Introduction
Cirrhosis is a disease with high morbidity and
mortality in China, which is caused by various factors, including
chronic viral hepatitis, autoimmune hepatitis, fatty liver disease
or hereditary metabolic disorders (1–4).
Cirrhosis is characterized by the excessive accumulation of
extracellular matrix (ECM), disrupting normal liver architecture
and hepatic function (5–7). Liver fibrosis is the middle stage in
the progression from chronic liver diseases to cirrhosis. Hepatic
stellate cells (HSCs) as a non-parenchymal liver cell population,
are the most important cell type for the production of collagens
(8–10). Therefore, it is a possible
therapeutic strategy to treat liver fibrosis by inhibiting the
activation of HSCs.
Previous studies have indicated that microRNAs
(miRNAs) are important in regulating the activitiy of HSCs
(11–14). The molecular mechanisms underlying
the biogenesis of miRNAs in mammalian cells has been studied
extensively (15,16). In brief, mature miRNAs are derived
from RNA molecules that are selectively cleaved by the ribonuclease
Drosha, exported into the cytoplasm and cleaved again by Dicer.
Consequently, Dicer is essential for the processing of miRNAs.
Dysregulation of Dicer globally impairs miRNA processing and
activity. Currently, Dicer is extensively studied in development
and cancer. Loss of Dicer in mice disrupts embryonic stem cell
differentiation and is lethal during early development (17). Additionally, Dicer is upregulated
in prostate adenocarcinoma, ovarian serous carcinomas, pleomorphic
adenomas of the salivary gland and acute myeloid leukemia (18–21).
By contrast, it has been demonstrated that Dicer is downregulated
in hepatocellular carcinoma (HCC). Reduced Dicer expression is
associated with the poor prognosis of patients with HCC (22). In summary, these findings imply
that the knockdown of Dicer may be involved in tumorigenesis.
However, whether Dicer expression is associated with liver fibrosis
is yet to be determined.
Our previous experiment demonstrates that the
expression of Dicer is upregulated in liver fibrosis. The purpose
of the present study was to utilize an RNA interfering (RNAi)
technique to explore the effect of Dicer gene silencing on the
activity of HSCs. Our results demonstrated that Dicer
downregulation inhibits the activity of HSCs through the reduction
of miRNAs associated with liver fibrosis. Our findings reveal that
Dicer is pivotal in the progression of liver fibrosis.
Materials and methods
Design of shRNA
Target gene Dicer (accession no. XM_001068155.3) was
searched from GenBank. According to Ambion's principles of short
hairpin RNA (siRNA) design, three sequences of 19 nucleotides
containing 30–50% GC were selected and used as the target sites.
Their sequences are listed in Table
I. Another unrelated sequence was used as a negative control.
No homology sequence was identified on BLAST analysis. The shRNA
was as follows: BamHI + sense + loop + antisense + stop
signal + BbsI. Single-stranded DNA oligonucleotide was
synthesized and annealed to form double strands by Shanghai
GenePharma Co., Ltd. (Shanghai, China).
 | Table IPredesigned siRNAs for rat Dicer. |
Table I
Predesigned siRNAs for rat Dicer.
| Name | Sequence (DNA) |
|---|
| siRNA1 |
GCTACACAGGAAGTTCTTA |
| siRNA2 |
GGGAAAGTCTGCAGAACAA |
| siRNA3 |
CCTCATAACCAAGCACCTT |
Lentiviral preparation and transfection
of HSC-T6 cells
Three lentiviral pGLV1-1 shRNA vectors and a
negative vector (shRNA non-related vector) were constructed by
Shanghai GenePharma Co., Ltd. With 293T cells, the pGLV1-1 and
negative shRNA vectors were packaged into concentrated lentiviruses
by a three plasmid transfection procedure. Viral titers were
estimated with a Lenti-X qRT-PCR kit (Chemicon, Billerica, MA, USA)
and typically averaged 1×109 viral genomes/ml. Rat
HSC-T6 cells (Chinese Academy of Medical Science, Beijing, China)
were cultured in Dulbecco's modified Eagle's medium supplemented
with 10% fetal bovine serum (Invitrogen Life Technologies,
Carlsbad, CA, USA). Then cells were transfected with pGLV1-1 shRNA
vectors according to the manufacturer's instructions. In brief,
~1×105 cells were plated on a 6-well dish 24 h prior to
transfection. HSCs were transfected at a multiplicity of infection
(MOI) of 5. Transfection efficiency was determined to be >90% by
direct visualization of green fluorescent protein (GFP) expression
following transfection for 48 h. The transfected cells were
selected by adding 2 μg/ml of puromycin to the media for 1–2
weeks.
Quantitative real-time PCR
Total RNA was isolated from HSC-T6 cells using the
mirVana miRNA extraction kit (Ambion, Austin, TX, USA) according to
the manufacturer's instructions. With the ReverTra Ace qPCR RT kit
(Toyobo Co., Ltd., Osaka, Japan), 50 ng of total RNA was reverse
transcribed to cDNA. The mRNA expression was determined by
real-time PCR using cDNA, SYBR-Green real-time PCR Master mix
(Toyobo Co., Ltd.) and a set of gene-specific oligonucleotide
primers (Table II). To detect
miRNA expression, the reverse transcription and real-time PCR
reaction was performed using the TaqMan MicroRNA Assay (Applied
Biosystem, Foster, CA, USA) according to the manufacturer's
instructions. The glyceraldehyde 3-phosphate dehydrogenase (GAPDH)
and U6 shRNA levels were measured and used to normalize the
relative abundance of mRNAs and miRNAs, respectively. The
expression levels of mRNAs and miRNAs were calculated by the
2−ΔΔCt method.
 | Table IIList of primer sequences. |
Table II
List of primer sequences.
| Gene | Sequence (5′-3′) |
|---|
| Dicer | Forward
CGATAACTTTATTGGAGATTTAC
Reverse ATTGGGTGTCCCGAAGAGTT |
| Type I
collagen | Forward
CCTGGCAAAGACGGACTCAAC
Reverse GCTGAAGTCATAACCGCCACTG |
| α-SMA | Forward
TCCCTGGAGAAGAGCTACGAACT
Reverse AAGCGTTCGTTTCCAATGGT |
| TIMP-1 | Forward
CCTCTGGCATCCTCTTGTTGCTAT
Reverse CATTTCCCACAGCGTCGAATCCTT |
| PTEN | Forward
GTGGTCTGCCAGCTAAAGGT
Reverse TGTCACCACACACAGGCAAT |
| RASAL1 | Forward
CTTCCTGGGCATGGTGGAAT
Reverse CACGGACTCCAGAAGCAGTT |
| ACSL1 | Forward
TGTGGGGTGGAAATCATCGG
Reverse CATTGCTCCTTTGGGGTTGC |
| p27 | Forward
AGATACGAGTGGCAGGAGGT
Reverse ATGCCGGTCCTCAGAGTTTG |
| GAPDH | Forward
TGCACCACCAACTGCTTAG
Reverse GGATGCAGGGATGATGTTC |
Protein extraction and western blot
analysis
Proteins were separated on sodium dodecyl
sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then
transferred onto nitrocellulose membranes. Following blocking, the
membranes were incubated with primary antibodies, followed by the
appropriate secondary antibodies (Maixin, Fuzhou, Fujian, China).
