Introduction
Hepatitis B virus (HBV) infection is a worldwide
health problem and more than two billion people have been infected
with HBV (1). Although many
individuals eventually achieve a state of non-replicative
infection, the prolonged immunological response to infection leads
to the development of cirrhosis, liver failure or hepatocellular
carcinoma (HCC) in up to 40% of patients (2). In China, ~120 million people are HBV
chronic carriers, and 50–80% of cirrhosis patients are infected
with HBV (1). Persistent HBV
infection has been considered a multifactorial and polygenic
disequilibrium among viral, environmental and host genetic
components (3). Single-nucleotide
polymorphisms (SNPs) are the most abundant form of DNA variation in
the human genome and contribute to human phenotypic differences
(4). A number of studies have
demonstrated the role of host genetic factors and their
interactions with environmental factors leading to various outcomes
following HBV infection (5–8).
Understanding the key factors that influence the clinical outcomes
of HBV infection is crucial for early diagnosis and optimal
treatment (9).
Cytotoxic T lymphocyte-associated antigen 4
(CTLA-4), mapped to chromosome 2q33 (10) and expressed in the activated and
regulatory T cells, regulates T-cell activation and tolerance
(11), Th1/Th2 differentiation and
cytokine production (12)
following B7 engagement. CTLA-4 has been established as a
significant negative regulator of T-cell responses, leading to the
preservation of T-cell homeostasis (13). Previously, it was reported that
CTLA-4 may downregulate the immune response strength of tumor
immunity (14), allergy (15), autoimmune disease (16), infection (17) and vaccination (18). In addition to inhibiting T-cell
activation and proliferation, CTLA-4 may also induce
Fas-independent apoptosis of activated T cells (19). Cytokines including TNF-α and IFN-β
are critical, not only for viral clearance, but also for the
immunopathogenesis of HBV infection (20,21).
Therefore, the interaction of CTLA-4 with B7 molecules in
regulating T-cell activation and Th1/Th2 cytokine production is
involved in the immune response against HBV infection (22).
SNPs in CTLA-4 have been connected to the
susceptibility to autoimmune disease and various types of cancer
(23). Findings from recent
studies have demonstrated that CTLA-4 gene polymorphisms may affect
the susceptibility and chronicity of the disease in patients with
HBV infection (24–29). However, the results remain
controversial and whether CTLA-4 gene polymorphisms are associated
with the status of HBV infection including chronic carriers and
cirrhosis, and hepatitis B e antigen (HBeAg) positivity in the
infected patients remains to be determined. In addition, the
criteria for the control and case groups were not completely equal
among these studies. Thus, whether there is an association between
CTLA-4 genetic variations and HBV infection in the Chinese
population using equal criteria remains to be clarified. In the
present study, three SNPs were selected from the CTLA-4 gene:
rs5742909 (−318 C/T) at the promoter region, rs231775 (+49 A/G), a
non-synonymous at the exon 1 and rs3087243 (+6230 G/A) at the
downstream in the 3′ untranslated region (UTR). The three
polymorphisms were genotyped in a hospital-based case-control
study, including 1,119 unrelated Han Chinese subjects from the
Hubei province (Central China).
Materials and methods
Study subjects
In this hospital-based case-control study, a total
of 1,119 unrelated Han Chinese individuals were recruited from
Tongji Hospital and Union Hospital, Wuhan, China, between July 2007
and September 2009. All the subjects were divided into four groups:
i) the HBV clearance group (clear); ii) the chronic active
hepatitis B group (CHB); iii) the HBV-related liver cirrhosis group
(LC) and iv) the HBV-related hepatocellular carcinoma group (HCC).
The diagnostic criteria for study inclusion were described
previously (6). Moreover, patients
with positive laboratory tests for human immunodeficiency virus
(HIV), alcoholic liver disease, suspected autoimmune diseases or
schistosomiasis were excluded from the study.
All the study subjects were of unrelated ethnic
background, Han Chinese who lived in Wuhan or the surrounding
region. Informed consent was obtained from each participant for
study enrollment. An information questionnaire was used and the
demographic information included gender, age and place of origin.
The study was approved by the local Research Ethics Committee at
the Tongji Hospital, Huazhong University of Science and Technology
in accordance with the principle of the Helsinki Declaration
II.
DNA isolation and genotyping
Genomic DNA was isolated from the peripheral whole
blood using a TIANamp blood DNA kit [Tiangen Biotech (Beijing) Co.,
Ltd., Beijing, China]. The concentration and purity of the DNA were
determined with a NanoDrop spectrophotometer (Thermo Fisher
Scientific, Wilimington, DE, USA), then diluted to a final
concentration of 8 ng/ml and distributed to a 96-well plate.
Genotyping of genetic polymorphisms was performed via the TaqMan
method according to the instructions of TaqMan®R SNP
Genotyping Assays (Applied Biosystems, Carlsbad, CA, USA). To
detect the three SNPs (rs5742909, rs231775 and rs3087243),
TaqMan®R MGB Probes and the primers for PCR
amplification (Table I) were
customized. The probes were labeled with FAM and VIC dyes to denote
the two different alleles, respectively, and the allelic category
was measured automatically using the Sequence Detection System 2.3
software (Applied Biosystems) according to the intensity of the VIC
and FAM dyes.
 | Table ITaqMan® probe and primer
for three SNPs. |
Table I
TaqMan® probe and primer
for three SNPs.
