Introduction
The presence of lymph node metastases is the main
prognostic factor in early-stage cervical cancer patients. Although
the International Federation of Gynecology and Obstetrics (FIGO)
staging system for cervical carcinoma does not take into account
the lymphadenopathy (LN) status, it is the most significant
prognostic factor for patients with stages IB or IIA of the
disease. In addition, the 5-year survival rate for patients
declines markedly from ~80–95% in patients without lymph node
metastasis to ~50–65% in patients with lymph node metastases
(1). Radical hysterectomy and
pelvic lymphadenectomy is the standard surgical treatment for early
stage cervical cancer. However, in a clinical study by Selman et
al (2) it was confirmed that
84% patients did not exhibit pathology following a lymph node
biopsy. Numerous studies have focused on strategies for detecting
lymph node metastasis; however, the accuracy of these approaches,
such as MRI and PET-CT, was identified to be only 50% (3). Therefore, the identification of
sensitive, accurate and non-invasive molecular markers may
facilitate the selection of patients with lymph node negative
cervical cancer with an unfavorable prognosis for adjuvant
treatment and allow the identification of targets for
patient-tailored therapy.
MicroRNA (miR) has an essential role in malignant
tumor development and progression (4,5).
Previous studies have demonstrated that certain miRs control tumor
cell invasion and metastasis. For example, miR-200c initiated
metastasis mediated by the loss of cell-cell adhesion caused by
E-cadherin repression, in a process commonly termed
epithelial-to-mesenchymal transition (EMT) (6). In addition, miR-26a enhanced the
metastatic potential of lung cancer cells via the AKT pathway by
targeting phosphatase and tensin homologue (PTEN) (7). Furthermore, enforced expression of
miR-148a suppressed gastric cancer cell invasion and metastasis
through directly targeting ROCK1 (8). The role of miR-10a in cancer
metastasis has recently attracted notable attention. Microarray
analysis data by Pereira et al (9) and Lee et al (10) demonstrated that miR-10a is
upregulated in cervical cancer. It was also demonstrated that
miR-10a triggered the metastatic properties of hepatocellular
carcinoma by directly targeting EphA4 (11). Furthermore, the expression of
miR-10a was at least 10-fold higher in three gastric cancer cell
lines compared with gastric mucosal cell lines, and was involved in
the development of gastric cancer and lymph node metastasis
(12). In addition, circulating
miR-10a was reported to be higher in the plasma of patients with
non-small cell lung cancer compared with that in healthy volunteers
(13). By contrast, controversial
results have emerged reporting miR-10a to be a tumor suppressor by
controlling cell migration/invasion in esophageal squamous cell
carcinoma (14). However, the
function of miR-10a in cervical cancer remains unknown and further
study is warranted to elucidate this.
Therefore, the present study investigated the
associated between the expression level of miR-10a and lymph node
metastasis in cervical cancer, and further investigated the
possible function of miR-10a in cervical cancer cell lines. The
results revealed that the overexpression of miR-10a is
significantly associated with metastasis and invasion in cervical
cancer. Furthermore, it was able to negatively regulate PTEN by
binding to the 3′-untranslated region (UTR) regions to affect
cervical cancer cell migration and invasion.
Materials and methods
Tissue samples and cell lines
Clinical data and pathological tissue samples were
retrieved from the Cancer Center of Wuhan Union Hospital (Wuhan,
China). The biopsy specimens from 40 unrelated patients who were
diagnosed with FIGO IIb~IIIb stage cervical cancer, were collected
in 2012, and the pathology of these samples were confirmed as
squamous cell carcinoma. None of the patients had previously
received chemotherapy or radiotherapy. The present study was
approved by the Institutional Review Board of the Wuhan Union
Hospital. Written informed consent was obtained from all of the
patients. Human tissues were frozen in liquid nitrogen and stored
at −80°C. The Hela and Siha human cervical cancer cell lines were
provided by the Gynaecology and Obstetric Laboratory of Wuhan Union
Hospital (Wuhan, China). Hela cells were cultured in RPMI-1640
medium (Hyclone, Logan, UT, USA) complemented with 10% fetal bovine
serum (FBS; Hyclone). The Siha cells were cultured in Dulbecco’s
modified Eagle’s high glucose medium (Hyclone) complemented with
10% FBS (Hyclone). All of the cell lines were cultured in a
humidified incubator (5% CO2) at 37°C.