The primary antibodies included: rabbit polyclonal anti-type I
collagen, anti-GAPDH, mouse monoclonal anti-α-smooth muscle actin
(α-SMA; Abcam, Cambridge, MA, USA) and rabbit polyclonal anti-Dicer
(Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA).
Immunoreactive bands were visualized by the enhanced
chemiluminescence substrate using the Fujifilm Image Reader
LAS-3000 (Fuji Medical Systems, Stamford, CT, USA).
Cell proliferation assays
The cell proliferation was measured by the WST-1
assay (Beyotime Institute of Biotechnology, Shanghai, China) based
on changes in absorbance at 450 nm. Cells were seeded in a 24-well
plate at a density of 1×104 cells/well. Then cells were
transfected with Dicer shRNAs and incubated for 48 h prior to the
assessment of cell proliferation. The optical density (OD) at 450
nm was determined using a microplate reader and the proliferation
ability was calculated as the experimental OD value/control OD
value. The experiments were implemented in triplicate.
Luciferase activity assay
3′UTRs containing putative miRNA target regions of
phosphatase and tension homolog deleted on chromosome 10 (PTEN),
Ras GTPase activating-like protein 1 (RASAL1), acyl-CoA synthetase
long-chain family member 1 (ACSL1) and p27 genes were obtained by
PCR using rat HSCs cDNA as a template. Primers were as follows:
PTEN-miR-29c, forward 5′-GGACTAGTTGGTGCTAGAAAAGGCAGCTA-3′, and
reverse 5′-AGCTTTGTTTAAACAACGGCTGACAGCTATTGAA-3′; RASAL1-miR-143,
forward 5′-GGACTAGTATTCCAGAAACGGGGCTAGG-3′ and reverse
5′-AGCTTTGTTTAAACGTATATTGTCTAGATTAA-3′; ACSL1-miR-214, forward,
5′-GGACTAGTTCTTAACCCCTTCCCCTTGT-3′ and reverse
5′-AGCTTTGTTTAAACGGGAGGAATCACAGTTGAGC-3′; and p27-miR-199a-5p,
forward 5′-GGACTAGTGATTCTGTTATGTAGCAAGAAATGG-3′ and reverse
5′-AGCTTTGTTTAAACAGTAGGGTGAGACACTTTGAATTT-3′. Each of the forward
and reverse primers carried the Spe1 and Pme1 sites
at their 5′-ends. The obtained DNA fragments were inserted into the
pMIR-reporter vector. The pMIR-reporter vector without the insert
was used as a negative control. pMIR-reporter β-gal control plasmid
was used for transfection normalization. HSCs were cultured in
24-well plates and transfected with 800 ng of reporter vector along
with 100 ng of pMIR-β-gal and 20 pmol of miRNA precursors.
Lipofectamine™ 2000 (Invitrogen Life Technologies) was used as the
transfection reagent. Following transfection 48 h later, luciferase
and β-gal activity were measured using the Dual-Light System.
Statistical analysis
Data from three independent experiments are
expressed as the mean ± SD. The difference between groups was
calculated using the Student's t-test when two groups were
compared. All tests performed were two sided. P<0.05 was
considered to indicate a statistically significant difference.
Results
Inhibition of Dicer expression by
lentivirus
We constructed a recombinant lentivirus to generate
shRNAs targeting Dicer. The Dicer expression was determined by
real-time RT-PCR and western blotting following transfection of
HSCs with pGLV1-1 shRNA vectors. Following 48 h, green fluorescence
was observed in the transfected cells under the fluorescence
microscope, the transfection efficiency being ~91%. It was
identified that the shRNA groups had significantly lower mRNA
expression of Dicer than the control group (Fig. 1A), of which the shRNA1 group
exhibited the strongest inhibitory effect. In addition, western
blot analysis demonstrated a marked decrease in the protein
expression of Dicer, of which the shRNA1 group exhibited the
strongest inhibitory effect (Fig. 1B
and C). However, there was no change in the negative group. It
was concluded that the mRNA and protein expression of Dicer was
effectively inhibited by shRNA vectors. Based on its inhibitory
effect, we selected the shRNA1 group as the object of the next
experiments.