| SNP (NCBI reference
no.) | rs5742909a | rs231775 | rs3087243 |
|---|
| Forward primer | Commercialized |
GCACAAGGCTCAGCTGAAC |
CCATCCTCTTTCCTTTTGATTTCTTCAC |
| Reverse primer | Commercialized |
CAGAAGACAGGGATGAAGAGAAGAA |
TGTGTTAAACAGCATGCCAATTGATT |
| MGB probe 1 | Commercialized |
VIC-CCAGGTCCTGGTAGCCA-MGB |
VIC-TCTGTGTTAACCCATGTTATA-MGB |
| MGB probe 2 | Commercialized |
FAM-CAGGTCCTGGCAGCCA-MGB |
FAM-TGTGTTAACCCACGTTATA-MGB |
Statistical analysis
A statistical analysis was conducted using Arlequin
3.5 (30), haploview 4.2
(Cambridge, MA, USA) and SPPS 17.0 (SPSS, Inc., Chicago, IL, USA)
softwares. The Hardy-Weinberg equilibrium was tested separately for
cases and controls by Arlequin 3.5. Linkage disequilibrium (LD) and
haplotypes were assessed by the haploview 4.2 software. Genotypic
analyses included allele, dominant, recessive and additive genetic
models. The χ2 test or Fisher’s-exact test was applied
to a row-by-column contingency table in the four genetic models.
Age- and gender-adjusted odds ratios (ORs) and 95% confidence
intervals (CIs) were calculated on the basis of the unconditional
binary logistic regression model. The strength of association
between the genotypes or alleles and HBV infection was estimated by
the SPSS17.0. ORs and 95% CIs were calculated using the major
allele as a reference. All the tests were two sided with P<0.05
considered to indicate a statistically significant difference.
Results
Clinical and demographic
characteristics
The clinical and demographic characteristics of the
case-control study included gender, age, drinking habits, serum
α-fetoprotein level, serum total bilirubin level, alanine
transaminase, HBV-DNA load and serum markers of hepatitis B virus
(Table II). Although an effort
was made to obtain a good match on the age and gender, there were
more male subjects in the three HBV infection groups (CHB+LC+HCC;
average 78.7%) compared with those in the clear group (59.3%,
P<0.05). The individuals in the CHB group were younger compared
with those in the LC and HCC groups (P<0.001). However,
populations in the LC and HCC groups demonstrated no significant
difference with the clear group with regard to age (P>0.05). No
significant difference was observed in the percentage of hepatitis
B e antigen (HBeAg)-positive (P>0.05) between the patients in
the LC group (11.74%) and those in the HCC group (10.24%). In
addition, there was more alcohol consumption in patients
(P<0.05) with the LC (29.11%) and HCC (34.19%) groups compared
with those in the clear (13.17%) and CHB (18.63%) groups. The
difference in the alcohol consumption status may be due to the
limited number of drinkers within the Chinese female
population.
 | Table IIClinical characteristics of the study
subjects. |
Table II
Clinical characteristics of the study
subjects.
|
Characteristics | Clear, n=205 | CHB, n=467 | LC, n=213 | HCC, n=234 |
|---|
| Gender, no.
(%) |
| Male | 121 (59.3) | 343 (73.4) | 184 (86.4) | 192 (82.1) |
| Female | 83 (40.7) | 124 (26.6) | 29 (13.6) | 42 (17.9) |
| Age in years, mean
± SD | 48.85±12.52 | 35.42±12.89 | 46.21±10.83 | 47.84±13.23 |
| Drinkers, no.
(%) | 27 (13.17) | 87 (18.63) | 62 (29.11) | 80 (34.19) |
| HBsAg |
All− |
All+ |
All+ |
All+ |
| Anti-HBs IgG |
All+ | All |
All− |
All− |
| HBeAg-positive, no.
(%) |
All− | 94 (20.13) | 25 (11.74) | 24 (10.24) |
| Anti-HBc IgG |
All+ |
All+ |
All+ |
All+ |
| Family history, no.
(%) | No | 69 (14.78) | 27 (12.68) | 47 (20.09) |
| ALT, U/l | No | 420.33±399.23 | 110.46±115.24 | 90.63±75.17 |
| TBil, mmol/l | No | 180.23±171.45 | 132.28±120.86 | 64.36±60.74 |
| HBV-DNA,
copy/ml | No | (3.05±3.67) E7 | (2.34±4.02) E6 | (5.59±2.34) E6 |
Hardy-Weinberg equilibrium test
Hardy-Weinberg equilibrium was estimated by the
Fisher’s exact test via Arlequin 3.5 software. The allele and
genotype distributions are shown in Tables III and IV. No significant difference was
revealed between the observed and expected frequencies of each
genotype in these groups (P>0.05). All the genotype
distributions of the three SNPs (rs231775, rs5742909 and rs3087243)
conformed to the Hardy-Weinberg equilibrium (P>0.05) in all the
groups, making it suitable for subsequent statistical analysis.
 | Table IIIAssociation of three SNPs (rs5742909,
rs231775 and rs3087243) with HBV infection progression and
clearance in the Han Chinese populations (dominant model). |
Table III
Association of three SNPs (rs5742909,
rs231775 and rs3087243) with HBV infection progression and
clearance in the Han Chinese populations (dominant model).