Cell transfection
The miR-10a gain and loss-of-function studies were
performed using miR-10a mimics (100 nM) and its negative control
(mimic nc; 100 nM) and the loss-of-function study were performed
with miR-10a inhibitor (150 nM) and its negative control (inhibitor
nc; 150 nM) in Hela and Siha cell lines (all obtained from
Guangzhou RiboBio Co., Ltd., Guangzhou, China). For each cell line,
there was a blank control without any transfection. The cells were
transfected using Lipofectamine™ 2000 reagent (Invitrogen Life
Technologies, Carlsbad, CA, USA) in Opti-minimal essential medium
(Gibco-BRL, Carlsbad, CA, USA), according to the manufacturer’s
instructions. The relative expression level of miR-10a in
transfected cells was examined by quantitative polymerase chain
reaction (qPCR).
Dual-luciferase reporter assay
The region of human PTEN-3′UTR, generated by PCR
amplification, was cloned into the pGL3 luciferase reporter
plasmid. The primers selected were as follows: Forward: hsa-miR-10a
5′-CTCGAGGAAATAAAACCAAAGCACTC-3′ and reverse:
5′-GGTACCGCCAGTCACCAGACTGTCCT-3′ for hsa-miR-10a-R; and forward:
5′-TCTAGAAATGGC AATAGGACATTGTG-3′ and reverse: 5′-TCTAGACCATCT
TTATTAATCCTAATTG-3′ for PTEN-3′UTR-wild type (wt). For the reporter
assay, 239 T cells were plated onto 24-well plates and transfected
with 500 ng pGL3-miR-PTEN-wt using Lipofectamine 2000. Following
transfection for 48 h, the cells were harvested and the Renilla and
Firefly luciferase were assayed by the Dual Luciferase Reporter
Assay system (GeneChem, Shanghai, China) according to the
manufacturer’s instructions. The tests were repeated in
triplicate.
RNA extraction, reverse transcription and
qPCR
Total RNA from the tissue samples and cells were
isolated using TRIzol reagent (Ambion, Invitrogen Life
Technologies). The relative expression levels of miR-10a were
examined by the altered stem-loop RT-PCR with specific RT and PCR
primers using U6 snRNA as a control. The primers for miR-10a were
as follows: Forward: 5′-GCCGTACTC TGTAGATCCGAA-3′ and reverse:
5′-CAGTGCAGGGTCCGAGGTAT-3′. The expression of PTEN mRNA was
detected by qPCR using paired primers. The GAPDH gene was used as a
control. The primers for PTEN mRNA were as follows: forward:
5′-AAGACCATAACCCACCACAGC-3′ and reverse: 5′-GAGCCCCAGCCTTCTCCAT-3′.
qPCR was performed on a Step One Plus Real-time PCR system (Applied
Biosystems, Foster City, CA, USA) using SYBR Premix Ex
Taq™ (Takara Biotechnology, Dalian, China) according to
the manufacturer’s instructions. The 2−ΔΔCt method,
where ΔΔCT = (Ct target gene-CtU6)N+-(Ct target
gene-CtU6)N0 was used for analysis. Where N+
represents the patients positive for lymph node metastasis and
N0 represents the patients negative for lymph node
metastasis.
In vitro transwell invasion and migration
assay
Transwell membranes (polycarbonic membrane;
diameter, 6.5 mm; pore size, 8 μm; Corning Costar, Inc., Corning,
NY, USA) coated with Matrigel (BD BioSciences, USA) were used to
assay the cell invasion and migration in vitro. At 48 h
post-transfection, the cells were resuspended into serum-free
medium. The transfected cells (10×104 in 200 μl
serum-free medium) were reseeded into the upper chamber, and 600 μl
medium with 10% FBS was added to the lower chamber as a
chemoattractant. Following 24 h incubation, non-invading cells on
the upper surface of the membrane were removed with a cotton swab.
The invasive cells, which penetrated to the lower surface, were
fixed with 4% paraformaldehyde and stained with 0.1% crystal
violet. The number of cells invading the membrane was counted from
five randomly selected visual fields with an inverted microscope
(Olympus, Tokyo, Japan) at a magnification of ×200. Data were
obtained from three independent experiments.
Wound healing assay
The Hela and Siha cells were seeded onto 6-well
plates. When the cell confluence reached ≥80% and above at ~48 h
post-transfection, the scratch wounds were made by scraping the
cell layer across each culture plate using the tip of 10 μl
pipette. Then five fields were randomly selected from each scratch
wound and visualized by microscopy. The experiments were performed
in triplicate.
Immunocytochemistry
The Hela and Siha cells were seeded onto 13-mm glass
coverslips on a 24-well plate. At 48 h following transfection with
the non-specific control, miR-10a mimic (100 nM) cells were
immunostained with anti-β-catenin antibody (Abnova, Taipei, Taiwan)
according to the immunofluorescence staining instructions provided
by the manufacturer. The labeled cells were observed with a A1Si
confocal microscope (Nikon, Tokyo, Japan).
