Introduction
Survivin, encoded by the human baculoviral IAP
repeat containing 5 (BIRC5) gene, is the smallest member of the
inhibitor of apoptosis (IAP) family and is involved in several
distinct molecular processes, including cell division,
intracellular signaling and apoptosis (1). Survivin is rarely expressed in normal
human tissues, but is widely expressed in the majority of cancer
types, including neuroblastoma, melanoma and hepatocellular
carcinoma, as well as brain, esophageal, laryngeal, gastric,
colorectal, bladder, renal, uterine, ovarian, lung, breast and
pancreatic cancer. Moreover, survivin is associated with increased
tumor aggression and poorer clinical outcome (2,3). Due
to the high global expression of survivin in cancer, survivin has
been proposed to be a molecular target for anticancer therapy
(4–7).
The expression of survivin in breast cancer has
attracted much research attention. Over the past year, several
studies have investigated survivin mRNA and protein expression in
breast cancer, as well as the correlation between survivin
expression and clinicopathological factors and prognosis in
patients with breast cancer. Survivin has been reported to be
expressed in numerous types of breast cancer and overexpression of
survivin has been associated with a poor overall prognosis
(8–14).
The mechanisms underlying survivin overexpression in
cancer have yet to be elucidated; however, survivin expression has
been proposed to be regulated primarily at the transcriptional
level (15). Genetic variation in
the survivin promoter region may be responsible for survivin
expression, particularly the −31G/C single-nucleotide polymorphism
(SNP), which has been shown to affect survivin expression, thereby
influencing overall susceptibility to cancer (16,17).
The −31G/C SNP is located at the cell cycle-dependent element/cell
cycle homology (CDE/CHR) repressor region and mediates survivin
expression through functional disruption of binding at the CDE/CHR
repressor motif (18). However,
−31 G/C polymorphism has not been found to be associated with
survivin expression in breast cancer (19). Analysis of the putative promoter
region of BIRC5 has revealed that the ubiquitous transcription
factor specificity protein (Sp) 1 may be responsible for the
constitutive survivin expression in tumors (20,21).
Furthermore, numerous other transcription factors, including
retinoblastoma protein, p53, signal transducer and activator of
transcription 3, c-Myc, E2F, Krüppel-like factor (KLF) 5 and KLF4,
may be involved in the regulation of survivin expression (22,23).
Among these transcription factors, Sp1 has attracted the most
research attention. Sp1 has been proposed to be the predominant
reason for constitutive survivin expression in tumors (20,21).
There are at least seven potential Sp1 binding sites in the core
promoter region of survivin, according to the canonical or close to
canonical sequence (G/T)(G/A)GGCG(G/T)(G/A)(G/A)(C/T), with at
least three of the Sp1 binding sites reported to be functionally
active (20–22). Furthermore, other transcription
factors, including KLF4, have been found to regulate survivin
expression through direct interaction with Sp1 (23,24).
Thus, Sp1 may be the key to understanding survivin
expression in cancer. In vitro assays have revealed that Sp1
mutation may decrease survivin promoter activity, as well as
survivin expression (22). The
present study aimed to investigate whether Sp1 binding sites vary
in breast cancer tissues and to analyze the correlation between Sp1
binding site variations and survivin expression.
Materials and methods
Tissue sample collection and
immunohistochemistry
Breast cancer tissue samples were obtained from
Loudi Central Hospital (Loudi, China). All samples were confirmed
by pathologists. Paraffin-embedded samples were cut into 4-mm
sections and immunohistochemistry was performed using a standard
streptavidin peroxidase (SP) method. Slides were dewaxed using
xylene and rehydrated with serial dilutions of ethanol. Antigen
retrieval was then performed and endogenous peroxidase activity was
blocked using 3% hydrogen peroxide and nonspecific staining was
blocked using 10% normal goat serum (Vector Laboratories Inc.,
Burlingame, CA, USA). Slides were incubated with anti-survivin
antibodies diluted 1:100 (Santa Cruz Biotechnology, Inc., Santa
Cruz, CA, USA) at 4°C in a humidified chamber overnight, followed
by incubation with biotin-conjugated secondary antibodies
(ZSGB-BIO, Beijing, China) for 30 min and incubation with
streptavidin peroxidase for 15 min. 3,3′-diaminobenzidine
tetrachloride (DAB; ZSGB-BIO) was added to the slides to detect
peroxidase activity. Sections were then dehydrated using alcohol
and cleared using xylene. The slides were assessed by two
individual investigators. Survivin immunoreactivity was scored
between 0 and 4 based on the color and percentage of cells showing
distinct nuclear and/or diffuse cytoplasmic immunohistochemical
staining. Informed consent was obtained from all patients prior to
sample collection and the protocol was approved by the
Institutional Review Board of Loudi Central Hospital (Loudi,
China).