| | | | | Dominant model |
|---|
| | | | |
|
|---|
| SNPs | Sample | Allele
distribution | P-valuea | Genotypes | P-value | OR (95%CI)b |
|---|
| rs5742909 | (−318C>T) | C/T | | CC/CT/TT | CC vs. CT+TT | CC vs. CT+TT |
| Clear | 358/42 | Ref. | 160/38/2 | Ref. | Ref. |
| CHB+LC | 1126/206 | 0.013 | 478/170/18 | 0.012 | 1.694
(1.124–2.553) |
| rs231775 | (+49A>G) | A/G | | AA/GA/GG | GG vs. AG+AA | GG vs. AG+AA |
| CH | 320/610 | Ref. | 53/214/198 | Ref. | Ref. |
| LC | 189/229 |
1.53*10E4 | 46/97/66 | 0.009 | 1.659
(1.137–2.421) |
| HCC | 184/284 | 0.071 | 42/100/92 | 0.537 | 1.119 (0.782,
1.602) |
| HCC+LC | 373/513 | 0.001 | 88/198/158 | 0.049 | 1.353
(1.001–1.829) |
| rs3087243 | (+6230G>A) | G/A | | GG/GA/AA | GG vs. GA+AA | GG vs. GA+AA |
| CH | 750/184 | Ref. | 301/148/18 | Ref. | Ref. |
| LC | 314/108 | 0.015 | 121/72/18 | 0.167 | 1.294
(0.898–1.866) |
| HCC | 349/113 | 0.041 | 134/81/16 | 0.235 | 1.247
(0.866–1.771) |
| LC+HCC | 663/221 | 0.007 | 255/153/34 | 0.191 | 1.224
(0.904–1.656) |
 | Table IVAlleles and genotypes distribution of
the three SNPs in the cases and controls. |
Table IV
Alleles and genotypes distribution of
the three SNPs in the cases and controls.
| SNP ID | Clear | CHB | LC | HCC |
|---|
| rs5742909 |
| C | 358 (89.5) | 796 (85.8) | 330 (81.7) | 400 (89.3) |
| T | 42 (10.5) | 132 (14.2) | 74 (18.3) | 48 (10.7) |
| CC | 160 (80.0) | 342 (73.7) | 136 (67.3) | 176 (78.6) |
| CT | 38 (19.0) | 112 (24.1) | 58 (28.7) | 48 (21.4) |
| TT | 2 (1.0) | 10 (2.2) | 8 (4.0) | 0 (0.0) |
| Hardy-Weinberg
equilibrium | 0.88 | 0.82 | 0.57 | 0.07 |
| rs231775 |
| A | 142 (34.8) | 320 (34.4) | 189 (45.2) | 184 (34.4) |
| G | 266 (65.2) | 610 (65.6) | 229 (54.8) | 284 (60.7) |
| AA | 20 (9.8) | 53 (11.4) | 46 (22.0) | 42 (17.9) |
| GA | 102 (50.0) | 214 (46) | 97 (46.4) | 100 (42.7) |
| GG | 82 (40.2) | 198 (42.6) | 66 (31.6) | 92 (39.3) |
| Hardy-Weinberg
equilibrium | 0.15 | 0.67 | 0.36 | 0.11 |
| rs3087243 |
| G | 311 (76.6) | 750 (80.3) | 314 (74.4) | 349 (75.5) |
| A | 95 (23.4) | 184 (19.7) | 108 (25.6) | 113 (24.5) |
| GG | 116 (57.1) | 301 (64.5) | 121 (64.5) | 134 (58.0) |
| GA | 79 (38.9) | 148 (31.7) | 72 (34.1) | 81 (35.1) |
| AA | 8 (3.9) | 18 (3.9) | 18 (8.5) | 16 (6.9) |
| Hardy-Weinberg
equilibrium | 0.22 | 0.97 | 0.13 | 0.44 |
Associations of the CTLA-4 polymorphisms
with HBV progression
To investigate which genotypic models were
significantly associated with various outcomes, a comparison of the
four models was conducted (multiplicative, additive, dominant and
recessive models) in the Hubei Han Chinese population (data not
shown). The best-fitting genotypic effect of the three SNPs
(rs5742909, rs231775, rs3087243) was observed in the dominant model
(Table III). Distributions of
the CTLA-4 polymorphisms in the case and control groups are
summarized in Tables III and
IV. At the SNP site rs231775, the
significant difference in allele distribution was observed only
between the CHB and LC groups (P=1.53*10E4). Following adjustment
for age and gender and analysis by unconditional binary logistic
regression, the difference remained significant (P=0.009; OR=1.659
and 95% CI=1.137–2.421). At SNP site rs3087243, compared with the
CHB group, the allele distributions revealed significant
differences in the LC and HCC groups (P=0.015 and 0.041,
respectively). However, no differences were evident following the
adjustment for age and gender. Although there was no difference
between the CHB and HCC groups at the SNPs rs231775 and rs3087243
following adjustment for age and gender, a trend for the
correlation was observed between the polymorphisms and HCC
susceptibility. The present study found that subjects with an A
allele of the two polymorphisms appeared to have a greater
susceptibility to HBV-related LC and HCC compared with those with a
G allele.
According to the clinical considerations, LC and HCC
could be lumped together since they are involved in different steps
in HBV progression. In the present study, these two HBV infection
populations were combined into one group by using the CHB group as
the reference. At the SNP site rs231775, a significant difference
was found, not only in the allele frequencies (P<0.05), but also
in the genotype distributions (P<0.05) when the combined group
(LC+HCC) was compared with the CHB group (P=0.049, OR=1.353 and 95%
CI=1.001–1.829). However, for the SNP site rs3087243, the
significant difference only appeared in the χ2 test for
allele (P=0.007) and genotype distribution (P=0.017; data not
shown). Although they were adjusted for age and gender and analysed
by unconditional binary logistic regression, the difference in the
CHB and the combined group (LC+HCC) was not significant in the site
rs3087243, and a trend (OR=1.224 and CI=0.904–1.656)) was still
observed. From these data, the subjects with A allele appeared to
have a greater susceptibility to HBV progression compared with
those with G allele in the two CTLA-4 polymorphisms, in particular
in the rs231775. In addition, no association of the CTLA-4
polymorphism rs5742909 was found with HBV progression.