Western blotting
The Hela and Siha cells were harvested after 72 h
following transfection with the miR-10a mimic, mimic nc, inhibitor
or inhibitor nc. Equal quantities of the denatured protein sample
were separated by SDS-PAGE and then transferred to polyvinylidene
difluoride membranes (Wuhan Goodbio Technology Co., Ltd., Wuhan,
China). The following primary antibodies were used: Anti-PTEN
(dilution, 1:100; Abnova) and anti-β-actin (dilution, 1:400; Santa
Cruz Biotechnology, Inc., Santa Cruz, CA, USA). Following overnight
incubation at 4°C and washing, the membranes were probed with
horseradish peroxidase-conjugated goat anti-rabbit antibody (Guge)
at 1:5,000 dilution. The images were captured with the exposure
machine (UVP OptiCam600; UVP Inc., Upland, CA, USA).
Statistical analysis
The results were analyzed using SPSS 17.0 software
(SPSS, Inc., Chicago, IL, USA). Statistical analysis was performed
using a Mann-Whitney test and one-way analysis of variance.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Expression level of miR-10a is
upregulated in cervical cancer tissues with pelvic lymph node
metastasis
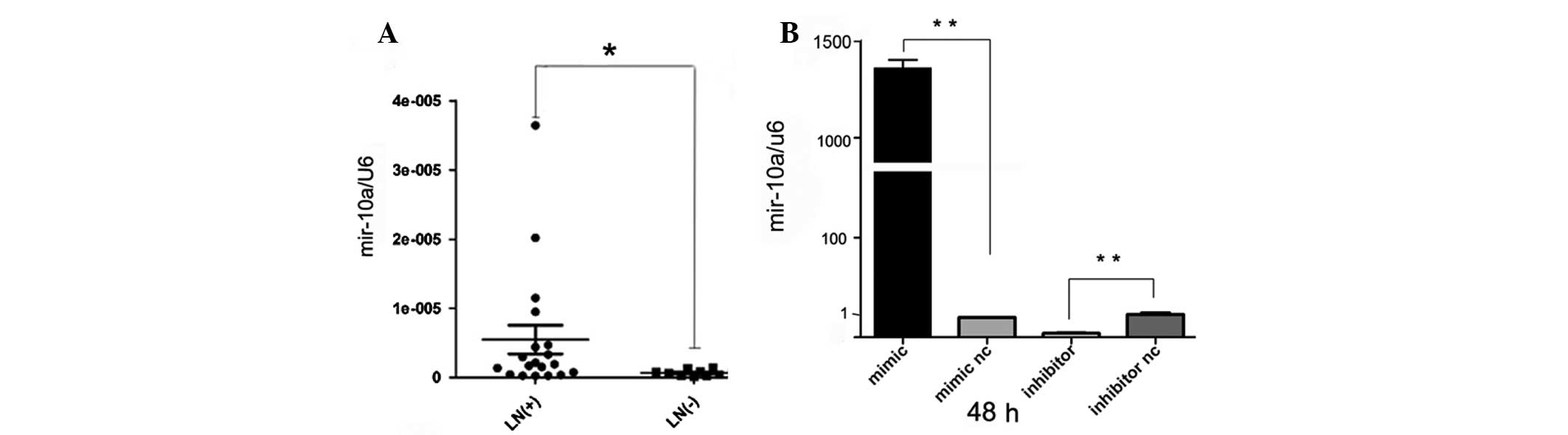
To identify the miR-10a expression in the clinical
samples, qPCR was performed and it was identified that following
normalization to RNU6B expression levels, the expression level of
miR-10a in cervical cancer tissues with LN+ [mean ±
standard deviation (SD): 0.000003448+0.00008553] was significantly
higher than LN− (mean ± SD: 0.000001109+0.000002271;
P=0.0053), as demonstrated in Fig.
1A. This data suggested that miR-10a may directly function in
cervical cancer cell metastasis.
Upregulation of miR-10a expression
promotes invasion and migration of cervical cancer cells
As the results above demonstrated that the
upregulation of miR-10a in cervical cancer is closely associated
with cervical cancer metastasis, it was hypothesized that the
overexpression of miR-10a in cervical cancer cells exerted effects
of cell invasion and metastasis. In gain-of-function and
loss-of-function studies, Hela and Siha cells were separately
transfected with mimic and mimic nc, inhibitor and inhibitor nc.
