Introduction
Physalis alkekengi var. francheti of
the Solanaceae family is a well-known edible and medicinal plant
used in Northeastern regions of China. Its fruit calyx has been
used in traditional Chinese medicine as a therapeutic agent for
removing fever or inflammation and toxic materials and relieving
sore throats (1). It has great
potential and development of a stable form of P. alkekengi
var. francheti as a product may fulfill the requirements of
the health food market. Studies on P. alkekengi var.
francheti have attracted increased attention in recent years
due to its potential biological functions. In addition, the active
components of this plant, including physalins, polysaccharides and
flavones, have been widely studied (2–5).
However, to the best of our knowledge, no studies
have analyzed the improvement of intestinal microflora balance
using P. alkekengi var. francheti. Therefore, the
present study focused on the structure of the polysaccharide
fraction [designated P. alkekengivar. francheti
polysaccharide B (PPSB) hereafter], which may improve the
intestinal microflora balance and evaluated the effects of PPSB on
intestinal bacteria in vitro and in vivo. The
objectives of this study were to investigate the isolation and
characterization of PPSB from the fruit calyx of P.
alkekengi var. francheti and to analyze its effects on
intestinal microflora in vitro and in vivo. PPSB
showed stimulatory effects on the growth of probiotic bacteria and
inhibitory effects on that of pathogenic bacteria. Denaturing
gradient gel electrophoresis (DGGE) was combined with image
analysis to provide insight into the microbial similarities and
diversity of the sites, while construction of an unweighted pair
group method with arithmetic mean (UPGMA) dendrogram (6,7) and
sequencing (8) were conducted to
test for DGGE motifs and taxa.
Materials and methods
Materials and chemicals
Calyces of P. alkekengi var. francheti
were purchased from Dalian Traditional Chinese Medicine Market
(Dalian, China) in October 2011 and identified according to the
identification standard of Pharmacopeia of the People’s Republic of
China (9). A voucher specimen (no.
LP201101) was deposited in the Department of Biotechnology, Dalian
Medical University, Dalian, China.
DEAE-52 Cellulose was purchased from Whatman
International Ltd. (Maidstone, Kent, UK). Sephadex G-200 was
purchased from Pharmacia Co. (Stockholm, Sweden) and the
E.Z.N.A® Stool DNA kit was purchased from Omega Bio-tek,
Inc. (Norcross, GA, USA). Polymerase chain reaction primers,
GC-357f, 518r and 357f (10)
(Table I), were synthesized by
Takara Biotechnology (Dalian) Co., Ltd. (Dalian, China). All other
chemical reagents used were of analytical grade.
 | Table INucleotide sequences of primers. |
Table I
Nucleotide sequences of primers.
| Primer | Sequence 5′-3′ |
|---|
| GC-357fa |
CGCCCGGGGCGCGCCCCGGGCGGGGCGGGGGACGGGGGGCCTACGGGAGGCAGCAG |
| 518rb |
ATTACCGCGGCTGCTGG |
| 357fa |
CCTACGGGAGGCAGCAG |
Isolation and purification of PPSB
The air-dried calyces of P. alkekengi var.
francheti (3.0 kg) were extracted four times at 85ºC using
distilled water (30 liters), for 3 h each time. Each extract was
filtered, concentrated and centrifuged (HC-3018 R High speed
refrigerated centrifuge; Zhongke Zhongjia Scientific Instrument
Co., Ltd., Anhui, China), prior to treating the supernatant with
three volumes of ethanol at 4ºC overnight. The crude
polysaccharides precipitated by ethanol were washed with dehydrated
alcohol and diethyl ether, collected by centrifugation and dried
under reduced pressure. The sample was dissolved in distilled water
and frozen at −20ºC, thawed at room temperature and centrifuged at
9,000 × g for 20 min to remove insoluble materials. Crude
polysaccharide was precipitated with 50% ethanol and the
supernatant was recovered by centrifugation. The polysaccharide
fraction, termed PPS, was obtained from the supernatant by
precipitation using 70% ethanol (4,11).
P. alkekengivar. francheti polysaccharide A (PPSA) was
deproteinated by a combination of proteinase and the Sevag method
(12). Dissolved PPSA was
fractionated on a DEAE-52 cellulose column (1.5×90 cm) (13). The column was eluted first with
distilled water and then with a gradient solution (0.1, 0.25 and
0.5 mol/l NaCl; 0.5 mol/l NaOH) at a flow rate of 0.5 ml/min. The
major polysaccharide fractions were collected and concentrated and
residues were loaded onto a Sephadex G-200 gel column (1.5×90 cm).
The column was eluted with 0.1 mol/l NaCl at a flow rate of 0.5
ml/min and the main polysaccharide fraction (PPSB) was collected,
dialyzed (MD 34 dialysis tube; Solarbio Co., Ltd. Beijing, China)
and lyophilized (FD-1A-50 lyophilizer; Bilang Instrument Co., Ltd.,
Shanghai, China) for further analysis. Each fraction was monitored
with the phenol sulfuric acid method using glucose as a standard
(14).
Physicochemical properties of PPSB
The total carbohydrate content of PPSB was
determined by the phenol-sulfuric acid colorimetric method and
protein content was quantified using the Bradford method (15).
