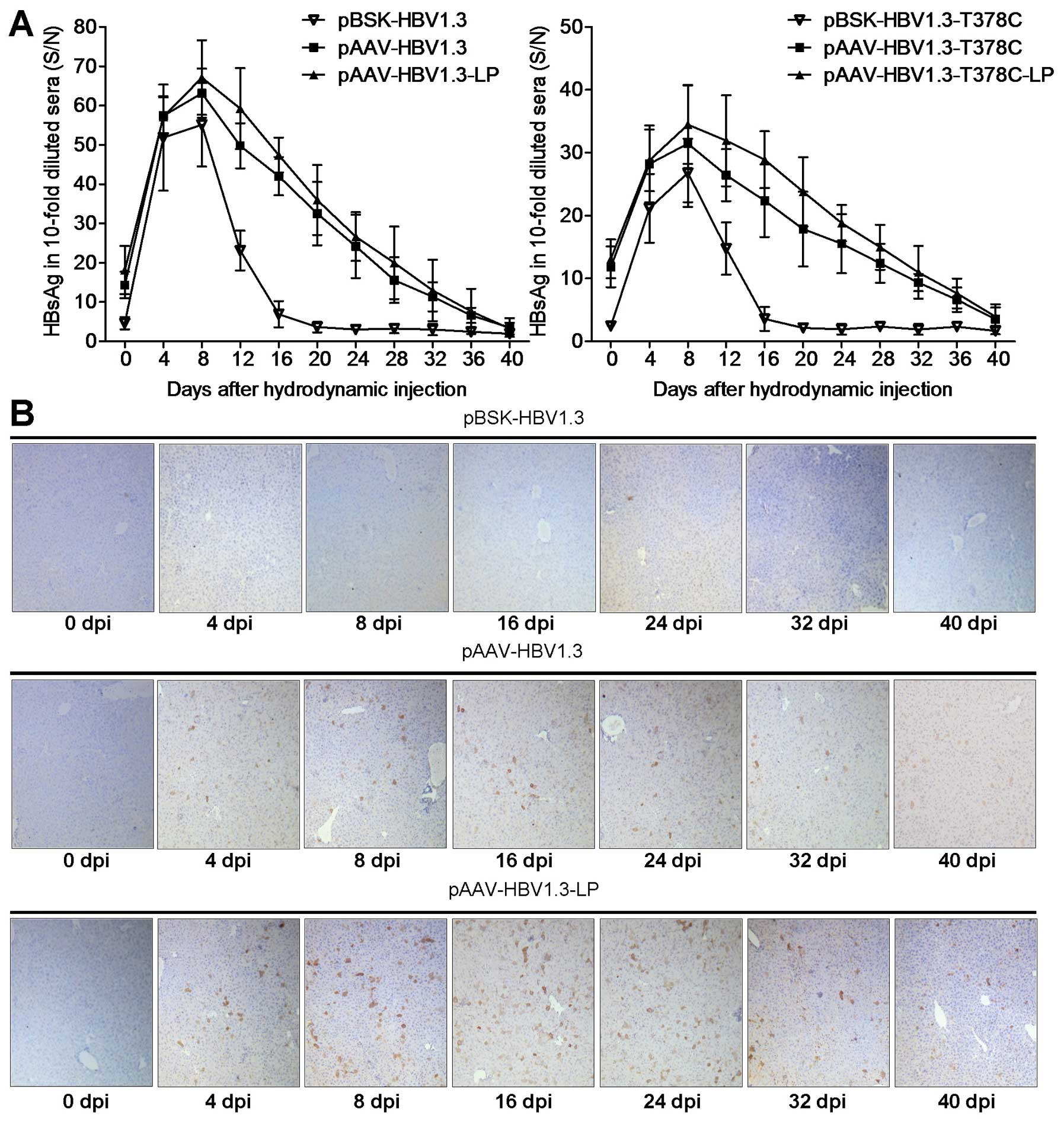
LP enhances HBsAg and HBcAg expression in
mice
The HBV HBsAg level peaked at 8 dpi and then
decreased gradually in mouse sera following HI with
pBSK-HBV1.3/-sM75T or pAAV-HBV1.3/-sM75T (Fig. 3A), which was comparable with
previously published data (10,17).