Transfection with 100 nM of miR-10a mimics in Hela cells led to an
~1328-fold increase in the miR-10a expression detected 48 h
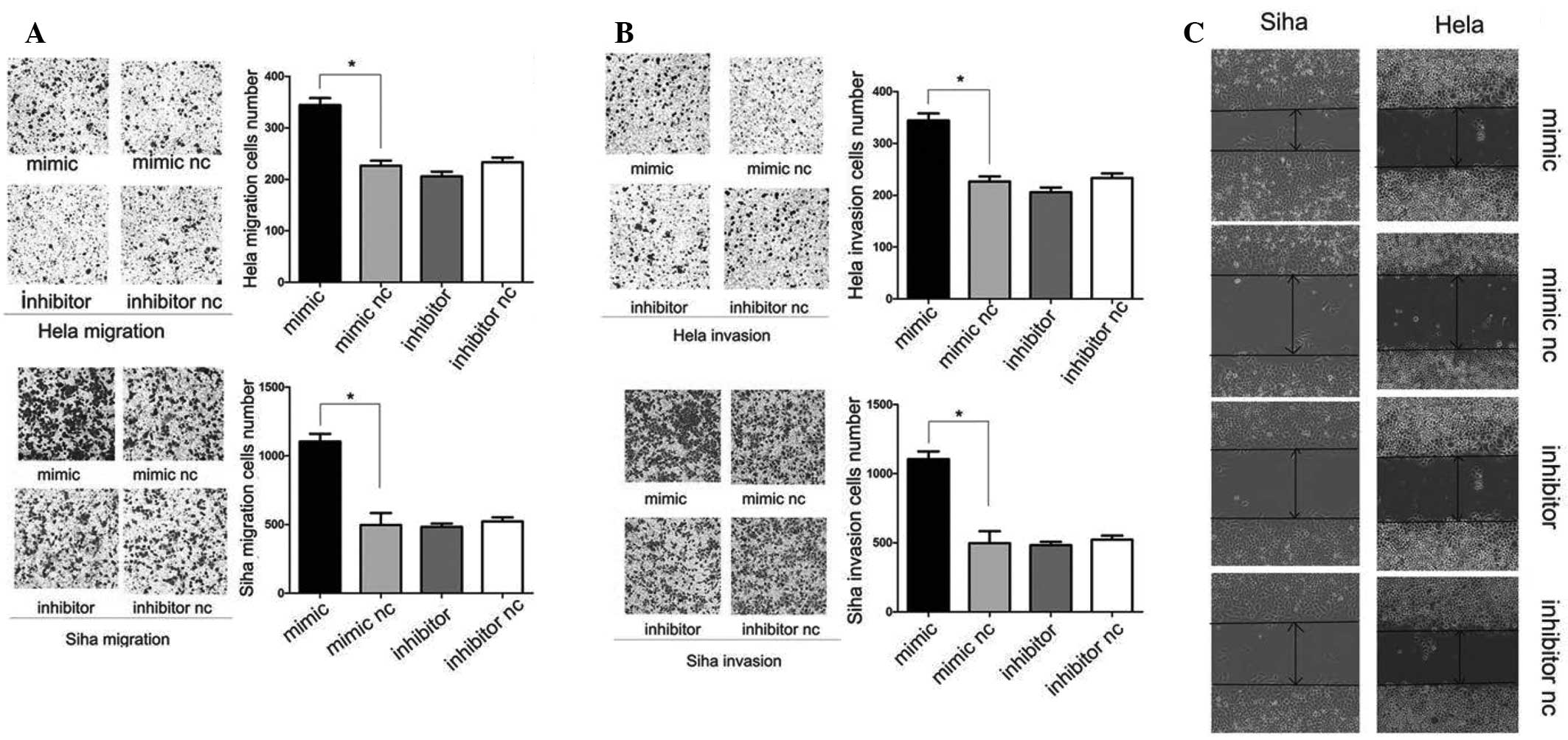
post-transfection, as examined by qPCR (Fig. 1B). It was identified that the
overexpression of miR-10a induced Hela and Siha cell migration,
while downregulated miR-10a could inhibit the migratory ability of
Hela and Siha cells (Fig. 2A and
C). Consistent with this finding, the matrigel invasion assay
results demonstrated that miR-10a overexpression significantly
enhanced the invasion capacity of Hela and Siha cells, while
decrease of miR-10a could inhibit invasion in Hela and Siha cells,
however, this was not statistically significant (Fig. 2B). These observations demonstrated
that miR-10a had an important role in promoting migration and the
invasive potential of cervical cancer cells.