In vitro effects on the intestinal
bacterium
Experimental bacterial strains
Strains of L. delbrueckii ATCC 7830 and E.
coli ATCC 25922 were supplied by the Department of
Biotechnology, Dalian Medical University (Dalian, China).
Growth curves
L. delbrueckii ATCC 7830 and E. coli
ATCC 25922 belong to the families Lactobacillaceae and
Enterobacteriaceae, respectively, which are common and important
intestinal microflora. The strains were cultured in
DeMan-Rogosa-Sharpe (MRS) and LB mediums, respectively, and the
mediums were autoclaved at 115ºC for 15 min.
The effects of PPSB on the growth of L.
delbrueckii were analyzed using 96-well microplates (16). Subsequent to this, 100 μl MRS
inoculated broth (2.25×106 cfu/ml) was added to each
well and 100 μl PPSB was added to the first well of each column for
testing. A 2-fold serial dilution was performed using a
multichannel micropipette to produce final concentrations of
0.20–25.0 mg/ml. The microplates were incubated at 37ºC for 28 h
and the optical density values were calculated at intervals of 2 h
at 595 nm (Thermo Labsystem MK3 Microplate Reader; Thermo Fisher
Scientific, Waltham, MA, USA).
The effects of PPSB on the growth of E. coli
were analyzed using the same procedure but with 100 μl LB
inoculated broth (1.25×106 cfu/ml) added to each
well.
In vivo effects on the intestinal
microflora
Experimental animals
Female BALB/c mice weighing 20±2 g, purchased from
the Animal Experimental Center of Dalian Medical University
[certificate of quality number, SCXK (Liao) 2008–0002], were used
for this study. The mice were kept under standardized conditions at
a temperature of 22–24ºC, 20% humidity and 12 h light/dark cycle.
The mice had free access to a standard diet and water (ad
libitum) and were allowed to acclimatize for five days prior to
the start of the experiments. The study was approved by the Ethics
Committee of Dalian Medical University (Dalian, China).
Antibiotic-induced intestinal
microflora imbalance model in mice
Intestinal microflora imbalance was induced by
intragastric (i.g.) administration of 65 mg/kg levofloxacin
dissolved in water for five days (17).
Experimental design
Sixty mice were randomly divided into six groups (10
mice per group): 1, levofloxacin control (LC) with normal mice
treated with levofloxacin; 2, PPSB-L with levofloxacin-treated mice
with 50 mg/kg PPSB; 3, PPSB-H with levofloxacin-treated mice with
100 mg/kg PPSB; 4, normal control (NC) with normal mice treated
with water; 5, N-PPSB-L with normal mice with 50 mg/kg PPSB and 6,
N-PPSB-H with normal mice with 100 mg/kg PPSB.
All groups received i.g. injection once per day.
Food and water consumption and body weight were recorded daily.
Following seven days of treatment, fecal matter was collected and
preserved at −80ºC.
Deoxyribonucleic acid (DNA)
extraction
DNA was extracted from fecal samples using an
E.Z.N.A® Stool DNA kit (Omega Bio-tek, Inc.) in
accordance with the manufacturer’s instructions. The quantity and
type of DNA extracts were analyzed by electrophoresis of 1% agarose
gel containing ethidium bromide and compared with a molecular
weight standard (1 kb). The DNA concentration was measured
spectrophotometrically using the BioPhotometer plus (NanoVue Plus,
GE Healthcare, Cleveland, OH, USA) and DNA extracts were preserved
at 20ºC.
PCR amplification
Primers GC-357f and 518r were used to amplify the V3
region of bacterial 16S rRNA. PCR amplification was performed using
an automated thermocycler (Thermo Fisher Scientific) as follows: 3
μl purified genomic DNA as template (~300 ng), 2.5 μl 10X Ex
Taq buffer (Mg2+), 4 μl dNTP mixture, 2.5 μl bovine
serum albumin (1 mg/ml), 10 pmol each primer and 1.25 units Ex
Taq polymerase [Takara Biotechnology (Dalian) Co., Ltd.],
filled up to a volume of 25 μl with sterile Milli-Q water. The
thermal program consisted of an initial denaturation at 94ºC for 5
min, followed by 30 cycles of 94, 54 and 72ºC for 30 sec each, in
which the annealing temperature was 72ºC for 7 min (18). Amplification products were analyzed
initially by electrophoresis of a 1% agarose gel containing
ethidium bromide and compared with a molecular weight standard (100
bp).
DGGE analysis
DGGE was performed using the D-Code™ Universal
Mutation Detection system (Bio-Rad, Hercules, CA, USA). PCR
products were electrophoresed on 8% polyacrylamide
(acylamide/bisacrylamide, 37.5:1) gels containing a linear
denaturant gradient of 25–50%, with 100% denaturant defined as a
solution of 7 M urea and 40% (v/v) deionized formamide.
Electrophoresis was performed for 10 min at 200 V and subsequently
for 16 h at 70 V in a 1X TAE buffer at a constant temperature of
60ºC (10). Gels were stained with
AgNO3 (19).