DNA extraction, nested polymerase chain
reaction (PCR) and DNA sequencing
Genomic DNA was extracted from three
paraffin-embedded sections representative of breast cancer tissues.
The DNA was then isolated using a DNA Extraction from
Paraffin-Embedded Tissue kit (Takara Bio, Inc., Shiga, Japan)
according to the manufacturer’s instructions. The concentration and
purity of the extracted DNA was assessed using spectroscopy with a
NanoDrop® ND-1000 spectrophotometer (Nanodrop
Technologies Inc., Wilmington, DE, USA). Nested PCR analysis was
performed to detect Sp1 binding sites in the survivin gene promoter
region. The primer sequences were as follows: External primers:
SurO1, forward 5′-CGCGTTCTTTGAAAGCAGTCGAG-3′ and SurO2, reverse
5′-GAAGGGCCAGTTCTTGAATGTAGAG-3′, which were used for the first
round of PCR; and internal primers: SurI1, forward
5′-GCTAGGTGTGGGCAGGGACGAGCTG-3′ and SurI2, reverse
5′-GATTCAAATCTGGCGGTTAATGG-3′, which were used to amplify a 207 bp
fragment in a final volume of 50 μl. A total of 5 μl PCR product
was separated on a 1.5% agarose gel and stained with ethidium
bromide, followed by analysis under ultraviolet (UV) light.
Amplified products which were observed as clear bands were
sequenced in duplicate in the forward and reverse directions using
a BigDye® Terminator sequencing kit (Applied Biosystems,
Inc., Foster City, CA, USA) and an ABI Prism 3730XL DNA Analyzer
(Applied Biosystems, Inc.) according to the manufacturer’s
instructions. Sequences were compared against the archived sequence
of the human survivin gene in GenBank (http://www.ncbi.nlm.nih.gov/genbank/).
Statistical analysis
All statistical analyses were performed using SPSS
13.0 (SPSS, Inc., Chicago, IL, USA) for windows. The χ2
test was performed to analyze the correlation between Sp1 binding
site mutations in the survivin gene promoter region and survivin
expression in breast cancer tissues. P<0.05 was considered to
indicate a statistically significant difference.
Results
Survivin expression in 42 breast cancer
tissues detected using immunohistochemistry
Survivin expression was analyzed in 42 patients with
breast cancer using immunohistochemistry. Samples were scored
between 0 and 4 based on survivin expression. The numbers of
samples exhibiting survivin expression with a score of 0, 1, 2, 3
and 4 were 7 (16.7%), 7 (16.7%), 12 (28.6%), 12 (28.6%) and 4
(9.5%), respectively. Scores of 0 and 1 were defined as ‘low
expression’ and scores of 2–4 were defined as ‘high expression’.
Among the 42 tissue samples, 14 tissues exhibited low survivin
expression, while 28 samples had high survivin expression.
Analysis of Sp1 binding site variation in
the survivin gene promoter region
In order to compare nucleotide sequences, survivin
gene promoter fragments were amplified from the genomic DNA
isolated from the 42 breast cancer tissue samples using nested PCR.