Associations of the CTLA-4 polymorphisms
with viral persistence
According to the clinical considerations of this
study and our previous study (6),
CHB and LC could be lumped together, since both of them were
chronic HBV carriers. The populations of the CHB and LC were
combined in a similar manner to the HBV persistence group by using
the clear group as the reference. Following a series of statistical
analyses, the associations of the CTLA-4 polymorphisms with viral
persistence were noted only at the SNP site rs5742909 of these
three SNPs (Table III). In the
HBV persistence group (including CHB and LC), the proportions of
the C and T alleles were 89.5 and 10.5%, respectively, which were
significantly different from those observed in the clear group
(P=0.013). Following adjustment for age and gender and analysis by
unconditional binary logistic regression, the statistical level
remained significant (P=0.012, OR=1.694 and 95% CI=1.124–2.553)
compared with the clear group under the best-fitting model
(dominant model). In addition, under the additive model, the
frequencies of the C/T and T/T genotypes in HBV persistence
patients were higher compared with those in the clear subjects
(25.53 vs. 19% and 2.7 vs. 1%, respectively). Their ORs reached
1.636 (95% CI=1.074–2.492 and P=0.022) and 2.695 (95%
CI=0.599–12.131 and P=0.196) compared with the C/C genotype
(Table V). These results indicated
that the T/T and C/T genotypes and T allele of the polymorphism
rs5742909 may increase the risk of HBV persistence.
 | Table VAssociations of the three SNPs
(rs5742909, rs231775 and rs3087243) with HBV infection progression
and clearance in the Han Chinese populations (recessive and
additive models). |
Table V
Associations of the three SNPs
(rs5742909, rs231775 and rs3087243) with HBV infection progression
and clearance in the Han Chinese populations (recessive and
additive models).
| | Recessive
model | Additive model |
|---|
| |
|
|
|---|
| SNP ID | Group | P-value | OR (95%CI) | Genotypes | P-value | OR (95%CI) |
|---|
| rs5742909 | | CC+CT vs TT | CC+CT vs TT | CC/CT/TT | | CC/CT/TT |
| Clear | Ref. | Ref. | Ref. | | Ref. |
| CHB+LC | 0.25 | 2.412, (0.537,
10.824) | (CC vs. CT) | 0.022 | 1.636, (1.074,
2.553) |
| | | | (CC vs. TT) | 0.196 | 2.695, (0.599,
12.131) |
| rs231775 | | GG+GA vs. AA | GG+GA vs. AA | GG/GA/AA | | GG/GA/AA |
| CHB | Ref. | Ref. | Ref. | | Ref. |
| LC | 0.017 | 1.78
(1.111–2.852) | (GG vs. GA) | P1=0.054 | 1.487
(0.993–2.226); |
| | | | (GG vs. AA) | P1=0.003 | 2.212
(1.310–3.735) |
| HCC+LC | 0.017 | 1.78
(1.111–2.852) | (GG vs. GA) | P2=0.172 | 1.252
(0.907–1.728) |
| | | | (GG vs. AA) | P2=0.022 | 1.673
(1.079–2.595) |
| rs3087243 | | GG+GA vs. AA | GG+GA vs. AA | GG/GA/AA | | GG/GA/AA |
| CHB | Ref. | Ref. | Ref. | | Ref. |
| LC | 0.18 | 1.667
(0.789–3.519) | (GG vs. GA) | P1=0.432 | 1.169
(0.791–1.728) |
| | | | (GG vs. AA) | P1=0.146 | 1.757
(0.822–3.754) |
| LC+HCC | 0.303 | 1.410