Stained gels were analyzed using Quantity One 4.6.2
gel analysis software (Bio-Rad). Similarities were displayed
graphically as a dendrogram. The clustering algorithm used to
construct the dendrograms was an unweighted pair group method with
arithmetic mean (UPGMA) (20).
The Shannon-Wiener index of diversity (H′)
(21) was used to determine the
diversity of the bacterial community. This index was calculated
using the following formula: H′ =
−∑(pi)(lnpi),
where pi was the proportion of bands in
the track and was calculated as follows:
pi =
ni/∑ni, where
ni was the average density of peak
i in the densitometric curve. The evenness (E), which
reflected uniformity of bacterial species distribution, was also
calculated. This index was calculated by: E = H′/lnS, where
S was the number of bands.
Sequence analysis
To identify any separated and strong bands, the
bands were cut from the polyacrylamide gel using a sterile scalpel.
Gel fragments were eluted in 20 μl distilled water overnight at
4ºC. Subsequent to this, the 4 μl eluted DNA was reamplified by PCR
following the aforementioned program but with 357f as the forward
primer. Each PCR product was also subjected to DGGE analysis to
confirm whether the band had been successfully purified. Following
this, idiographic sequences were attained by Takara Biotechnology
(Dalian) Co., Ltd. and the sequences were compared directly with
those in GeneBank by basic local alignment search tool (BLAST;
http://blast.ncbi.nlm.nih.gov/Blast.cgi) search
(NCBI).
Statistical analysis
In vitro experiments to produce growth curves
were performed in triplicate and the DGGE analyses were repeated at
least three times. Statistical software SPSS version 17.0 (SPSS,
Inc., Chicago, IL, USA) was used for analysis. P-values were
determined by a t-test and P<0.01 was considered to indicate a
statistically significant difference.
Results
Isolation and purification of PPSB
Crude polysaccharide from the calyces of P.
alkekengi var. francheti was extracted by hot water and
ethanol precipitation with a yield of 13.6%. Following
deproteination using a combination of proteinase and the Sevag
method, the crude polysaccharide sample was purified by a DEAE-52
Cellulose and Sephadex G-200 gel column. The main fraction (PPSB)
was isolated for further analysis of physicochemical properties and
intestinal microflora balance activities.
Physicochemical properties and chemical
compositions
PPSB appeared as a white powder and had a negative
response to the Bradford assay. No absorption was detected by UV
spectrophotometery at 260 or 280 nm. These observations showed an
absence of nucleic acid and proteins in PPSB. A high-performance
liquid chromatography profile indicated that PPSB had a single,
symmetrical and sharp peak, therefore identifying PPSB as a
homogeneous polysaccharide. In addition, the phenol sulfuric acid
assay showed that PPSB contained 92.5% carbohydrate.
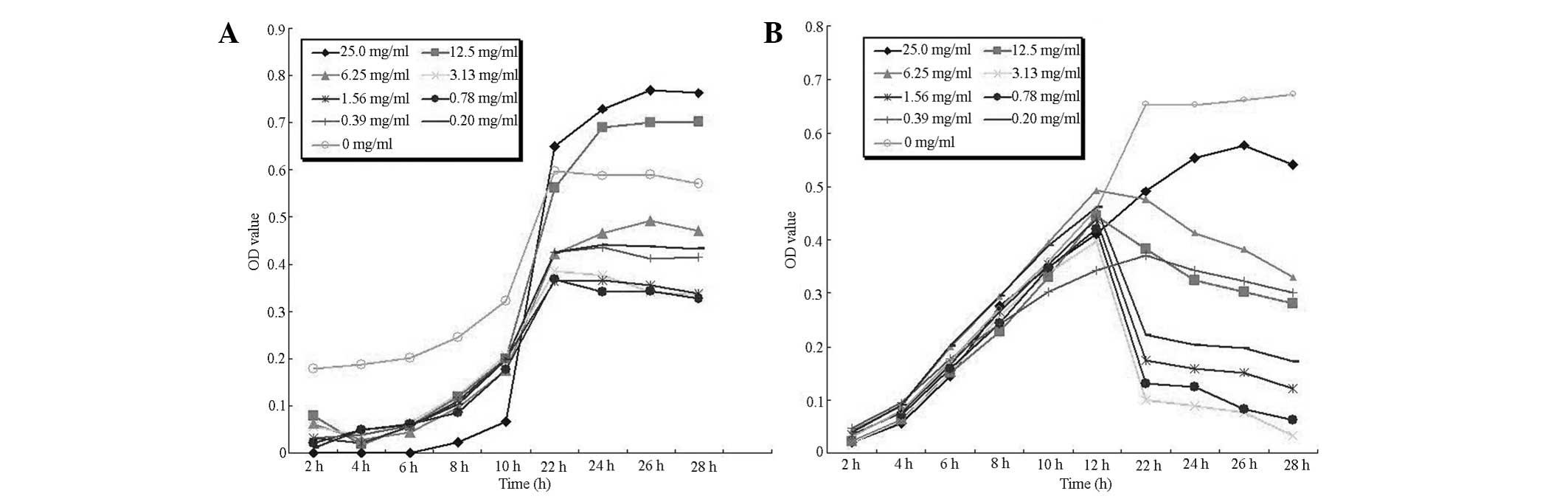
Growth curves
Fig. 1 shows the
growth curves for nine PPSB concentrations of L. delbrueckii
and Escherichia coli. Growth of L. delbrueckii was
promoted by high concentrations of PPSB (12.5–25.0 mg/ml) only.