The intended sequence was 207 bp long, containing seven Sp1 binding
sites from −34 to −240 upstream of the translation initiation site
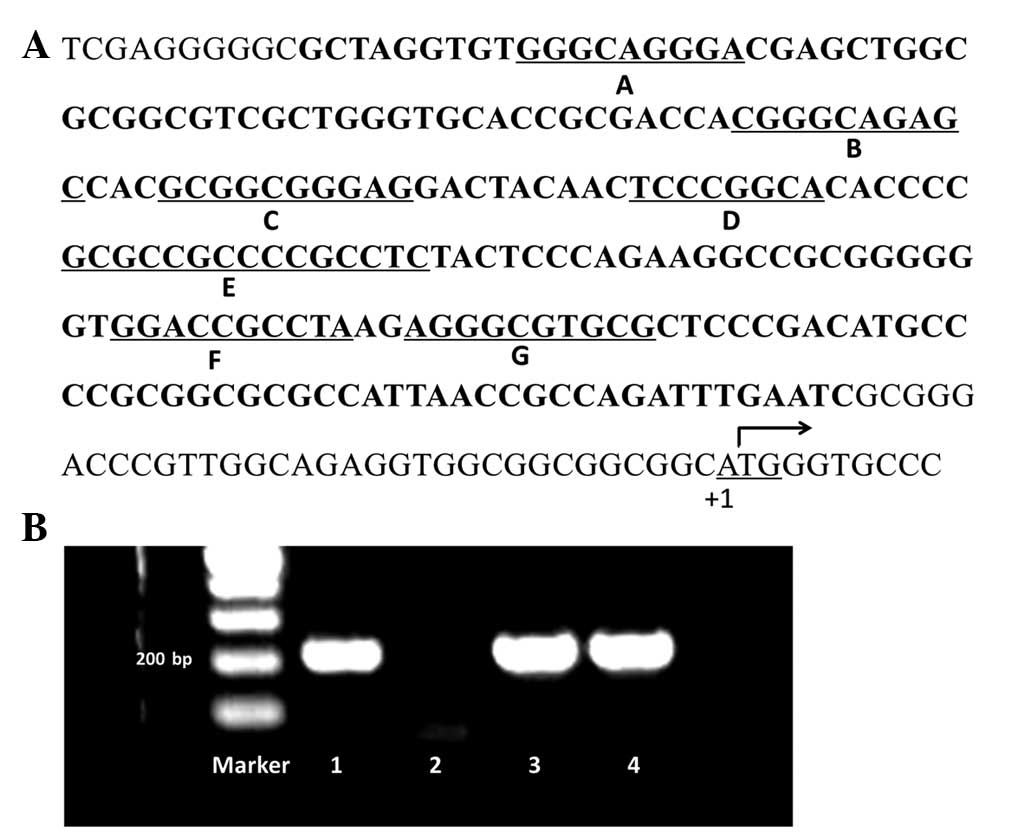
of the survivin gene (Fig. 1A).
The PCR products were separated on a 1.5% agarose gel. PCR
fragments which exhibited single clear bands under UV light were
sequenced (samples 1, 3 and 4; Fig.
1B). In total, 15 variations on seven Sp1 binding sites were
observed in 12 samples. No Sp1 binding site variations were found
in the other 30 samples. Among these variations, Sp1 binding site
E, which is located at the ‘Sp1-complex’ (21), was observed to have the greatest
mutation frequency, while the Sp1 binding site A exhibited no
variation (Table I).
 | Table IVariations in Sp1 binding sites in the
survivin gene promoter in 42 patients with breast cancer. |
Table I
Variations in Sp1 binding sites in the
survivin gene promoter in 42 patients with breast cancer.