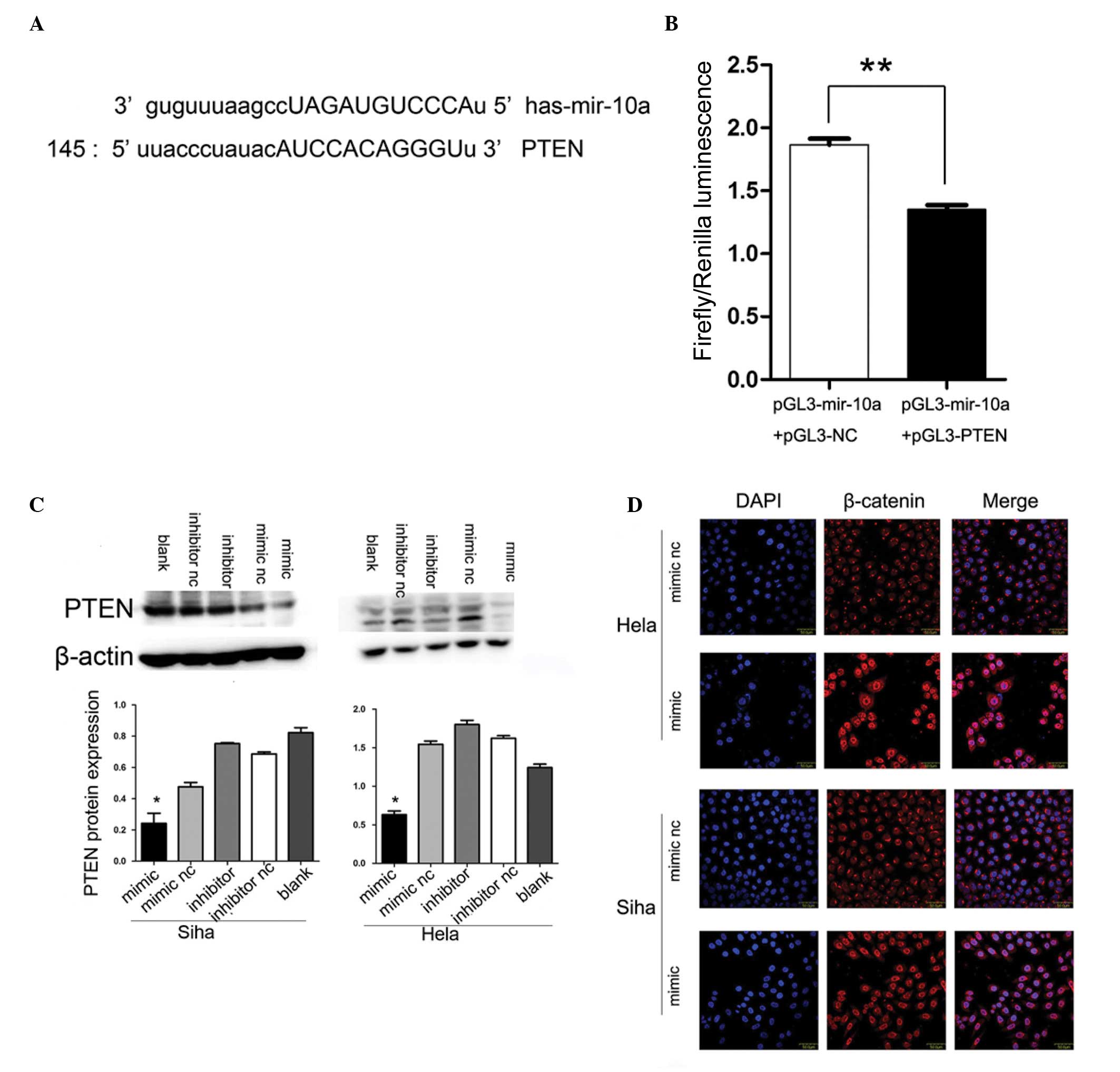
miR-10a targets PTEN
The analysis conducted using publicly available
programs, TargetScan (http://www.targetscan.org) and miRanda (http://www.microrna.org), indicated that PTEN is
theoretically the target gene of miR-10a (Fig. 3A). Next, a luciferase reporter
assay was performed to verify that miR-10a directly targets PTEN.
It was identified that co-transfection of pGL3-miR-10a and
pGL3-PTEN-wt, significantly decreased the luciferase activity in
239 T cells as compared with the control (Fig. 3B).
Alteration of miR-10a expression changes
PTEN protein but not mRNA expression levels
PTEN had been reported to be downregulated in
cervical cancer and closely associated with lymph node metastasis.