E. coli growth was inhibited by all tested concentrations
and bacteriostatic activity increased with increasing PPSB
concentrations.
L. delbrueckii and E. coli are members
of the Lactobacillaceae and Enterobacteriaceae families,
respectively, which are typical, important intestinal microflora.
Lactobacillaceae family members are probiotics (22), whereas Enterobacteriaceae family
members are mainly regarded as opportunistic pathogens (23). Therefore, results of the present
study demonstrate that PPSB has stimulatory effects on the growth
of probiotic bacteria but inhibitory effects on the growth of
pathogenic bacteria in vitro.
DGGE analysis
The dominant intestinal microflora of LC, PPSB-L,
PPSB-H, NC, N-PPSB-L and N-PPSB-H groups was examined by DGGE
analysis with universal primers targeting the V3 region of 16S rRNA
(Fig. 2). Lane 4 was the sample
from the NC group, lane 1 contained the LC group, lanes 2 and 3
contained samples from the 50 and 100 mg/kg levofloxacin-treated
groups, respectively, whilst lanes 5 and 6 contained samples from
the normal mice treated with 50 and 100 mg/kg of PPSB,
respectively. Band A was present in the LC and NC groups but
decreased markedly in intensity in other groups. The intensity of
bands C, H, O, E, K, L and M increased markedly in