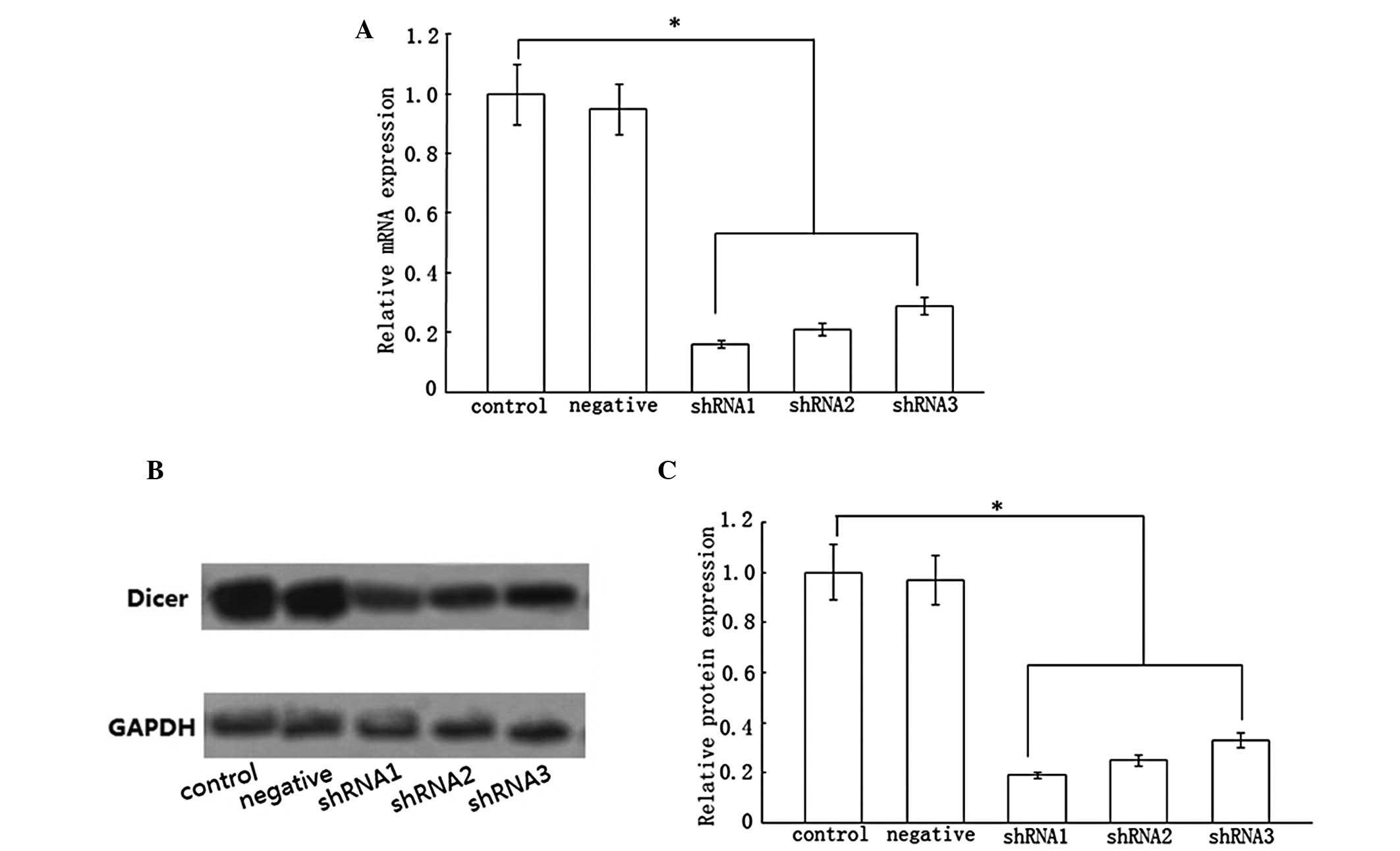 | Figure 1Effect of three pairs of shRNA vectors
on Dicer. Three pairs of shRNA vectors were transfected into HSCs
for 48 h. Compared with the control group, Dicer mRNA expression
was decreased in the shRNA1, shRNA2 and shRNA3 groups by ~84, 79
and 71%, respectively. Dicer protein expression was reduced in
shRNA1, shRNA2 and shRNA3 groups by ~81, 75 and 67%, respectively.
(A and C) Statistical analysis. (B) Results of western blotting.
Data represent the results from three independent experiments.
*P<0.05 compared with the control. shRNA, short
hairpin RNA; HSCs, hepatic stellate cells; GAPDH, glyceraldehyde
3-phosphate dehydrogenase. |
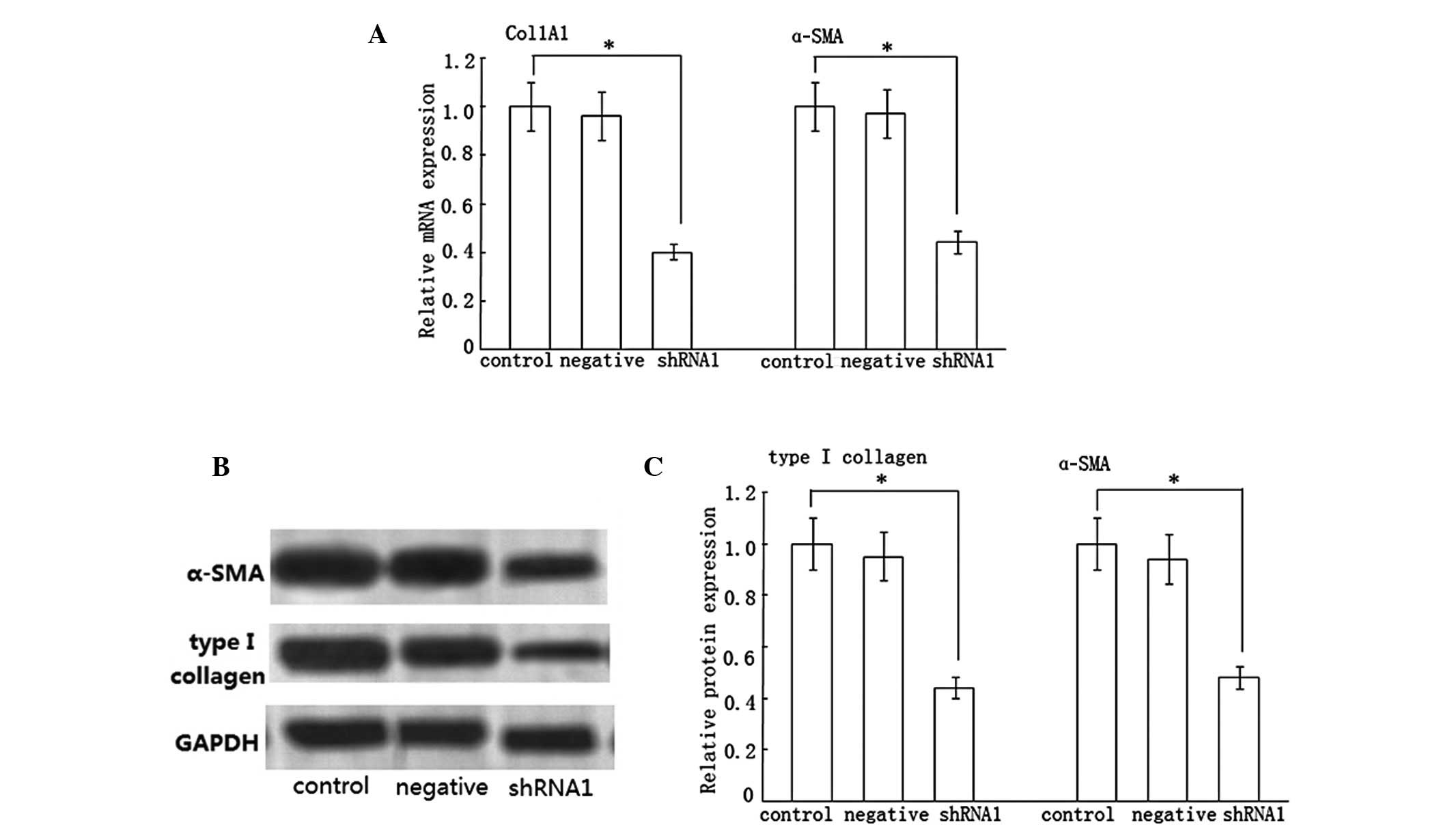
Effect of Dicer shRNA on type I collagen
and α-SMA expression
In the progression of liver fibrosis, HSCs can
express an abundance of type I collagen and α-SMA. Consequently, we
examined the effect of Dicer shRNA on type I collagen and α-SMA
expression. Compared with the control group, the shRNA1 group
presented the significant inhibition of Col1A1 and α-SMA mRNA
expression, which was decreased by ~60 and 56%, respectively
(Fig. 2A). By contrast, the
negative group had a negligible effect on them. Additionally, we
determined the effect of Dicer gene silencing on type I collagen
and α-SMA protein expression. Consistent with the real-time RT-PCR
results, the protein expression of type I collagen and α-SMA were
reduced by ~56 and 52%, respectively (Fig. 2B and C), while there was no decline
in the negative group.