To further confirm that PTEN was the downstream target of miR-10a,
upregulation and downregulation of miR-10a expression were
conducted with subsequent detection of PTEN mRNA and protein
changes. The increase in endogenous miR-10a levels significantly
decreased PTEN protein expression as determined by western blot
analysis (P<0.05; Fig. 3C),
while the mRNA expression of PTEN remained unchanged (data not
shown).
miR-10a mimic promotes nuclear
translocation of β-catenin
Considering that PTEN was able to restrain β-catenin
nuclear translocation and miR-10a inhibited PTEN expression, it was
determined whether miR-10a may affect the β-catenin nuclear
translocation activity. Notably, the immunofluorescence staining
assays demonstrated that the overexpression of miR-10a resulted in
marked nuclear accumulation of β-catenin in Hela and Siha cervical
cancer cell lines, suggesting that miR-10a may contribute to the
accumulation of β-catenin in the nuclei (Fig. 3D).
Discussion
Lymph node status is one of the most important
prognostic factors following cervical cancer diagnosis, which
dominates the majority of clinical decisions regarding treatment
(15,16). As the current imaging techniques
are not reliable to diagnose lymph node status, pelvic +/−
para-aortic lymphadenectomy remains the gold standard. These
surgical procedures are, however, responsible for specific
morbidity: Lymphocele and lymphedema. However, lymph node
metastasis is rarely identified by CT or MRI when they are <1
cm. Therefore, effective molecular markers for detecting lymph node
metastasis are urgently required (17,18).
Previously, a number of miRNAs were identified to be lymph node
metastasis biomarkers, including miRNA-10b and miRNA-373, which are
used for detecting lymph node metastasis of breast cancer (19). Furthermore, miR-21, miR-212 and
miR-195 were found to be promising novel biomarkers for lymph node
metastasis in gastric cancer (20,21),
and it appears miR-20a may be a potential biomarker for detecting
the lymph node status of cervical cancer patients (22). The miR-10 family containing five
miRNA genes, including miR-10a, miR-10b, miR-196a-1, miR-196a-2 and
miR-196b, has attracted notable attention due to its conservation
and the position of the miR-10 genes within the Hox clusters of
developmental regulators (23).
For miR-10a, it interacts with the 5′ untranslated region of mRNAs
encoding RPs (ribosomal proteins) to enhance their translation.
Accordingly, RPs are found to be deregulated in cancer (24). Furthermore, miR-10a was found
overexpressed in glioblastoma, hepatocellular carcinomas, colon
cancer and acute myeloid leukemia with NPM1 mutations (25–28).
In the present study, it was identified that miR-10a in primary
tumor tissue was markedly upregulated in N+ patient
samples. Next, the function and possible mechanisms of miR-10a in
regulating certain biological properties of Hela and Siha cervical
cancer cells was investigated. However, through MTT and flow
cytometry, it was verified that miR-10a had no effect on the cell
proliferation and apoptosis in 72 h (data not shown). The data also
suggested that a miR-10a mimic triggered the migration and invasion
capacities of cervical cancer cells by negatively regulating tumor
suppressor PTEN at the transcriptional level via binding to
non-coding regions of PTEN.
PTEN is a tumor suppressor that negatively regulates
(mTOR)/PI3K/Akt pathways. PTEN is one of the most mutated and
deleted tumor suppressors in human cancer and has an important role
in tumor metastasis (29–32). Loss of PTEN is associated with
lymph node metastasis of cervical cancer (33,34),
sentinel lymph node micro-metastasis in breast cancer (35) and cervical lymph node metastasis in
salivary gland cancer (36). The
present study found that an miR-10a mimic was able to inhibit PTEN
at the transcriptional level. The function of PTEN is involved in
the suppression of nuclear-catenin accumulation (37,38).
The results demonstrated that the miR-10a mimic was able to trigger
β-catenin nuclear translocation. In cancer cells, β-catenin within
the nucleus generates a positive and reinforcing feedback loop,
resulting in the nuclear accumulation of β-catenin (39). A strong correlation has been
identified between β-catenin nuclear translocation, tumor
metastasis and poor prognosis (40–42).
In cervical cancer, β-catenin nuclear translocation has been noted
for its function in lymph node metastasis, growth, invasion and
angiogenesis (41,43). Stabilization and nuclear
localization of β-catenin, which induces the expression of Wnt
target genes and subsequently triggers EMT, may be associated with
the induction of metastasis (44).
The present study suggests that the mechanisms of miR-10a promoting
the invasion and migration ability of Hela and Siha cells, may be
that miR-10a inhibits PTEN protein expression and then promotes
β-catenin accumulation in the nucleus. To the best of our
knowledge, this mechanism has not been reported prior to this study
and requires further investigation.
In conclusion, it was observed that miR-10a is
overexpressed in cervical tumor tissues with lymph node metastasis,
and miR-10a may significantly enhance the migration and invasion
capacities of cervical cancer cells by inhibiting PTEN at the
transcriptional level. To the best of our knowledge, these data
have not been documented in previous studies. The present data
suggests that miR-10a may be useful as a molecular marker for
detecting patients with lymph node metastasis of cervical cancer.