Serum HBsAg of the pAAV group remained positive for the complete
observation period of 5 weeks, but only for ~2 weeks in the pBSK
group. Similar results were observed in the -sM75T group (Fig. 3A). The intrahepatic HBcAg was
further analyzed by immunohistochemical staining with specific
antibodies (Fig. 3C). HBcAg was
expressed in hepatic cells in the pAAV-HBV1.3 group at 4 dpi and
persisted for at least 5 weeks, particularly in the pAAV-HBV1.3-LP
group, while no obvious HBcAg expression was identified in the
pBSK-HBV1.3 group at any of the time points. HBcAg expression was
not detected in livers of the mice that were administered with
pAAV-HBV1.3-sM75T and pBSK-HBV1.3-sM75T via HI (data not
shown).
Discussion
Approximately 2 billion people worldwide are
infected with human HBV, of which >350 million are chronically
infected and at high risk of progression to cirrhosis, liver
failure or cancer. Over 50% of liver cancers worldwide are
attributable to HBV infection. Thus, HBV-related liver diseases
remain a major public health concern, with ~1 million mortalities
annually. Individuals co-infected with HBV and HDV are at greater
risk for rapid progression and severe disease.
LP is a proprietary formulation for transfecting
nucleic acids (DNA, RNA and mRNA) into a wide range of eukaryotic
cells and was used in our previous study to package HBV plasmid DNA
and allowing entry into Huh7 cells (11). However, it is not clear whether LP
promoted HBV replication when mixed and co-injected with
pAAV-HBV1.3 into mice via the tail vein. According to the results
of the present study, the pAAV vector was confirmed to be more
suitable than pBSK for in vivo studies of HBV
replication.
The HBV life cycle is a complicated process that is
regulated by various host and viral factors (21,22).
Although the surface proteins predominantly assist viral
envelopment and secretion, there are also studies that have
indicated that the surface proteins may be involved in replication
regulation (23). In our previous
study, the T378C substitution led to sM75T within the HBsAg
encoding gene and was identified to reduce cellular HBsAg
expression and HBV replication (19), which was also demonstrated in our
in vitro experiments. Thus, our previous and present results
confirmed that surface proteins are not essential for HBV DNA
replication in vivo and in vitro.
The in vivo HBV model based on the
hydrodynamic injection of an engineered, replication-competent HBV
DNA into the tail veins of C57BL/6 mice was used to evaluate the
anti-HBV effect of nucleos(t)ide analog and study resistance
mutation of HBV in vivo in our previous study (14). The greatest problem is that the HBV
replication capacity in this mice model was too low for southern
blot analysis. In the present study, we modified the mice model by
injecting the mixture of LP and replication-competent HBV plasmid
DNA. Based on our data, LP significantly enhanced the HBV
replication capacity, the level of HBsAg and HBcAg in mouse liver.
This improved model is more conducive to HBV study in vivo.
In the future, we aim to achieve an improved standardization in
vivo model and use it to perform HBV resistant phenotype
analysis.
Acknowledgements
The authors would like to thank the participants for
their work. This study was supported by the Qianjiang Talent
Project of Zhejiang Province, China (grant no. 2012R10084).
References
|
1
|
Lavanchy D: Hepatitis B virus
epidemiology, disease burden, treatment, and current and emerging
prevention and control measures. J Viral Hepat. 11:97–107. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lee WM: Hepatitis B virus infection. N
Engl J Med. 337:1733–1745. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yan H, Zhong G, Xu G, et al: Sodium
taurocholate cotransporting polypeptide is a functional receptor
for human hepatitis B and D virus. Elife. 1:e000492012.
|
|
4
|
Paganelli M, Dallmeier K, Nyabi O, et al:
Differentiated umbilical cord matrix stem cells as a new in vitro
model to study early events during hepatitis B infection.