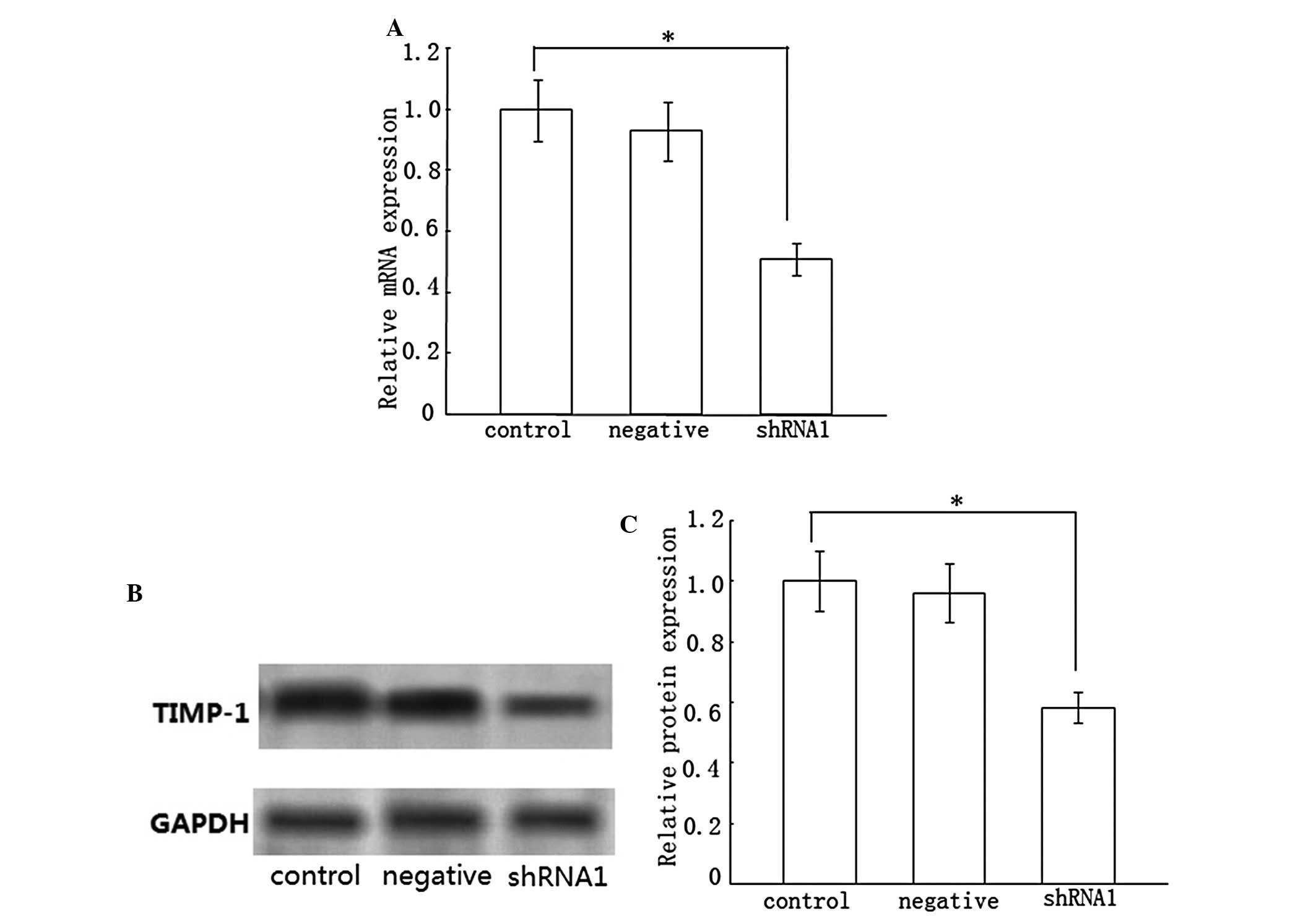
Effect of Dicer gene silencing on the
expression of tissue inhibitor of metalloproteinases-1
(TIMP-1)
A high level of TIMP-1 in liver fibrosis accounts
for the slow degradation of ECM. Accordingly, we monitored TIMP-1
mRNA and protein expression in HSC-T6 cells following transfection
with Dicer shRNA. Compared with the control group, loss of Dicer
markedly inhibited TIMP-1 mRNA and protein expression, which was
suppressed by ~49 and 42%, respectively (Fig. 3). Nevertheless, the negative group
did not demonstrate a statistical change in TIMP-1 expression.
These results suggested that the knockdown of Dicer may treat liver
fibrosis by degrading the deposition of ECM.
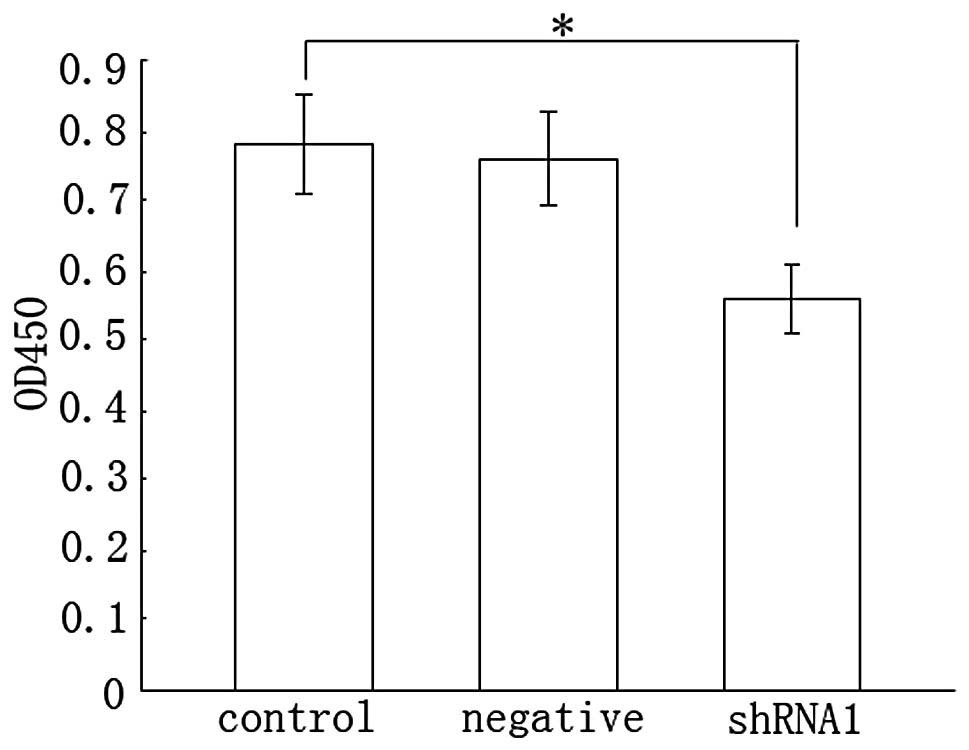
Cell proliferation evaluated by WST-1
assay
Activated HSCs are known to acquire proliferation
ability. We considered the possibility that the inhibition of Dicer
was able to regulate the proliferation of HSCs. As shown by the
WST-1 assay, shRNA1 inhibition of HSC proliferation was decreased
by 28% relative to the control (Fig.