(0.734–2.709) | (GG vs. GA) | P2=0.427 | 1.138
(0.827–1.568) |
| | | | (GG vs. AA) | P2=0.251 | 1.474
(0.760–2.858) |
Results of the haplotype analysis
In order to understand the contributions of these
loci to the HBV susceptibility, three-locus haplotypes were
constructed for the SNPs rs5742909, rs231775 and rs3087243
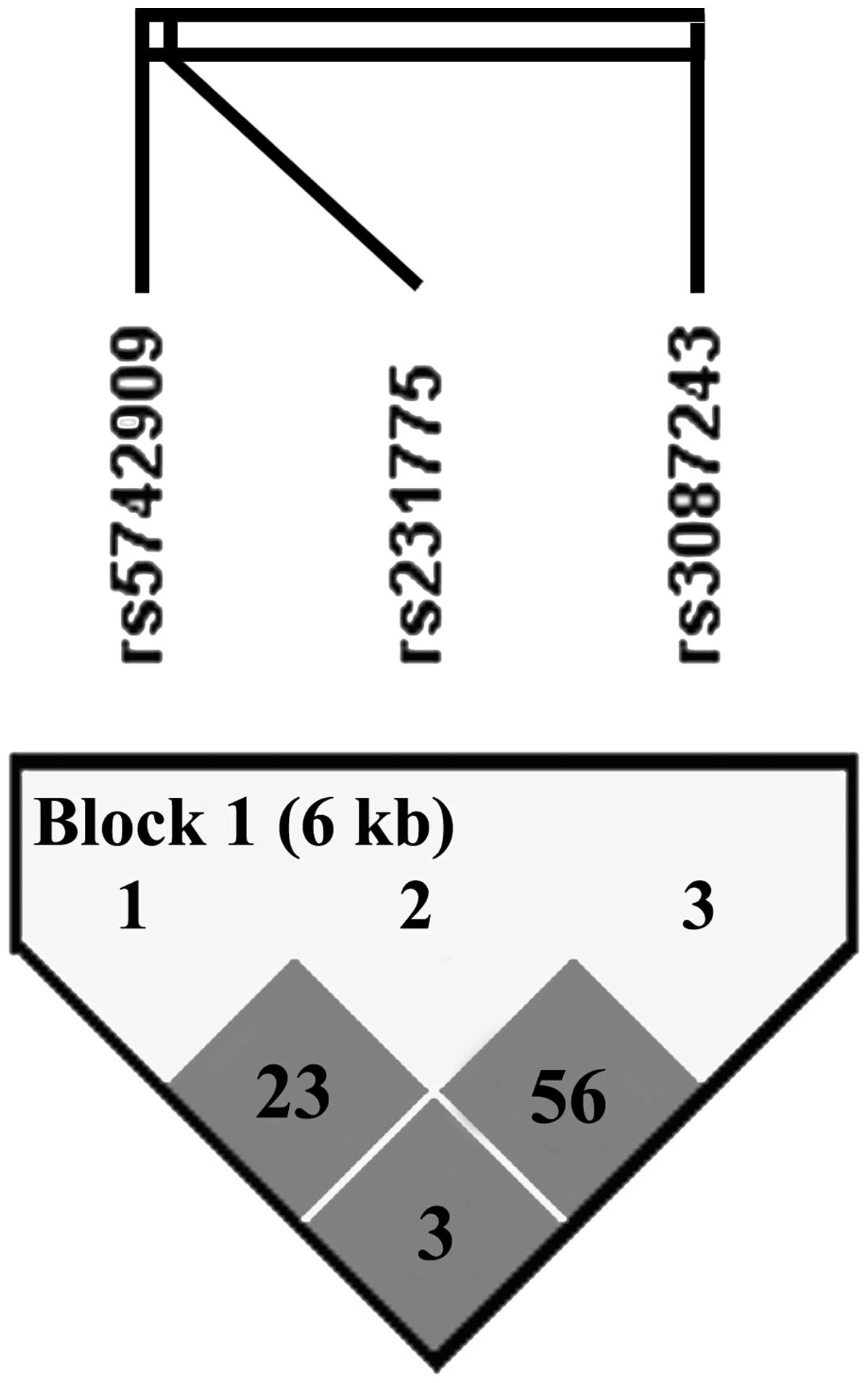
(Table VI). Pairwise LD analyses
(Fig. 1) were performed using all
the individuals from the clear group. Results of the analyses
revealed that the SNPs rs5742909, rs231775 and rs3087243 were in LD
with each other (D′=1, r2=0.233 between rs5742909 and
rs231775; D′=1, r2=0.564 between rs231775 and rs3087243;
D′=1, r2=0.035 between rs5742909 and rs3087243). In
order to derive HBV infection-specific haplotypes, haplotypes with
frequencies <0.05 were not considered and three haplotypes were
observed. When a protective haplotype C-G-G was selected as a
baseline, haplotypes C-A-A and T-A-G exhibited an increased
susceptibility to progressed hepatitis and the haplotype T-A-G
revealed an increased risk for viral persistence (P-value and OR
are shown in Table VI).
Three-loci haplotyping was performed only for the subjects with
complete genotyping.
 | Table VIResults of the association test for
the three SNPs haplotypes in the Han Chinese populations. |
Table VI
Results of the association test for
the three SNPs haplotypes in the Han Chinese populations.
| Haplotype/SNP | −318C>T | +49A>G | +6230G>A | Clear (2n=400) | CH (2n=928) | LC (2n=404) | HCC+LC
(2n=1332) | CH+LC (2n=852) |
|---|
| 1 | C | G | G | 261 (65.2) | 609 (65.6) | 219 (54.3) | 828 (62.6) | 491 (59.9) |
| 2 | C | A | A | 93 (23.3) | 186 (20.0) | 103 (25.4) | 289 (21.9) | 211 (25.7) |
| 3 | T | A | G | 46 (11.5) | 133 (14.3) | 72 (17.9) | 205 (15.5) | 118 (14.4) |
| P1-valuea | | | | Ref. | 0.283 | 0.102 | 0.849 | |
| OR (95%CI) | | | | 1 | 0.854
(0.640–1.139) | 1.320
(0.946–1.841) | 0.974
(0.742–1.279) | |
| P2-valuea | | | | Ref. | 0.246 | 0.003 | 0.054 | |
| OR (95%CI) | | | | 1 | 1.246
(0.861–1.778) | 1.865
(1.236–2.814) | 1.406
(0.992–1.993) | |
| P1-valueb | | | | | Ref. | 0.003 | | 0.004 |
| OR (95%CI) | | | | | 1 | 1.546
(1.161–2.058) | | 1.403
(1.114–1.767) |
| P2-valueb | | | | | Ref. | 0.014 | | 0.512 |
| OR (95%CI) | | | | | 1 | 1.503
(1.085–2.082) | | 1.096
(0.833–1.443) |
Discussion
The outcomes of the HBV infection vary according to
the vigor of the immune response, a process that is regulated by a
number of molecules, including the cell surface receptor CTLA-4
(25). The CTLA-4 gene is
expressed by T-lymphocytes and functions as an inhibitory receptor,
acting as a negative regulator of T-cell responses (31). Mohammad Alizadeh et al
(26) and Schott et al
(27) demonstrated a significant
association between the genotypes or alleles of the mutation
rs5742909 and the susceptibility to chronic hepatitis B. In their
studies, Gu et al and Hu et al demonstrated that the
SNP site rs231775 was associated with HCC (23,29).