These findings warrant further studies with a large cohort of
patients to validate and develop the tissue biomarker as a critical
tool for cervical cancer care.
References
|
1
|
Sakuragi N: Up-to-date management of lymph
node metastasis and the role of tailored lymphadenectomy in
cervical cancer. Int J Clin Oncol. 12:165–175. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Selman TJ, Luesley DM, Murphy DJ and Mann
CH: Is radical hysterectomy for early stage cervical cancer an
outdated operation? BJOG. 112:363–365. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Monteil J, Maubon A, Leobon S, et al:
Lymph node assessment with (18)F-FDG-PET and MRI in uterine
cervical cancer. Anticancer Res. 31:3865–3871. 2011.PubMed/NCBI
|
|
4
|
van Kouwenhove M, Kedde M and Agami R:
MicroRNA regulation by RNA-binding proteins and its implications
for cancer. Nat Rev Cancer. 11:644–656. 2011.PubMed/NCBI
|
|
5
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ceppi P, Mudduluru G, Kumarswamy R, Rapa
I, Scagliotti GV, Papotti M and Allgayer H: Loss of miR-200c
expression induces an aggressive, invasive, and chemoresistant
phenotype in non-small cell lung cancer. Mol Cancer Res.
8:1207–1216. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liu B, Wu X, Liu B, Wang C, Liu Y, Zhou Q
and Xu K: MiR-26a enhances metastasis potential of lung cancer
cells via AKT pathway by targeting PTEN. Biochim Biophys Acta.
1822:1692–1704. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zheng B, Liang L, Wang C, et al:
MicroRNA-148a suppresses tumor cell invasion and metastasis by
downregulating ROCK1 in gastric cancer. Clin Cancer Res.
17:7574–7583. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pereira PM, Marques JP, Soares AR, Carreto
L and Santos MA: MicroRNA expression variability in human cervical
tissues. PLoS One. 5:e117802010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lee JW, Choi CH, Choi JJ, et al: Altered
microRNA expression in cervical carcinomas. Clin Cancer Res.
14:2535–2542. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yan Y, Luo YC, Wan HY, et al: MicroRNA-10a
is involved in the metastatic process by regulating Eph tyrosine
kinase receptor A4-mediated epithelial-mesenchymal transition and
adhesion in hepatoma cells. Hepatology. 57:667–677. 2013.
View Article : Google Scholar
|
|
12
|
Chen W, Tang Z, Sun Y, et al: miRNA
expression profile in primary gastric cancers and paired lymph node
metastases indicates that miR-10a plays a role in metastasis from
primary gastric cancer to lymph nodes. Exp Ther Med. 3:351–356.
2012.
|
|
13
|
Markou A, Sourvinou I, Vorkas PA, Yousef
GM and Lianidou E: Clinical evaluation of microRNA expression
profiling in non small cell lung cancer. Lung Cancer. 81:388–396.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Matsushima K, Isomoto H, Kohno S and Nakao
K: MicroRNAs and esophageal squamous cell carcinoma. Digestion.
82:138–144. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gouy S, Morice P, Narducci F, et al:
Nodal-staging surgery for locally advanced cervical cancer in the
era of PET. Lancet Oncol. 13:e212–e220. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kato T, Watari H, Takeda M, et al:
Multivariate prognostic analysis of adenocarcinoma of the uterine
cervix treated with radical hysterectomy and systematic
lymphadenectomy. J Gynecol Oncol. 24:222–228. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
He L, Thomson JM, Hemann MT, et al: A
microRNA polycistron as a potential human oncogene. Nature.
435:828–833. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang H, Li Y and Lai M: The microRNA
network and tumor metastasis. Oncogene. 29:937–948. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen W, Cai F, Zhang B, Barekati Z and
Zhong XY: The level of circulating miRNA-10b and miRNA-373 in
detecting lymph node metastasis of breast cancer: potential
biomarkers. Tumour Biol. 34:455–462. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xu Y, Sun J, Xu J, Li Q, Guo Y and Zhang
Q: miR-21 is a promising novel biomarker for lymph node metastasis
in patients with gastric cancer. Gastroenterol Res Pract.
2012:6401682012.PubMed/NCBI
|
|
21
|
Wu WY, Xue XY, Chen ZJ, et al: Potentially
predictive microRNAs of gastric cancer with metastasis to lymph
node. World J Gastroenterol. 17:3645–3651. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhao S, Yao D, Chen J and Ding N:
Circulating miRNA-20a and miRNA-203 for screening lymph node
metastasis in early stage cervical cancer. Genet Test Mol
Biomarkers. 17:631–636. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lund AH: miR-10 in development and cancer.