4). Meanwhile, there was no difference between the negative and
control group. Therefore, we concluded that the knockdown of Dicer
inhibited the proliferation of HSCs.
Effect of Dicer shRNA on miRNAs
Dicer is critical for the biogenesis of miRNAs. To
determine whether the inhibition of Dicer in HSC-T6 cells could
affect miRNAs biogenesis, we performed real-time RT-PCR on RNA
isolated from cultures transfected with the shRNA1 vector. We
decided to test a set of specific miRNAs that were markedly
different between the activated and quiescent HSCs, as previously
described from microarray profiles (23). The inhibition of Dicer resulted in
a decrease in the expression of miR-214, −143, −29c and −199a-5p,
of which miR-214 exhibited the strongest decline (Fig. 5). In comparison, we did not detect
a statistically significant change in miR-19b or miR-99a,
indicating that not all miRNAs were affected by the reduction of
Dicer. These data revealed that the antifibrotic effect of Dicer
was mainly implemented by the majority of fibrosis-related
miRNAs.
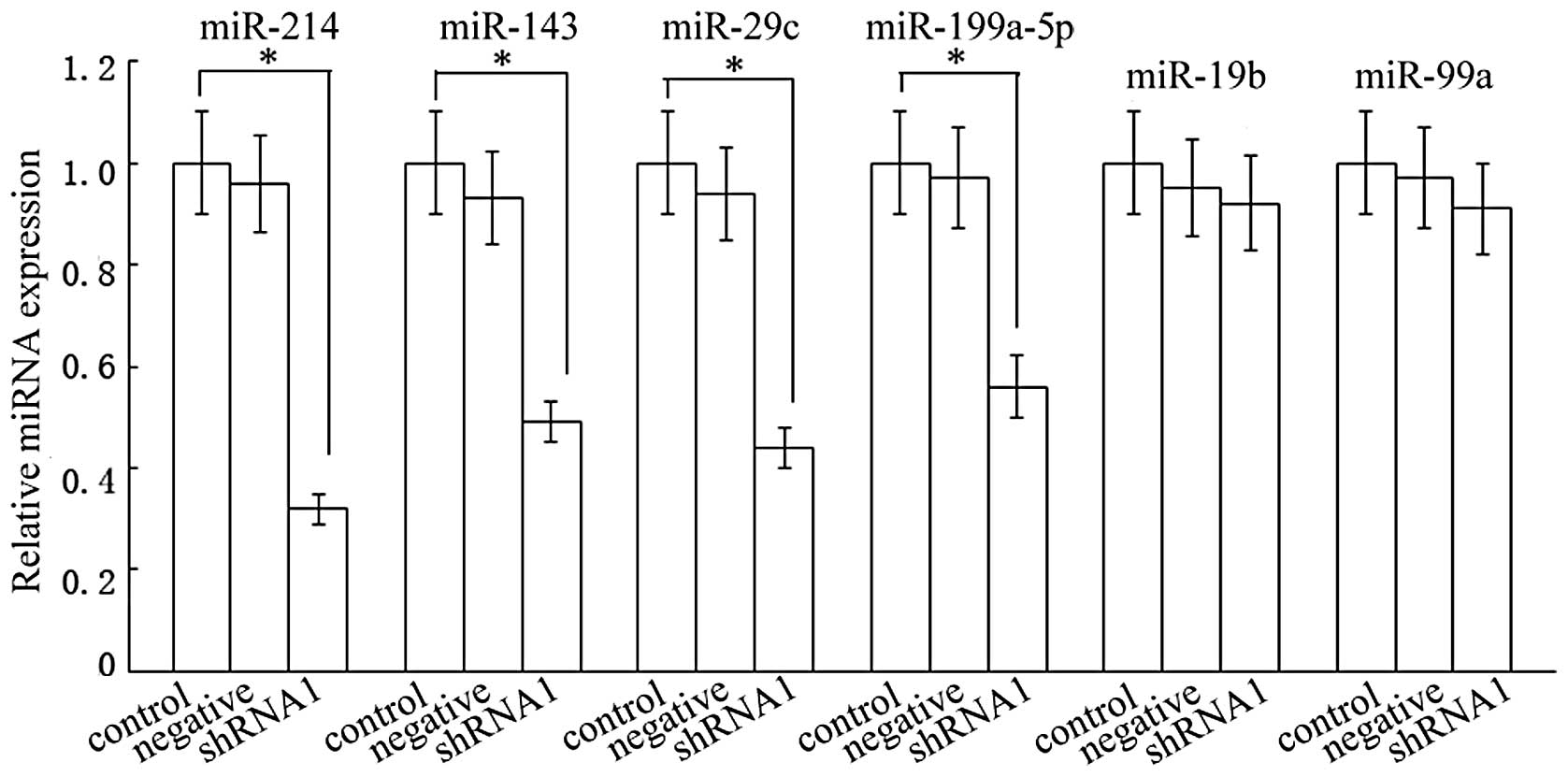 | Figure 5Effect of the shRNA1 vector on miRNA
expression. Real-time RT-PCR was performed on RNA isolated from
shRNA transfected HSCs. Compared with the control group, the levels
of miR-214, −143, −29c and −199a-5p in the shRNA1 group were
reduced by ~68, 51, 56 and 44%, respectively. (n=3,
*P>0.05). The levels of miR-19b or miR-99a were not
significantly affected (n=3, P>0.05). shRNA, short hairpin RNA;
RT-PCR, reverse transcription polymerase chain reaction; HSCs,
hepatic stellate cells. |
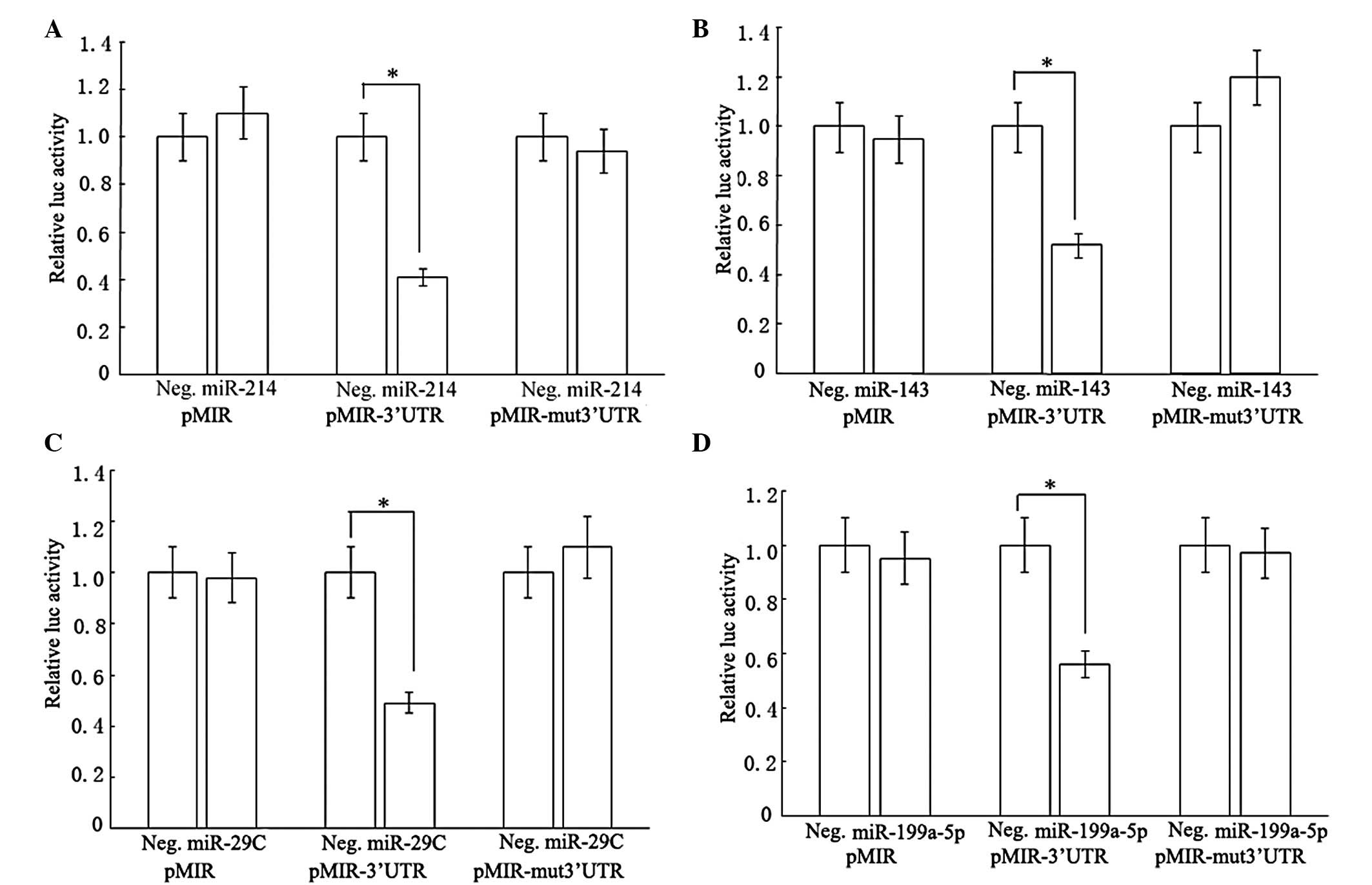
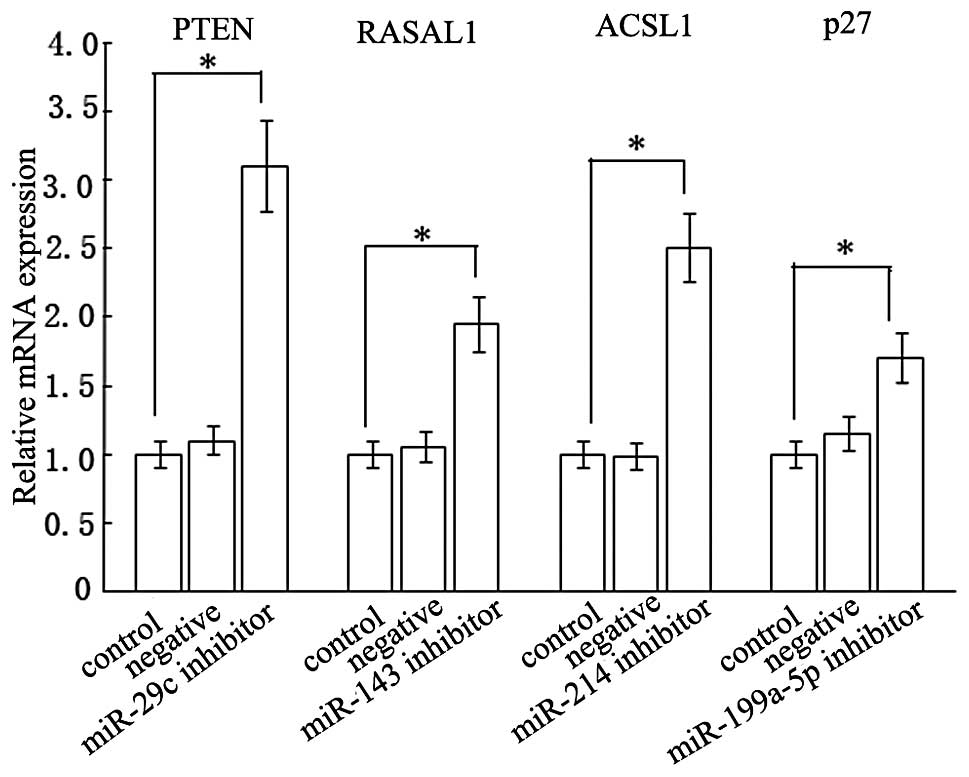