groups PPSB-H and N-PPSB-H but were not present in the LC group.
Other bands were observed in the majority of the groups, the
intensities of which were similar between groups.
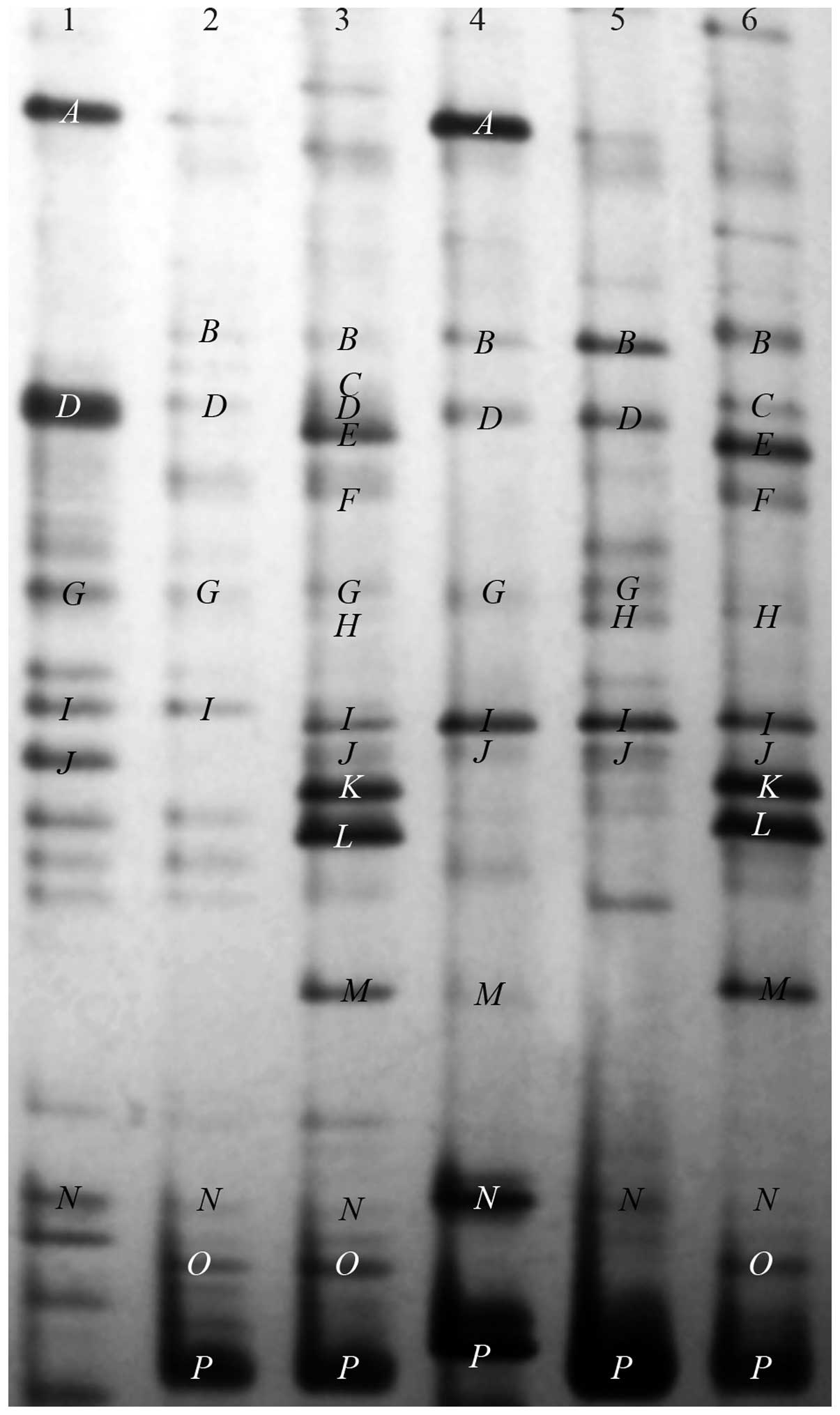 | Figure 2Representative denaturing gradient
gel electrophoresis profiles of normal, levofloxacin-treated and
P. alkekengivar. francheti polysaccharide B (PPSB)-treated
treated mice. Lane 1, levofloxacin control, normal mice treated
with levofloxacin; lane 2, PPSB-L, levofloxacin-treated mice with
50 mg/kg PPSB; lane 3, PPSB-H, levofloxacin-treated mice with 100
mg/kg PPSB; lane 4, normal control, normal mice treated with water;
lane 5, N-PPSB-L, normal mice with 50 mg/kg PPSB; lane 6, N-PPSB-H,
normal mice with 100 mg/kg PPSB. |
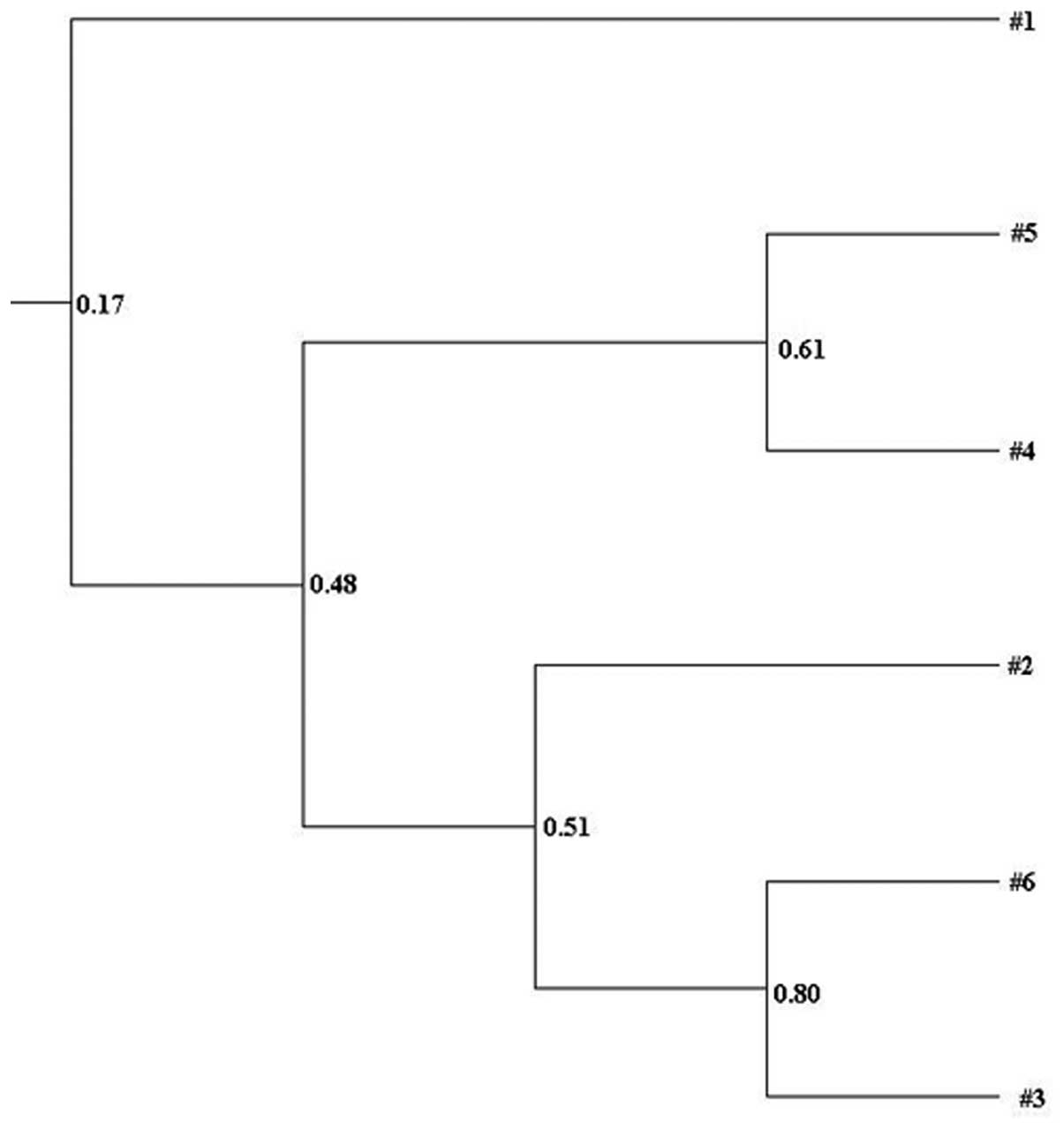
Clustering analysis based on Dice coefficient values
was depicted in a UPGMA dendrogram and the general patterns of
similarity among different groups were studied using Quantity One
software (Bio-Rad). Fig. 3
demonstrates the significantly different clustering profiles formed
by different groups. Three main clusters were identified in the
dendrogram, the first involved the lane 1 LC group, the second
involved the lanes 4 and 5 NC and N-PPSB-L groups and the third
involved the lanes 2, 3 and 6 PPSB-L, PPSB-H and N-PPSB-H
groups.