| Sp1 binding site | Variations |
|---|
|
|---|
| 1 | 2 | 3 | 4 | 5 |
|---|
| A | | | | | |
| B | −184G/A | | | | |
| C | −171G/A | | | | |
| D | −150G/A | −151G/A | | | |
| E | −129C/T | −130G/A | −132C/T | −140G/A | −141C/T |
| F | −94C/T | −95C/T | −101G/A | | |
| G | −82G/A | −84G/A | −87G/A | | |
Correlation between variation in Sp1
binding sites in the survivin gene promoter region and survivin
expression
In order to analyze the correlation between
variation in Sp1 binding sites in the survivin gene promoter region
and survivin expression, χ2 tests were performed and a
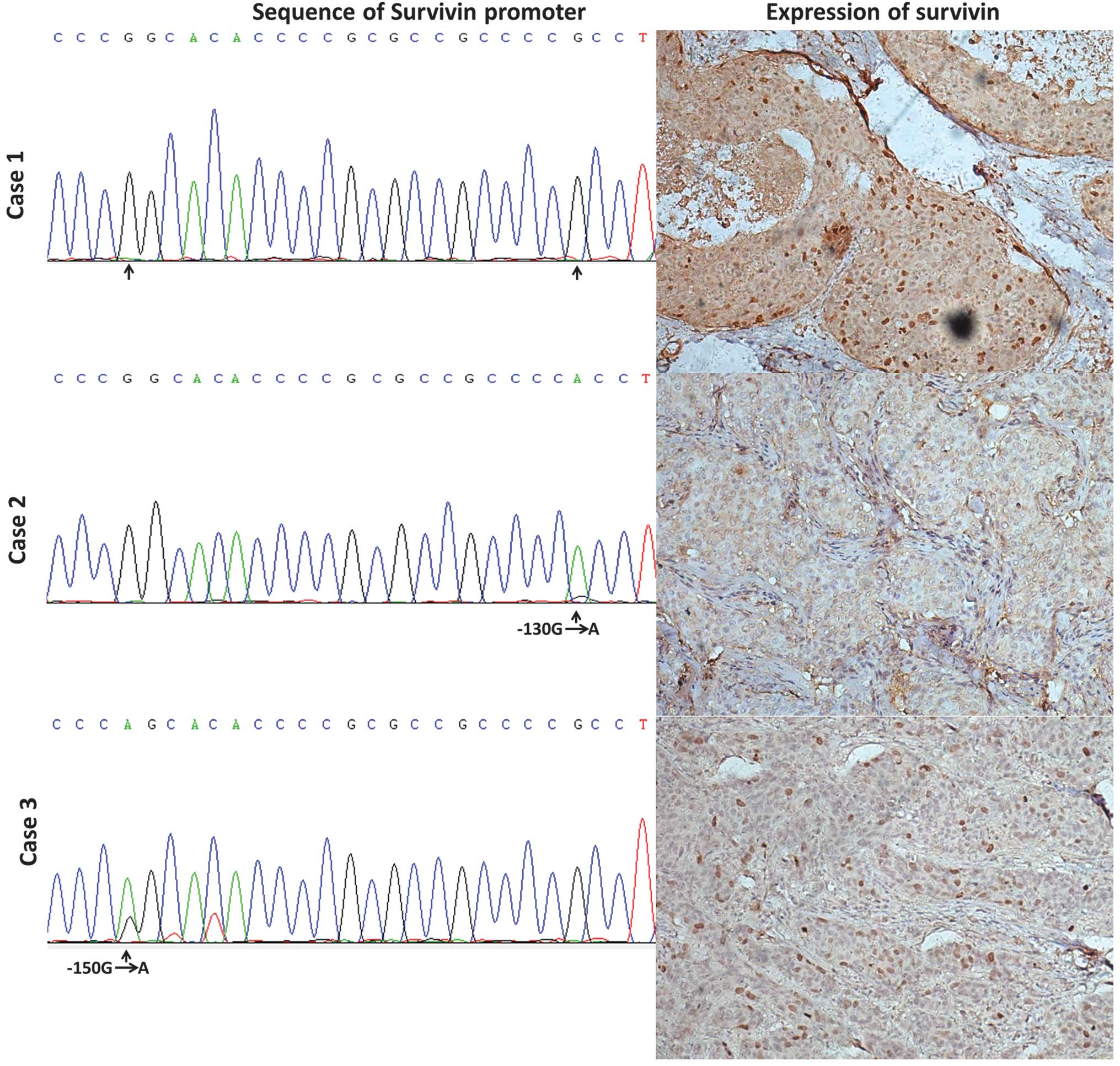
significant correlation was observed (P<0.05, Table II). As shown in Fig. 2, case 1 was a sample which
exhibited no Sp1 binding site variations in the survivin promoter
and had high survivin expression. Case 2 had a −130 G to A
homozygous variation and its survivin expression was low. Moreover,
case 3 had a −150 G to A heterozygous variation and low expression
of survivin (Fig. 2).