Interaction of miR-214, −143, −29c and
−199a-5p with 3′UTRs of ACSL1, RASAL1, PTEN and p27 mRNAs
The question of how miRNAs functioned in blocking
HSCs activation was also examined. We firstly investigated what the
targets of miR-214, −143, −29c and −199a-5p were. The prediction of
miRNA target regions by bioinformatics indicated that ACSL1,
RASAL1, PTEN and p27 3′UTR had one target region for miR-214, −143,
−29c and −199a-5p, respectively. To explore the direct targeting of
ACSL1 by miR-214, the sequence of the target region was cloned and
inserted into the downstream region of the firefly luciferase
reporter gene. Then the vector was cotransfected into rat HSCs with
miRNA precursors or negative control precursors. Compared with the
negative control precursors, the miR-214 precursors decreased
luciferase activity in pMIR containing the miR-214 target sequence
(pMIR-3′UTR), whereas there were no changes in pMIR empty vector
(pMIR) and pMIR with miR-214 mutant target sequence (pMIR-mut
3′UTR; Fig. 6A). In a similar way,
the miR-143, −29c and −199a-5p precursors also lowered luciferase
activities of the vectors carrying the respective miRNA target
sequence (Fig. 6B–D). Based on
these observations, we inferred that PTEN, RASAL1, ACSL1 and p27
3′UTR sequences could be targeted by miR-29c, −143, −214 and
−199a-5p, respectively. To confirm the results, we measured
endogenous PTEN, RASAL1, ACSL1 and p27 levels in HSC-T6 cells
expressing miR-29c, −143, −214 and −199a-5p inhibitors,
respectively. RT-PCR analyses of whole cell extracts suggested that
the steady-state levels of PTEN, RASAL1, ACSL1 and p27 were
elevated by ~210, 95, 150 and 70%, respectively (Fig. 7).