Hepatology. 57:59–69. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yan Z, Tan W, Dan Y, et al: Estrogen
receptor alpha gene polymorphisms and risk of HBV-related acute
liver failure in the Chinese population. BMC Med Genet. 13:492012.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zoulim F and Locarnini S: Hepatitis B
virus resistance to nucleos(t)ide analogues. Gastroenterology.
137:1593–1608. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liu F, Chen L, Yu DM, et al: Evolutionary
patterns of hepatitis B virus quasispecies under different
selective pressures: correlation with antiviral efficacy. Gut.
60:1269–1277. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Takehara T, Suzuki T, Ohkawa K, et al:
Viral covalently closed circular DNA in a non-transgenic mouse
model for chronic hepatitis B virus replication. J Hepatol.
44:267–274. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lim W, Kwon SH, Cho H, et al: HBx
targeting to mitochondria and ROS generation are necessary but
insufficient for HBV-induced cyclooxygenase-2 expression. J Mol Med
(Berl). 88:359–369. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Schinazi RF, Ilan E, Black PL, Yao X and
Dagan S: Cell-based and animal models for hepatitis B and C
viruses. Antivir Chem Chemother. 10:99–114. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tang N, Huang AL, Zhang BQ, et al:
Construction of recombinant eukaryotic expression plasmid
containing 1.3-fold-overlength genome of HBV and its expression in
HepG2 cells. Zhonghua Gan Zang Bing Za Zhi. 11:464–466. 2003.(In
Chinese).
|
|
12
|
Huang LR, Wu HL, Chen PJ and Chen DS: An
immunocompetent mouse model for the tolerance of human chronic
hepatitis B virus infection. Proc Natl Acad Sci USA.
103:17862–17867. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yin Y, Wu C, Song J, et al: DNA
immunization with fusion of CTLA-4 to hepatitis B virus (HBV) core
protein enhanced Th2 type responses and cleared HBV with an
accelerated kinetic. PloS One. 6:e225242011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Qin B, Budeus B, Cao L, et al: The amino
acid substitutions rtP177G and rtF249A in the reverse transcriptase
domain of hepatitis B virus polymerase reduce the susceptibility to
tenofovir. Antiviral Res. 97:93–100. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mizokami M, Orito E, Ohba K, Ikeo K, Lau
JY and Gojobori T: Constrained evolution with respect to gene
overlap of hepatitis B virus. J Mol Evol. 44(Suppl 1): S83–S90.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gao W and Hu J: Formation of hepatitis B
virus covalently closed circular DNA: removal of genome-linked
protein. J Virol. 81:6164–6174. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Miller RH, Kaneko S, Chung CT, Girones R
and Purcell RH: Compact organization of the hepatitis B virus
genome. Hepatology. 9:322–327. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Carman WF: The clinical significance of
surface antigen variants of hepatitis B virus. J Viral Hepat.
4(Suppl 1): 11–20. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Qiu J, Qin B, Rayner S, et al: Novel
evidence suggests Hepatitis B virus surface proteins participate in
regulation of HBV genome replication. Virol Sin. 26:131–138. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Qin B, He T, Chen Z, Xu W, Pan G and Tu C:
A novel method for the analysis of drug-resistant phenotypes of
hepatitis B virus. Int J Mol Med. 31:975–981. 2013.PubMed/NCBI
|
|
21
|
Hu J and Boyer M: Hepatitis B virus
reverse transcriptase and epsilon RNA sequences required for
specific interaction in vitro. J Virol. 80:2141–2150. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hu J, Flores D, Toft D, Wang X and Nguyen
D: Requirement of heat shock protein 90 for human hepatitis B virus
reverse transcriptase function. J Virol. 78:13122–13131. 2004.
View Article : Google Scholar
|
|
23
|
Chua PK, Wang RY, Lin MH, Masuda T, Suk FM
and Shih C: Reduced secretion of virions and hepatitis B virus
(HBV) surface antigen of a naturally occurring HBV variant
correlates with the accumulation of the small S envelope protein in
the endoplasmic reticulum and Golgi apparatus. J Virol.
79:13483–13496. 2005. View Article : Google Scholar
|