 | Table IICorrelation between Sp1 binding site
variation and survivin expression in 42 breast cancer samples. |
Table II
Correlation between Sp1 binding site
variation and survivin expression in 42 breast cancer samples.
| Sp1 binding
site | Low survivin
expression, n (%) | High survivin
expression, n (%) | Total n (%) | Value |
|---|
|
|---|
| χ2 | P-value |
|---|
| Mutated | 8 (19.0) | 4 (9.5) | 12 (28.6) | | |
| Not mutated | 6 (14.3) | 24 (57.1) | 30 (71.4) | | |
| Total | 14 (33.3) | 28 (66.7) | 42 (100) | 8.400 | 0.004 |
Discussion
Breast cancer is a heterogeneous group of tumors
which differ in molecular characteristics, prognosis and response
to therapy (25,26). Somatic mutations occurring at any
stage of cancer development may affect the properties of the cancer
cells. Certain recurrent somatic alterations in breast cancer have
been reported, including human epidermal growth factor receptor 2
amplifications, which were the first successful therapeutic targets
defined by a genomic aberration (27). Several studies have investigated
mutations across different breast cancer subtypes (28–31)
and recurrent somatic gene mutations, particularly in coding
regions, including core-binding factor, beta subunit (28), TP53 (28) and GATA3 (32), have been identified in breast
cancer. Genomic alterations which affect survivin expression in
breast cancer cells have been reported and loss of heterozygosity
in breast cancer has been associated with survivin expression
(19). In addition, the survivin
gene is located at 17q25, a hotspot for mutations in breast cancer
and other tumors (33–35). Thus, the high mutation rate at the
survivin gene promoter may be associated with genomic
instability.
In addition to somatic gene mutations, SNPs have
been a focus of cancer research. The −31 G/C SNP of the survivin
gene promoter has been reported to be associated with gastric
(36), colorectal (17), lung (37) and urothelial cancer (38) as well as esophageal squamous cell
carcinoma (39), and may be a risk
factor for cancer development. However, the −31 G/C polymorphism
has not been associated with cervical (40) or breast (32) cancer. Based on the central role of
survivin in multiple cellular networks, it is important to
elucidate the factors regulating and influencing survivin
expression (6).
The transcription factor Sp1 is one of the most
important regulators of survivin. Sp1 binds to GC-rich motifs in
various promoters and is involved in numerous cellular processes,
including cell differentiation, growth and apoptosis, as well as
immune responses, DNA damage responses and chromatin remodeling
(41). Polymorphisms of Sp1
binding sites which affect protein expression have been
investigated in several studies (42–45),
particularly in patients with cancer. For example, the −216 G/T
polymorphism of the Sp1 binding site at the epidermal growth factor
receptor promoter region has been reported to be associated with
altered promoter activity and gene expression in vitro and
in vivo (43). Furthermore,
SNPs of the Sp1 binding site at the mouse double minute 2 homolog
promoter have been associated with risk of breast and ovarian
cancer, and Sp1 binding site polymorphisms at the matrix
metalloproteinase-2 promoter have been correlated with risk of
gastric (44) and lung cancer
(45). Prior to the identification
of Sp1 binding site polymorphisms at the survivin promoter,
survivin expression was shown to be regulated by Sp1 through
several Sp1 binding sites in the survivin promoter, which were
close to the translation initiation site, and mutations in these
binding sites were found to decrease survivin expression in
vitro (20–22,46,47).
In the present study, low survivin expression was observed to
correlate with Sp1 binding site variation in the survivin promoter
in vivo. While the mechanism underlying the high mutation
rate in the survivin gene promoter region and the functional
significance of such mutations in breast cancer remain to be fully
elucidated, the phenomenon suggests that survivin promoter
genotypes may influence survivin expression, thus influencing
individual susceptibility to the initiation and development of
certain types of cancer.