Discussion
Dicer, the key enzyme in the RNAi pathway, is
required for the processing of miRNA precursors into mature miRNA
molecules. In the present study, we investigated the effect of
reducing the miRNA biogenesis enzyme Dicer on the activity of HSCs.
In the present study, we investigated several lead proteins that
may represent the activation of HSCs. In response to inflammatory
stimuli, HSCs activate and become myofibroblastic cells that
express α-SMA as a representative marker (24). In the progression of liver
fibrosis, the leading ECM proteins become type I collagen instead
of type III collagen. Therefore, type I collagen is a good
indicator representing the deposition of ECM. Our results indicate
that the mRNA and protein expression of type I collagen and α-SMA
are markedly decreased by Dicer gene silencing. These findings
raise the possibility that a reduction in Dicer contributes to the
downregulation of fibrosis-related genes. Taken together, these
results suggest that Dicer inhibition is able to suppress HSC
activation as well as ECM expression.
A second major biological consequence of Dicer
knockdown is the inhibition of TIMP1. Matrix metalloproteinases
(MMPs) are a family of enzymes responsible for the proteolytic
processing of ECM structural proteins, an essential step in the
treatment of liver fibrosis (25).
TIMP-1 can inhibit the activity of MMPs, thus accelerating the
progression of liver fibrosis. Our results suggested that the mRNA
and protein expression of TIMP1 was significantly lowered by Dicer
loss. Therefore, we conclude that impaired miRNA processing
enhances the metastasic potential. These findings may have
significant clinical value.
Although the disruption of Dicer has been associated
with the reduced activity of HSCs, the mechanisms of this event
remain elusive. In the present study, we began to investigate
several miRNAs that were associated with liver fibrosis.
Post-transcriptional regulation of gene expression by miRNAs is
pivotal in a wide variety of cellular processes. In the present
study, we identified an association between the downregulation of
Dicer and the expression of miRNAs. The present study demonstrates
that the downregulation of a key component in the miRNA processing
machinery decreases the expression of certain miRNAs. These
specific miRNAs were selected based on previously published miRNA
microarrays, which identified these miRNAs as of potential interest
in liver fibrosis (23). Notably,
not all tested miRNAs were reduced, presenting either high basal
expression levels or long half-lives. Taken together, these data
reveal that global miRNA loss is responsible for the antifibrotic
effect of Dicer knowdown.
The functions of miRNA have been gradually
uncovered. The prediction of miRNA target regions currently depends
mainly on bioinformatics programs. Since each miRNA is able to
control numerous target genes, its function can be explained as the
sum of the functions of the genes it regulates. The activation of
PI3K signaling is a pivotal event during HSC activation. PTEN is a
negative regulator of PI3K signaling and thus the elevated PTEN can
block PI3K activity and prevent the activation of HSCs into
fibrogenic cells (26,27). RASAL1, a member of the RAS-GAP
family, is critical in the constitutive activity of Ras signaling.
It has been previously demonstrated that the decreased expression
of RASAL1 contributes to liver fibrosis progression (28,29).
ACSL1 is crucial in lipid metabolism and fatty acid metabolism in
the liver. Li et al (29)
indicated that the reduced amounts of ACSL1 in the liver blocked
the partitioning of fatty acids to triglyceride storage and by
doing so contributed to the progression of hepatic fibrosis. p27, a
key inhibitor of the cell cycle, can lower the proliferation of
HSCs and cell cycle progression by increasing the number of S phase
cells (30). An assay of
luciferase activity in a luminometer using the Dual-Light System
indicates that ACSL1, RASAL1, PTEN and p27 are targets of miR-214,
−143, −29c and −199a-5p, respectively. We observed that, when HSCs
were transfected with the respective miRNA inhibitor, the levels of
ACSL1, RASAL1, PTEN and p27 were increased accordingly. In
agreement with these studies, we revealed a significant reduction
in these miRNAs upon Dicer shRNA transfection. It is hypothesized
that the upregulation of target mRNA levels induced by
corresponding miRNA inhibitors presumably blocks the activation of
HSCs, which explains the antifibrotic effect of Dicer shRNA.
Although α-SMA and TIMP-1 are not predicted targets
of the above miRNAs, their mRNA and protein levels were suppressed.
Thus, this effect was thought to be a secondary action of miRNA
inhibition. That is, it is suggested that miRNAs can not only
impede the expression of target regions, but also suppress the
activation of HSCs by regulating other unidentified mechanisms,
resulting in the inhibition of α-SMA and TIMP-1.
In summary, the present study suggests for the first
time, to the best of our knowledge, that Dicer gene silencing can
inhibit collagen synthesis in HSCs. Importantly, our study
contributes important new findings for the role of Dicer-mediated
miRNA processing in the collagen synthesis of HSCs. Targeted
delivery of Dicer to activated HSCs in the liver may become a new
therapeutic strategy for liver fibrosis in the future.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (nos. 81000176/H0317, 81100292/H0317
and 81200350), the Zhejiang Provincial Natural Science Foundation
of China (nos. Y2090326, Y2110634 and LY12H08003), the Wang Bao-En
Liver Fibrosis Foundation (nos. 20100002 and 20120127), the Wenzhou
Municipal Science and Technology Bureau (no. Y20110033), the
Zhejiang Extremely Key Subject of Surgery and the Key Disciplines
in Colleges and Universities of Zhejiang Province.