However, Gu et al only identified the association in a male
Chinese population and Hu et al included HCC subjects with a
virus infection as well as with HBV. Thio et al (25) reported that presence of allele
+6230A in the 3′UTR of the CTLA4 gene (at SNP site rs3087243) was
suspected to have viral persistence and that allele +49G (at SNP
site rs231775) was detected more often in individuals who recovered
from HBV infection. Although the association between various
outcomes of HBV infection and the CTLA-4 gene has become
increasingly evident (24,28), their diagnosis criteria were not
completely equal and the ethnic background difference should be
considered. Different results are therefore likely to arise with
different diagnosis criteria and ethic backgrounds although the
control and case groups are similar.
In the analysis of the present study, three SNPs
sites (rs5742909, rs231775 and rs3087243) in the gene CTLA-4 were
confirmed to be significantly associated with HBV infection.
Additionally, CTLA-4 haplotypes are significant determinants of the
HBV infection in the Hubei Han Chinese population. Haplotypes
containing the +49G allele were protective against HBV progression
and viral persistence. Since these haplotypes may alter the ability
of CTLA-4 to downregulate the immune response, these data indicated
that the vigor of counter-regulatory mechanisms contributes to HBV
infection. Although the present study may not have indicated that
the genotypes distribution in all these three sites (rs5742909,
rs231775 and rs3087243) was significantly different between the
control and case groups, the frequencies of susceptible alleles
were similar compared with those in other Chinese populations.
Thus, it may be confirmed that the polymorphisms of the CTLA-4 gene
play a crucial role in HBV progression and viral persistence in the
Hubei Han Chinese population.
Following HBV infection, the inflammatory immune
response of the host induces hepatocellular damage and is followed
by the pathogenesis of liver cirrhosis and cancer (32). Liver cancer arises most frequently
in the setting of chronic liver inflammation (33). In the present study, the allele
+49A was identified as a risk factor for HBV progression while the
mutation allele +49G may be protective against HBV progression.
Inherited changes or SNPs in CTLA-4 expression that presumably
alter T-cell self-reactivity have been found to be associated with
autoimmune disorders (10,34,35)
including autoimmune liver disease (36–39).
The A→G mutation which exists at position +49 in exon 1 (rs231775)
leads to a non-synonymous amino acid change from threonine to
alanine, thus changing the polarity of the amino acid and
potentially altering the function of the protein. The +49A allele
has been associated with a decreased risk for diseases resulting
from a downregulated vigorous immune response (36,40–42).
Similarly, in an earlier study, the allele +49G was also associated
with improved clearance of an HCV infection resulting from
α-interferon-based therapy (43).
Lymphocytes from donors carrying +49G appear to express less CTLA-4
on their surfaces, proliferate more under conditions of suboptimal
activation and exhibit less CTLA-4-mediated inhibition of the
T-cell responses (44).
The SNP rs5742909 is an A→G mutation in the promoter
region at position −318 of the CTLA-4 gene. It has been
demonstrated that the −318T allele of CTLA-4 may result in
increased levels of CTLA-4 mRNA compared with the −318C/−318C
homozygote (45) and an increased
expression of CTLA-4 following activation (46). Although in the present study, only
the association between the mutation and viral persistence was
observed, the haplotype containing −318C or −318T is associated
with viral persistence and HBV progression, as mentioned in
previous studies (24,27). In addition, individuals with the
allele +6230A in the 3′ UTR (at the site rs3087243) were also found
to be more likely to have HBV progression. Ueda et al
(10) reported that the SNP site
rs3087243 determines the levels of the soluble isoform of CTLA-4
(sCTLA-4), which has been demonstrated in vitro to inhibit
T-cell proliferation. This may partially account for the
association of this haplotype containing the +6230A allele with HBV
progression.
In the presence of two functional polymorphisms on
the same LD block, when the predisposing allele of one is in LD
with the protective allele of the other, the genetic effects of
individual SNPs are likely to be blunted by an increase in the
frequency of haplotypes that carry one predisposing and one
protective allele (47).
Chistiakov et al (48)
suggested that whether the haplotype are likely to be susceptible,
protective or neutral depends on a ratio between predisposing and
protective alleles constituting a haplotype and an interaction
between their functional significance and strength of their
functional effects. Therefore, it is possible that the effects of
the three SNPs in chronic HBV patients involved in the present
study may be due to the linkage to each other, in particular to the
linkage to +49 A/G.
In summary, in the present case-control study, the A
allele of the rs231775 and rs3087243 SNPs sites in CTLA-4 was
confirmed to be significantly associated with HBV progression in
the Han Chinese population, and allele T in rs5742909 revealed a
strong risk effect on viral persistence. Although HBV disease is
not determined solely by genetic factors, the experimental results
offer the foundation for further study of genetic variations in
CTLA-4 for the prevention and therapy of chronic HBV infection.
However, following adjustment for age and gender, the association
among the three SNPs and the HBV-related HCC, excluding the
(LC+HCC) group, was not observed. This may be due to the difference
in criteria and ethnic background. Future investigations with a
larger sample size, multi-center study and functional studies in
this gene are required to confirm the results of the present
study.
Acknowledgements
This study was financially supported by the National
Basic Research Program of China (no. 2007CB512903) and the National
Natural Science Foundation of China (no. 81101824 and
30872237).