Acknowledgements
The authors would like to thank Lanping Quan and
Jinfeng Bai from the Laboratory of Cell and Molecular Biology for
their technical assistance, as well as Sangon Biotech (Shanghai)
Co., Ltd. (Shanghai, China) for the DNA sequencing. The present
study was supported by grants from the National Natural Science
Foundation of China (nos. 31171322 and 81021061).
Abbreviations:
|
IAP
|
inhibitor of apoptosis
|
|
BIRC5
|
human baculoviral IAP repeat
containing 5
|
|
Sp1
|
specificity protein 1
|
References
|
1
|
Li F and Ling X: Survivin study: an update
of ‘what is the next wave’? J Cell Physiol. 208:476–486. 2006.
|
|
2
|
Li F: Survivin study: what is the next
wave? J Cell Physiol. 197:8–29. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jha K, Shukla M and Pandey M: Survivin
expression and targeting in breast cancer. Surg Oncol. 21:125–131.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Altieri DC: New wirings in the survivin
networks. Oncogene. 27:6276–6284. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Altieri DC: Survivin, cancer networks and
pathway-directed drug discovery. Nat Rev Cancer. 8:61–70. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Guha M and Altieri DC: Survivin as a
global target of intrinsic tumor suppression networks. Cell Cycle.
8:2708–2710. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Waligórska-Stachura J, Jankowska A, Waśko
R, et al: Survivin - prognostic tumor biomarker in human neoplasms
- review. Ginekol Pol. 83:537–540. 2012.PubMed/NCBI
|
|
8
|
Sarti M, Pinton S, Limoni C, et al:
Differential expression of testin and survivin in breast cancer
subtypes. Oncol Rep. 30:824–832. 2013.PubMed/NCBI
|
|
9
|
Dedić Plavetić N, Jakić-Razumović J, Kulić
A and Vrbanec D: Prognostic value of proliferation markers
expression in breast cancer. Med Oncol. 30:5232013.
|
|
10
|
Song J, Su H, Zhou YY and Guo LL:
Prognostic value of survivin expression in breast cancer patients:
a meta-analysis. Tumour Biol. 34:2053–2062. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xu C, Yamamoto-Ibusuki M, Yamamoto Y, et
al: High survivin mRNA expression is a predictor of poor prognosis
in breast cancer: a comparative study at the mRNA and protein
level. Breast Cancer. Sep 12–2012.(Epub ahead of print).
|
|
12
|
Li X, Dang X and Sun X: Expression of
survivin and VEGF-C in breast cancer tissue and its relation to
lymphatic metastasis. Eur J Gynaecol Oncol. 33:178–182.
2012.PubMed/NCBI
|
|
13
|
Adamkov M, Kajo K, Vybohova D, Krajcovic
J, Stuller F and Rajcani J: Correlations of survivin expression
with clinicomorphological parameters and hormonal receptor status
in breast ductal carcinoma. Neoplasma. 59:30–37. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kapoor S: Beyond pancreatic carcinoma: The
close relationship between survivin levels and prognosis in
systemic malignancies. World J Clin Oncol. 3:80–81. 2012.
View Article : Google Scholar
|
|
15
|
Johnson ME and Howerth EW: Survivin: a
bifunctional inhibitor of apoptosis protein. Vet Pathol.
41:599–607. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gazouli M, Tzanakis N, Rallis G, et al:
Survivin −31G/C promoter polymorphism and sporadic colorectal
cancer. Int J Colorectal Dis. 24:145–150. 2009.
|
|
17
|
Li XB, Li SN, Yang ZH, Cao L, Duan FL and
Sun XW: Polymorphisms of survivin and its protein expression are
associated with colorectal cancer susceptibility in Chinese
population. DNA Cell Biol. 32:236–242. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xu Y, Fang F, Ludewig G, Jones G and Jones
D: A mutation found in the promoter region of the human survivin
gene is correlated to overexpression of survivin in cancer cells.