DGGE profiles showed the typical characteristics of
general bacteria in the intestinal tract. Each band is likely to be
derived from one phylogenetically distinct community and thus the
total number of bands in the DGGE profile may provide an estimation
of species number (24). The
Shannon-Wiener index values of H′, reflecting the
structural diversity of the bacterial community (21), were calculated from the number and
relative intensities of bands on the gel (Table II).
 | Table IIMicroflora diversity index analyses
(mean ± standard deviation; n=10). |
Table II
Microflora diversity index analyses
(mean ± standard deviation; n=10).
| Group | S | H′ b | Ec |
|---|
| NC | 11.6±0.9 | 2.1607±0.0203 | 0.8720±0.0497 |
| LC | 18.4±1.1a |
2.8158±0.0145a | 0.9653±0.0176 |
| PPSB-L | 15.7±1.3a |
2.4160±0.0154a | 0.8621±0.0187 |
| PPSB-H | 19.8±0.9a |
2.6538±0.0773a | 0.9096±0.0011 |
| N-PPSB-L | 11.1±2.2 | 2.1556±0.0386 | 0.9198±0.0384 |
| N-PPSB-H | 16.8±1.5a |
2.7070±0.0098a | 0.9564±0.0247 |
LC, PPSB-L, PPSB-H and N-PPSB-H group diversity
indexes were shown to increase significantly compared with the NC
group. The number of bands present in LC, PPSB-L, PPSB-H and
N-PPSB-H groups was also greater compared with the NC group.
Data in Table III
shows the closest relatives based on results of BLAST searches
using DNA sequences obtained from DGGE gel bands identified by
cluster analysis. Bands in the same position but in different lanes
were excised and sequenced to confirm that they had the same
identity (data not shown). C, O and H were
sequenced and identified as Lactobacillus gasseri,
reuteri and amylolyticus of the Lactobacillus
genus with similarities of 100, 91 and 98%, respectively. Bands for
these three bacteria increased in number in the PPSB-H and N-PPSB-H
groups, while there was no band distinguishable at the
corresponding position in other groups, notably from the LC group.
A was sequenced and identified as Enterococcus
faecium of the Enterococcus genus with a similarity of 99%.
Band A was present in LC and NC groups and at a lower band
intensity in other groups. J, K, L, M
and N were sequenced and identified as Prevotella
micans, amnii, dentails, buccae and sp. of
the Prevotella genus with similarities of 98, 99, 97, 94 and
96%, respectively. D, F and I were sequenced
and identified as Helicobacter bilis, cinaedi and
hepaticus of the Helicobacter genus with similarities of 95,
96 and 97%, respectively. B, E, G and P
were sequenced and identified as Akkermansia muciniphila,
Alistipes putredinis, Odoribacter splanchnicus and
Bacteroides coprosuis with similarities of 94, 90, 96 and
98%, respectively.
 | Table IIISequences of PCR amplicons derived
from denaturing gradient gel electrophoresis gels and identities
based on the BLAST database. |
Table III
Sequences of PCR amplicons derived
from denaturing gradient gel electrophoresis gels and identities
based on the BLAST database.
| Selected band | Most similar
sequence relative (GenBank accession number) | Bacteria genus | Identity, % |
|---|
| A | Enterococcus
faecium (AJKH01000109.1) |
Enterococcus | 99 |
| B | Akkermansia
muciniphila (NC 010655.1) |
Akkermansia | 94 |
| E | Alistipes
putredinis (ABFK02000016.1) |
Alistipes | 90 |
| G | Odoribacter
splanchnicus (NC 015160.1) |
Odoribacter | 96 |
| C | Lactobacillus
gasseri (ADFT01000001.1) |
Lactobacillus | 100 |
| O | Lactobacillus
reuteri (CACS02000061.1) | | 91 |
| H | Lactobacillus
amylolyticus (ADNY01000006.1) | | 98 |
| D | Helicobacter
bilis (ACDN01000023.1) |
Helicobacter | 95 |
| F | Helicobacter
cinaedi (ABQT01000054.1) | | 96 |
| I | Helicobacter
hepaticus (NC 004917.1) | | 97 |
| J | Prevotella
micans (AGWK01000061.1) |
Prevotella | 98 |
| K | Prevotella
amnii (ADFQ01000002.1) | | 99 |
| L | Prevotella
dentails (AFPW01000057.1) | | 97 |
| M | Prevotella
buccae (AEPD01000042.1) | | 94 |
| N | Prevotella
sp. (ACZS01000106.1) | | 96 |
| P | Bacteroides
coprosuis (AFFW01000002.1) |
Bacteroides | 98 |
DGGE analysis indicated that levofloxacin has an
important effect on the intestinal microflora of mice. An increased
diversity of bacterial composition in LC, PPSB-L, PPSB-H and
N-PPSB-H groups was found and may be associated with bacterial
growth in levofloxacin-/PPSB-treated groups. Bacterial composition
and structure were divided into three clusters for each of the
different groups. The intestinal microflora communities of PPSB-H
and N-PPSB-H groups demonstrated a relatively high homology. The
results from sequencing provided more precise information to
confirm the intestinal bacterial compositions of different groups.