References
|
1
|
Hsu YC, Lin JT, Chen TT, Wu MS and Wu CY:
Long-term risk of recurrent peptic ulcer bleeding in patients with
liver cirrhosis: a 10-year nationwide cohort study. Hepatology.
56:698–705. 2012.PubMed/NCBI
|
|
2
|
Li L, Yu C and Li Y: Endoscopic band
ligation versus pharmacological therapy for variceal bleeding in
cirrhosis: a meta-analysis. Can J Gastroenterol. 25:147–155.
2011.PubMed/NCBI
|
|
3
|
Qin T, Xia J, Ren H, Zhou H, Tang B and
Shao Z: Liver cirrhosis as a predisposing condition for
Legionnaires' disease: a report of four laboratory-confirmed cases
from China. J Med Microbiol. 61:1023–1028. 2012. View Article : Google Scholar
|
|
4
|
Wang SB, Wang JH, Chen J, Giri RK and Chen
MH: Natural history of liver cirrhosis in south China based on a
large cohort study in one center: a follow-up study for up to 5
years in 920 patients. Chin Med J (Engl). 125:2157–2162.
2012.PubMed/NCBI
|
|
5
|
Giannone FA, Baldassarre M, Domenicali M,
et al: Reversal of liver fibrosis by the antagonism of
endocannabinoid CB1 receptor in a rat model of CCl(4)-induced
advanced cirrhosis. Lab Invest. 92:384–395. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Madro A, Czechowska G, Slomka M, Celinski
K, Szymonik-Lesiuk S and Kurzepa J: The decrease of serum MMP-2
activity corresponds to alcoholic cirrhosis stage. Alcohol.
46:155–157. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wirkowska A and Paczek L: Liver fibrosis
and cirrhosis--selected cytokines, growth factors and proteins.
Part II Przegl Lek. 68:228–230. 2011.(In Polish).
|
|
8
|
Oliveira-Junior MC, Monteiro AS,
Leal-Junior EC, et al: Low-level laser therapy ameliorates
CCl(4)-induced liver cirrhosis in rats. Photochem Photobiol.
89:173–178. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lee JK, Kim JH and Shin HK: Therapeutic
effects of the oriental herbal medicine Sho-saiko-to on liver
cirrhosis and carcinoma. Hepatol Res. 41:825–837. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cao S, Yaqoob U, Das A, et al:
Neuropilin-1 promotes cirrhosis of the rodent and human liver by
enhancing PDGF/TGF-beta signaling in hepatic stellate cells. J Clin
Invest. 120:2379–2394. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Iizuka M, Ogawa T, Enomoto M, et al:
Induction of microRNA-214-5p in human and rodent liver fibrosis.
Fibrogenesis Tissue Repair. 5:122012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
He Y, Huang C, Sun X, Long XR, Lv XW and
Li J: MicroRNA-146a modulates TGF-beta1-induced hepatic stellate
cell proliferation by targeting SMAD4. Cell Signal. 24:1923–1930.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Noetel A, Kwiecinski M, Elfimova N, Huang
J and Odenthal M: microRNA are central players in anti- and
profibrotic gene regulation during liver fibrosis. Front Physiol.
3:492012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ogawa T, Enomoto M, Fujii H, et al:
MicroRNA-221/222 upregulation indicates the activation of stellate
cells and the progression of liver fibrosis. Gut. 61:1600–1609.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Dueck A, Ziegler C, Eichner A, Berezikov E
and Meister G: microRNAs associated with the different human
Argonaute proteins. Nucleic Acids Res. 40:9850–9862. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gurtan AM, Lu V, Bhutkar A and Sharp PA:
In vivo structure-function analysis of human Dicer reveals
directional processing of precursor miRNAs. RNA. 18:1116–1122.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Andersson T, Rahman S, Sansom SN, et al:
Reversible block of mouse neural stem cell differentiation in the
absence of dicer and microRNAs. PLoS One. 5:e134532010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chiosea S, Jelezcova E, Chandran U, et al:
Up-regulation of dicer, a component of the MicroRNA machinery, in
prostate adenocarcinoma. Am J Pathol. 169:1812–1820. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Vaksman O, Hetland TE, Trope' CG, Reich R
and Davidson B: Argonaute, Dicer, and Drosha are up-regulated along
tumor progression in serous ovarian carcinoma. Hum Pathol.
43:2062–2069. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang X, Cairns M, Rose B, et al:
Alterations in miRNA processing and expression in pleomorphic
adenomas of the salivary gland. Int J Cancer. 124:2855–2863. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Martin MG, Payton JE and Link DC: Dicer
and outcomes in patients with acute myeloid leukemia (AML). Leuk
Res. 33:e1272009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wu JF, Shen W, Liu NZ, et al:
Down-regulation of Dicer in hepatocellular carcinoma. Med Oncol.
28:804–809. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lakner AM, Steuerwald NM, Walling TL, et
al: Inhibitory effects of microRNA 19b in hepatic stellate
cell-mediated fibrogenesis. Hepatology. 56:300–310. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Uyama N, Iimuro Y, Kawada N, et al:
Fascin, a novel marker of human hepatic stellate cells, may
regulate their proliferation, migration, and collagen gene
expression through the FAK-PI3K-Akt pathway. Lab Invest. 92:57–71.
2012. View Article : Google Scholar
|
|
25
|
Ramachandran P and Iredale JP: Liver
fibrosis: a bidirectional model of fibrogenesis and resolution.
QJM. 105:813–817. 2012. View Article : Google Scholar
|
|
26
|
Takashima M, Parsons CJ, Ikejima K,
Watanabe S, White ES and Rippe RA: The tumor suppressor protein
PTEN inhibits rat hepatic stellate cell activation. J
Gastroenterol. 44:847–855. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bian EB, Huang C, Ma TT, et al:
DNMT1-mediated PTEN hypermethylation confers hepatic stellate cell
activation and liver fibrogenesis in rats. Toxicol Appl Pharmacol.
264:13–22. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tao H, Huang C, Yang JJ, et al: MeCP2
controls the expression of RASAL1 in the hepatic fibrosis in rats.
Toxicology. 290:327–333. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li WQ, Chen C, Xu MD, et al: The
rno-miR-34 family is upregulated and targets ACSL1 in
dimethylnitrosamine-induced hepatic fibrosis in rats. FEBS J.
278:1522–1532. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang B, Li W, Guo K, Xiao Y, Wang Y and
Fan J: miR-181b promotes hepatic stellate cells proliferation by
targeting p27 and is elevated in the serum of cirrhosis patients.
Biochem Biophys Res Commun. 421:4–8. 2012. View Article : Google Scholar : PubMed/NCBI
|