Cell Death Differ. 17:209–214. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ørom UA, Nielsen FC and Lund AH:
MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and
enhances their translation. Mol Cell. 30:460–471. 2008.PubMed/NCBI
|
|
25
|
Gaur A, Jewell DA, Liang Y, et al:
Characterization of microRNA expression levels and their biological
correlates in human cancer cell lines. Cancer Res. 67:2456–2468.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Varnholt H, Drebber U, Schulze F, et al:
MicroRNA gene expression profile of hepatitis C virus-associated
hepatocellular carcinoma. Hepatology. 47:1223–1232. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Volinia S, Calin GA, Liu CG, et al: A
microRNA expression signature of human solid tumors defines cancer
gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Garzon R, Garofalo M, Martelli MP, et al:
Distinctive microRNA signature of acute myeloid leukemia bearing
cytoplasmic mutated nucleophosmin. Proc Natl Acad Sci USA.
105:3945–3950. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kotelevets L, van Hengel J, Bruyneel E,
Mareel M, van Roy F and Chastre E: The lipid phosphatase activity
of PTEN is critical for stabilizing intercellular junctions and
reverting invasiveness. J Cell Biol. 155:1129–1135. 2001.
View Article : Google Scholar
|
|
30
|
Li J, Yen C, Liaw D, et al: PTEN, a
putative protein tyrosine phosphatase gene mutated in human brain,
breast, and prostate cancer. Science. 275:1943–1947. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tamura M, Gu J, Matsumoto K, Aota S,
Parsons R and Yamada KM: Inhibition of cell migration, spreading,
and focal adhesions by tumor suppressor PTEN. Science.
280:1614–1617. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Carracedo A, Alimonti A and Pandolfi PP:
PTEN level in tumor suppression: how much is too little? Cancer
Res. 71:629–633. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Eijsink JJ, Noordhuis MG, ten Hoor KA, et
al: The epidermal growth factor receptor pathway in relation to
pelvic lymph node metastasis and survival in early-stage cervical
cancer. Hum Pathol. 41:1735–1741. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lu D, Qian J, Yin X, Xiao Q, Wang C and
Zeng Y: Expression of PTEN and survivin in cervical cancer:
promising biological markers for early diagnosis and prognostic
evaluation. Br J Biomed Sci. 69:143–146. 2012.PubMed/NCBI
|
|
35
|
Zhu L, Loo WT and Louis WC: PTEN and VEGF:
possible predictors for sentinel lymph node micro-metastasis in
breast cancer. Biomed Pharmacother. 61:558–561. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ettl T, Gosau M, Brockhoff G, et al:
Predictors of cervical lymph node metastasis in salivary gland
cancer. Head Neck. 36:517–523. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Damsky WE, Curley DP, Santhanakrishnan M,
et al: Beta-catenin signaling controls metastasis in Braf-activated
Pten-deficient melanomas. Cancer Cell. 20:741–754. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Persad S, Troussard AA, McPhee TR,
Mulholland DJ and Dedhar S: Tumor suppressor PTEN inhibits nuclear
accumulation of beta-catenin and T cell/lymphoid enhancer factor
1-mediated transcriptional activation. J Cell Biol. 153:1161–1174.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Jamieson C, Sharma M and Henderson BR: Wnt
signaling from membrane to nucleus: beta-catenin caught in a loop.
Int J Biochem Cell Biol. 44:847–850. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Giangreco A, Lu L, Vickers C, et al:
Beta-Catenin determines upper airway progenitor cell fate and
preinvasive squamous lung cancer progression by modulating
epithelial-mesenchymal transition. J Pathol. 226:575–587. 2012.
View Article : Google Scholar
|
|
41
|
Noordhuis MG, Fehrmann RS, Wisman GB, et
al: Involvement of the TGF-beta and beta-catenin pathways in pelvic
lymph node metastasis in early-stage cervical cancer. Clin Cancer
Res. 17:1317–1330. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Li XQ, Yang XL, Zhang G, et al: Nuclear
β-catenin accumulation is associated with increased expression of
Nanog protein and predicts poor prognosis of non-small cell lung
cancer. J Transl Med. 11:1142013.
|
|
43
|
Ramachandran I, Thavathiru E, Ramalingam
S, et al: Wnt inhibitory factor 1 induces apoptosis and inhibits
cervical cancer growth, invasion and angiogenesis in vivo.
Oncogene. 31:2725–2737. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Heuberger J and Birchmeier W: Interplay of
cadherin-mediated cell adhesion and canonical Wnt signaling. Cold
Spring Harb Perspect Biol. 2:a0029152010. View Article : Google Scholar : PubMed/NCBI
|