Bacteria belonging to the Bacteroides, Lactobacillus,
Helicobacter, Prevotella, Enterococcus,
Odoribacter, Alistipes and Akkermansia genera
were dominant organisms in the intestinal tract of mice when
analyzed by DGGE analysis. It is likely that the transformation of
dominant organisms may be a risk factor associated with the side
effects of antibiotics. Specifically, in the present study, the
levofloxacin-treated group showed decreased Lactobacillus
levels and increased Enterococcus levels. In addition, as
the dose of PPSB increased, Lactobacillus and
Prevotella noticeably increased in terms of number of
species and quantity. These observations indicate that
Lactobacillus growth may be promoted by PPSB in vivo,
consistent with the results in vitro.
Discussion
The abuse of antibiotics has become a worldwide
public health problem as unnecessary use of antibiotics is a major
cause of the development of antibiotic-resistant bacterial strains
(25). Side effects affecting the
gastrointestinal tract, including abdominal pain, diarrhea and
astriction, are common (26,17).
Non-pathogenic diarrhea is caused by an imbalance of intestinal
microflora in the majority of cases. Once the intestinal microflora
balance is disturbed, pathogenic bacteria, viruses and coccidia may
easily infect the host.
Probiotics are live microorganisms that positively
affect the colonization and composition of intestinal microflora
and have stimulatory effects on the digestive processes and the
immunity of the host (27).
Lactobacillus spp. (28,29),
B. subtilis and S. cerevisiae (30) are probiotics with positive effects
on gastrointestinal microflora. However, probiotics are not
currently administered with antibiotics, but various other methods
have been used for improving the intestinal microflora balance, for
example the use of prebiotics (31). An increasing number of studies
(31,32) have indicated that traditional
Chinese medicines have been used as prebiotics for improving host
immunity and are pivotal in the improvement of intestinal
microflora balance.
In the present study, PPSB compounds, the main
components of P. alkekengi var. francheti, were
identified as the active compounds, which promote the number of
species and quantity of Lactobacillus in vitro and in
vivo. Intestinal microflora imbalances caused by antibiotics
may induce pathogenic diseases and it has been suggested that an
improvement in the intestinal microflora, for example an increase
in the quantity of Lactobacillus and Bifidobacterium,
may aid in the recovery from pathogenic diseases. The intestinal
microflora composition produced from the present study indicates
that the side effects of antibiotics are associated with the entire
intestinal microflora composition rather than with the action of a
single bacteria. As the dose of PPSB increased,
Lactobacillus and Prevotella levels increased but
those of Enterococcus markedly decreased in terms of quality
and quantity, which was in accordance with known probiotic
characteristics. Thus PPSB may be considered as a potential
candidate for developing a novel agent that is able to improve the
intestinal microflora balance.
Acknowledgements
This study was supported by a 973 project from the
National Basic Research Program of China (project no.
2007CB513006).
References
|
1
|
Ge Y, Duan Y, Fang G, Zhang Y and Wang S:
Study on biological activities of Physalis alkekengi var.
francheti polysaccharide. J Sci Food Agr. 89:1593–1598.
2009.
|
|
2
|
Vessal M, Mehrani HA and Omrani GH:
Effects of an aqueous extract of Physalis alkekengi fruit on
estrus cycle, reproduction and uterine creatine kinase BB-isozyme
in rats. J Ethnopharmacol. 34:69–78. 1991.
|
|
3
|
Vessal M, Rasti M and Kooshesh F:
Modulation of the pituitary and basomedial hypothalamic
lysylaminopeptidase activities by beta-estradiol and/or an aqueous
extract of Physalis alkekengi fruits. Comp Biochem Physiol B
Biochem Mol Biol. 115:267–271. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tong H, Liang Z and Wang G: Structural
characterization and hypoglycemic activity of a polysaccharide
isolated from the fruit of Physalis alkekengi L. Carbohydr
Polym. 71:316–323. 2008. View Article : Google Scholar
|
|
5
|
Ge Y, Duan Y, Fang G, Zhang Y and Wang S:
Polysaccharides from fruit calyx of Physalis alkekengi var.
francheti: Isolation, purification, structural features and
antioxidant activities. Carbohydr Polym. 77:188–193. 2009.
|
|
6
|
Fromin N, Hamelin J, Tarnawski S, et al:
Statistical analysis of denaturing gel electrophoresis (DGE)
fingerprinting patterns. Environ Microbiol. 4:634–643. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
van der Gast CJ, Whiteley AS, Lilley AK,
Knowles CJ and Thompson IP: Bacterial community structure and
function in a metalworking fluid. Environ Microbiol. 5:453–461.