References
|
1
|
Guo XC and Wu YQ: A review: progress of
prevention and control on viral hepatitis in China. Biomed Environ
Sci. 12:227–232. 1999.PubMed/NCBI
|
|
2
|
Ganem D and Prince AM: Hepatitis B virus
infection - natural history and clinical consequences. N Engl J
Med. 350:1118–1129. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Thursz M: Genetic susceptibility in
chronic viral hepatitis. Antiviral Res. 52:113–116. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Harnprasopwat R, Ha D, Toyoshima T, Lodish
H, Tojo A and Kotani A: Alteration of processing induced by a
single nucleotide polymorphism in pri-miR-126. Biochem Biophys Res
Commun. 399:117–122. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li J, Yang D, He Y, et al: Associations of
HLA-DP variants with hepatitis B virus infection in southern and
northern Han Chinese populations: a multicenter case-control study.
PLoS One. 6:e242212011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
He XX, Chang Y, Jiang HJ, et al:
Persistent effect of IFNAR-1 genetic polymorphism on the long-term
pathogenesis of chronic HBV infection. Viral Immunol. 23:251–257.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liu L, Li J, Yao J, et al: A genome-wide
association study with DNA pooling identifies the variant
rs11866328 in the GRIN2A gene that affects disease progression of
chronic HBV infection. Viral Immunol. 24:397–402. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu L, Yao J, Li J, et al: Effects of
variant rs346473 in ARHGAP24 gene on disease progression of HBV
infection in Han Chinese population. J Huazhong Univ Sci Technolog
Med Sci. 31:482–487. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Szmaragd C, Nichols RA and Balloux F: A
novel approach to characterise pathogen candidate genetic
polymorphisms involved in clinical outcome. Infect Genet Evol.
6:38–45. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ueda H, Howson JM, Esposito L, et al:
Association of the T-cell regulatory gene CTLA4 with susceptibility
to autoimmune disease. Nature. 423:506–511. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Carreno BM, Bennett F, Chau TA, et al:
CTLA-4 (CD152) can inhibit T cell activation by two different
mechanisms depending on its level of cell surface expression. J
Immunol. 165:1352–1356. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kuchroo VK, Das MP, Brown JA, et al: B7-1
and B7-2 costimulatory molecules activate differentially the
Th1/Th2 developmental pathways: application to autoimmune disease
therapy. Cell. 80:707–718. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Dariavach P, Mattéi MG, Golstein P and
Lefranc MP: Human Ig superfamily CTLA-4 gene: chromosomal
localization and identity of protein sequence between murine and
human CTLA-4 cytoplasmic domains. Eur J Immunol. 18:1901–1905.
1988. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Leach DR, Krummel MF and Allison JP:
Enhancement of antitumor immunity by CTLA-4 blockade. Science.
271:1734–1736. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hellings PW, Vandenberghe P, Kasran A, et
al: Blockade of CTLA-4 enhances allergic sensitization and
eosinophilic airway inflammation in genetically predisposed mice.
Eur J Immunol. 32:585–594. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Karandikar NJ, Vanderlugt CL, Walunas TL,
Miller SD and Bluestone JA: CTLA-4: a negative regulator of
autoimmune disease. J Exp Med. 184:783–788. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kirman J, McCoy K, Hook S, et al: CTLA-4
blockade enhances the immune response induced by mycobacterial
infection but does not lead to increased protection. Infect Immun.
67:3786–3792. 1999.PubMed/NCBI
|
|
18
|
Espenschied J, Lamont J, Longmate J, et
al: CTLA-4 blockade enhances the therapeutic effect of an
attenuated poxvirus vaccine targeting p53 in an established murine
tumor model. J Immunol. 170:3401–3407. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Scheipers P and Reiser H: Fas-independent
death of activated CD4(+) T lymphocytes induced by CTLA-4
crosslinking. Proc Natl Acad Sci U S A. 95:10083–10088. 1998.
|
|
20
|
Ohta A, Sekimoto M, Sato M, et al:
Indispensable role for TNF-alpha and IFN-gamma at the effector
phase of liver injury mediated by Th1 cells specific to hepatitis B
virus surface antigen. J Immunol. 165:956–961. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Stoop JN, Woltman AM, Biesta PJ, et al:
Tumor necrosis factor alpha inhibits the suppressive effect of
regulatory T cells on the hepatitis B virus-specific immune
response. Hepatology. 46:699–705. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Han Q, Duan S, Zhang G, et al:
Associations between cytotoxic T lymphocyte-associated antigen-4
polymorphisms and serum tumor necrosis factor-α and interferon-γ
levels in patients with chronic hepatitis B virus infection.
Inflamm Res. 60:1071–1078. 2011.
|
|
23
|
Hu L, Liu J, Chen X, et al: CTLA-4 gene
polymorphism +49 A/G contributes to genetic susceptibility to two
infection-related cancers-hepatocellular carcinoma and cervical
cancer. Hum Immunol. 71:888–891. 2010.
|
|
24
|
Duan S, Zhang G, Han Q, et al: CTLA-4 exon
1 +49 polymorphism alone and in a haplotype with −318 promoter
polymorphism may confer susceptibility to chronic HBV infection in
Chinese Han patients. Mol Biol Rep. 38:5125–5132. 2011.
|
|
25
|
Thio CL, Mosbruger TL, Kaslow RA, et al:
Cytotoxic T-lymphocyte antigen 4 gene and recovery from hepatitis B
virus infection. J Virol. 78:11258–11262. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mohammad Alizadeh AH, Hajilooi M, Ranjbar
M, Fallahian F and Mousavi SM: Cytotoxic T-lymphocyte antigen 4
gene polymorphisms and susceptibility to chronic hepatitis B. World
J Gastroenterol. 12:630–635. 2006.PubMed/NCBI
|
|
27
|
Schott E, Witt H, Pascu M, et al:
Association of CTLA4 single nucleotide polymorphisms with viral but
not autoimmune liver disease. Eur J Gastroenterol Hepatol.
19:947–951. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen DQ, Zeng Y, Zhou J, et al:
Association of candidate susceptible loci with chronic infection
with hepatitis B virus in a Chinese population. J Med Virol.
82:371–378. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gu X, Qi P, Zhou F, et al: +49G > A
polymorphism in the cytotoxic T-lymphocyte antigen-4 gene increases
susceptibility to hepatitis B-related hepatocellular carcinoma in a
male Chinese population. Hum Immunol. 71:83–87. 2010.
|
|
30
|
Excoffier L and Lischer HE: Arlequin suite
ver 3.5: a new series of programs to perform population genetics
analyses under Linux and Windows. Mol Ecol Resour. 10:564–567.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Shevach EM: CD4+
CD25+ suppressor T cells: more questions than answers.
Nat Rev Immunol. 2:389–400. 2002.
|
|
32
|
Fattovich G, Bortolotti F and Donato F:
Natural history of chronic hepatitis B: special emphasis on disease
progression and prognostic factors. J Hepatol. 48:335–352. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yang HI, Lu SN, Liaw YF, et al: Hepatitis
B e antigen and the risk of hepatocellular carcinoma. N Engl J Med.
347:168–174. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Aguilar F, Torres B, Sánchez-Román J,
Núñez-Roldán A and González-Escribano MF: CTLA4 polymorphism in
Spanish patients with systemic lupus erythematosus. Hum Immunol.
64:936–940. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Parks CG, Hudson LL, Cooper GS, et al:
CTLA-4 gene polymorphisms and systemic lupus erythematosus in a
population-based study of whites and African-Americans in the
southeastern United States. Lupus. 13:784–791. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Agarwal K, Czaja AJ, Jones DE and
Donaldson PT: Cytotoxic T lymphocyte antigen-4 (CTLA-4) gene
polymorphisms and susceptibility to type 1 autoimmune hepatitis.
Hepatology. 31:49–53. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Agarwal K, Jones DE, Daly AK, et al:
CTLA-4 gene polymorphism confers susceptibility to primary biliary
cirrhosis. J Hepatol. 32:538–541. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Djilali-Saiah I, Ouellette P,
Caillat-Zucman S, Debray D, Kohn JI and Alvarez F: CTLA-4/CD 28
region polymorphisms in children from families with autoimmune
hepatitis. Hum Immunol. 62:1356–1362. 2001. View Article : Google Scholar
|
|
39
|
Fan LY, Tu XQ, Cheng QB, et al: Cytotoxic
T lymphocyte associated antigen-4 gene polymorphisms confer
susceptibility to primary biliary cirrhosis and autoimmune
hepatitis in Chinese population. World J Gastroenterol.
10:3056–3059. 2004.
|
|
40
|
Ahmed S, Ihara K, Kanemitsu S, et al:
Association of CTLA-4 but not CD28 gene polymorphisms with systemic
lupus erythematosus in the Japanese population. Rheumatology
(Oxford). 40:662–667. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ihara K, Ahmed S, Nakao F, et al:
Association studies of CTLA-4, CD28, and ICOS gene polymorphisms
with type 1 diabetes in the Japanese population. Immunogenetics.
53:447–454. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kouki T, Sawai Y, Gardine CA, Fisfalen ME,
Alegre ML and DeGroot LJ: CTLA-4 gene polymorphism at position 49
in exon 1 reduces the inhibitory function of CTLA-4 and contributes
to the pathogenesis of Graves’ disease. J Immunol. 165:6606–6611.
2000.PubMed/NCBI
|
|
43
|
Yee LJ, Perez KA, Tang J, van Leeuwen DJ
and Kaslow RA: Association of CTLA4 polymorphisms with sustained
response to interferon and ribavirin therapy for chronic hepatitis
C virus infection. J Infect Dis. 187:1264–1271. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Mäurer M, Loserth S, Kolb-Mäurer A, et al:
A polymorphism in the human cytotoxic T-lymphocyte antigen 4 (
CTLA4) gene (exon 1 +49) alters T-cell activation. Immunogenetics.
54:1–8. 2002.
|
|
45
|
Wang XB, Zhao X, Giscombe R and Lefvert
AK: A CTLA-4 gene polymorphism at position −318 in the promoter
region affects the expression of protein. Genes Immun. 3:233–234.
2002.
|
|
46
|
Ligers A, Teleshova N, Masterman T, Huang
WX and Hillert J: CTLA-4 gene expression is influenced by promoter
and exon 1 polymorphisms. Genes Immun. 2:145–152. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Anjos SM, Tessier MC and Polychronakos C:
Association of the cytotoxic T lymphocyte-associated antigen 4 gene
with type 1 diabetes: evidence for independent effects of two
polymorphisms on the same haplotype block. J Clin Endocrinol Metab.
89:6257–6265. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Chistiakov DA, Savost’anov KV, Turakulov
RI, Efremov IA and Demurov LM: Genetic analysis and functional
evaluation of the C/T(−318) and A/G(−1661) polymorphisms of the
CTLA-4 gene in patients affected with Graves’ disease. Clin
Immunol. 118:233–242. 2006.PubMed/NCBI
|