2003.PubMed/NCBI
|
|
8
|
McBain AJ, Bartolo RG, Catrenich CE,
Charbonneau D, Ledder RG and Gilbert P: Effects of
triclosan-containing rinse on the dynamics and antimicrobial
susceptibility of in vitro plaque ecosystems. Antimicrob Agents
Chemother. 47:3531–3538. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pharmacopeia of the People’s Republic of
China. Chemical Industry Press; Beijing: pp. 250–251. 2005
|
|
10
|
Muyzer G, de Waal EC and Uitterlinden AG:
Profiling of complex microbial populations by denaturing gradient
gel electrophoresis analysis of polymerase chain reaction-amplified
genes coding for 16S rRNA. Appl Environ Microbiol. 59:695–700.
1993.
|
|
11
|
Zhao W, Li JJ, Yue SQ, Zhang LY and Dou
KF: Antioxidant activity and hepatoprotective effect of a
polysaccharide from Bei Chaihu (Bupleurum chinense DC).
Carbohydr Polym. 89:448–452. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Staub AM: Removal of protein-Sevag method.
Meth Carbohydr Chem. 5:5–6. 1965.
|
|
13
|
Tan RX: Analysis of plant composition.
Science Press; China, Beijing: pp. 76–78. 2002, (In Chinese).
|
|
14
|
DuBois M, Gilles KA, Hamilton JK, Rebers
PA and Smith F: Colorimetric method for determination of sugars and
related substances. Anal Chem. 28:350–356. 1956. View Article : Google Scholar
|
|
15
|
Bradford MM: A rapid and sensitive method
for the quantitation of microgram quantities of protein utilizing
the principle of protein-dye binding. Anal Biochem. 72:248–254.
1976. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gutierrez J, Barry-Ryan C and Bourke P:
The antimicrobial efficacy of plant essential oil combinations and
interactions with food ingredients. Int J Food Microbiol.
124:91–97. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li XL, Wu DC, Zhang CL and Xin Y: Effects
of levofloxacin hydrochloride on the intestinal microbiota of
BALB/c mice by PCR-DGGE. Afr J Microbiol Res. 6:3455–3460.
2012.
|
|
18
|
Ledder RG, Gilbert P, Huws SA, et al:
Molecular analysis of the subgingival microbiota in health and
disease. Appl Environ Microbiol. 73:516–523. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Edenborn SL and Sexstone AJ: DGGE
fingerprinting of culturable soil bacterial communities complements
culture-independent analyses. Soil Biol Biochem. 39:1570–1579.
2007. View Article : Google Scholar
|
|
20
|
Du H, Jiao N, Hu Y and Zeng Y: Real-time
PCR for quantification of aerobic anoxygenic phototrophic bacteria
based on pufM gene in marine environment. J Exp Mar Biol
Ecol. 329:113–121. 2006. View Article : Google Scholar
|
|
21
|
Gafan GP, Lucas VS, Roberts GJ, Petrie A,
Wilson M and Spratt DA: Statistical analyses of complex denaturing
gradient gel electrophoresis profiles. J Clin Microbiol.
43:3971–3978. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cruywagen CW, Jordaan I and Venter L:
Effect of Lactobacillus acidophilus supplementation of milk
replacer on preweaning performance of calves. J Dairy Sci.
79:483–486. 1996.
|
|
23
|
Liu J, Wu D, Ahmed A, et al: Comparison of
the gut microbe profiles and numbers between patients with liver
cirrhosis and healthy individuals. Curr Microbiol. 65:7–13. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hu Q, Qi HY, Zeng JH and Zhang HX:
Bacterial diversity in soils around a lead and zinc mine. J Environ
Sci (China). 19:74–79. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bia P, Tong SL and Parton KA: Family
self-medication and antibiotics abuse for children and juveniles in
a Chinese city. Soc Sci Med. 50:1445–1450. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sprandel KA and Rodvold KA: Safety and
tolerability of fluoroquinolones. Clin Cornerstone. (Suppl 3):
S29–S36. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Choi JY, Shinde PL, Ingale SL, et al:
Evaluation of multi-microbe probiotics prepared by submerged liquid
or solid substrate fermentation and antibiotics in weaning pigs.
Livest Sci. 138:144–151. 2011. View Article : Google Scholar
|
|
28
|
Abe F, Ishibashi N and Shimamura S: Effect
of administration of bifidobacteria and lactic acid bacteria to
newborn calves and piglets. J Dairy Sci. 78:2838–2846. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Guerra NP, Bernárdez PF, Méndez J,
Cachaldora P and Pastrana Castro L: Production of four potentially
probiotic lactic acid bacteria and their evaluation as feed
additives for weaned piglets. Anim Feed Sci Tech. 134:89–107. 2007.
View Article : Google Scholar
|
|
30
|
Mathew AG, Chattin SE, Robbins CM and
Golden DA: Effects of a direct-fed yeast culture on enteric
microbial populations, fermentation acids, and performance of
weaning pigs. J Anim Sci. 76:2138–2145. 1998.PubMed/NCBI
|
|
31
|
Ishihara N, Chu DC, Akachi S and Juneja
LR: Improvement of intestinal microflora balance and prevention of
digestive and respiratory organ diseases in calves by green tea
extracts. Livest Prod Sci. 68:217–229. 2001. View Article : Google Scholar
|
|
32
|
Koropatkin NM, Cameron EA and Martens EC:
How glycan metabolism shapes the human gut microbiota. Nat Rev
Microbiol. 10:323–335. 2012.PubMed/NCBI
|