DNA Cell Biol. 23:419–429. 2004. View Article : Google Scholar
|
|
19
|
Boidot R, Vegran F, Jacob D, et al: The
expression of BIRC5 is correlated with loss of specific chromosomal
regions in breast carcinomas. Genes Chromosomes Cancer. 47:299–308.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li F and Altieri DC: Transcriptional
analysis of human survivin gene expression. Biochem J. 344:305–311.
1999. View Article : Google Scholar
|
|
21
|
Xu R, Zhang P, Huang J, Ge S, Lu J and
Qian G: Sp1 and Sp3 regulate basal transcription of the survivin
gene. Biochem Biophys Res Commun. 356:286–292. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mityaev MV, Kopantzev EP, Buzdin AA,
Vinogradova TV and Sverdlov ED: Functional significance of a
putative Sp1 transcription factor binding site in the survivin gene
promoter. Biochemistry (Mosc). 73:1183–1191. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang G, Zhu H, Wang Y, et al:
Krüppel-like factor 4 represses transcription of the survivin gene
in esophageal cancer cell lines. Biol Chem. 390:463–469. 2009.
|
|
24
|
Li Y, Xie M, Yang J, et al: The expression
of antiapoptotic protein survivin is transcriptionally upregulated
by DEC1 primarily through multiple Sp1 binding sites in the
proximal promoter. Oncogene. 25:3296–3306. 2006. View Article : Google Scholar
|
|
25
|
Sørlie T, Perou CM, Tibshirani R, et al:
Gene expression patterns of breast carcinomas distinguish tumor
subclasses with clinical implications. Proc Natl Acad Sci USA.
98:10869–10874. 2001.PubMed/NCBI
|
|
26
|
Chin K, DeVries S, Fridlyand J, et al:
Genomic and transcriptional aberrations linked to breast cancer
pathophysiologies. Cancer Cell. 10:529–541. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
King CR, Kraus MH and Aaronson SA:
Amplification of a novel v-erbB-related gene in a human mammary
carcinoma. Science. 229:974–976. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Banerji S, Cibulskis K, Rangel-Escareno C,
et al: Sequence analysis of mutations and translocations across
breast cancer subtypes. Nature. 486:405–409. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sjöblom T, Jones S, Wood LD, et al: The
consensus coding sequences of human breast and colorectal cancers.
Science. 314:268–274. 2006.
|
|
30
|
Wood LD, Parsons DW, Jones S, et al: The
genomic landscapes of human breast and colorectal cancers. Science.
318:1108–1113. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kan Z, Jaiswal BS, Stinson J, et al:
Diverse somatic mutation patterns and pathway alterations in human
cancers. Nature. 466:869–873. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Usary J, Llaca V, Karaca G, et al:
Mutation of GATA3 in human breast tumors. Oncogene. 23:7669–7678.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ly M, Valent A, Diallo G, et al: Gene copy
number variations in breast cancer of Sub-Saharan African women.
Breast. 22:295–300. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Orsetti B, Courjal F, Cuny M, Rodriguez C
and Theillet C: 17q21–q25 aberrations in breast cancer: combined
allelotyping and CGH analysis reveals 5 regions of allelic
imbalance among which two correspond to DNA amplification.
Oncogene. 18:6262–6270. 1999.
|
|
35
|
Kalikin LM, Frank TS, Svoboda-Newman SM,
Wetzel JC, Cooney KA and Petty EM: A region of interstitial 17q25
allelic loss in ovarian tumors coincides with a defined region of
loss in breast tumors. Oncogene. 14:1991–1994. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Cheng ZJ, Hu LH and Huang SJ: Correlation
of −31G/C polymorphisms of survivin promoter to tumorigenesis of
gastric carcinoma. Ai Zheng. 27:258–263. 2008.(In Chinese).
|
|
37
|
Jang JS, Kim KM, Kang KH, et al:
Polymorphisms in the survivin gene and the risk of lung cancer.
Lung Cancer. 60:31–39. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang YH, Chiou HY, Lin CT, et al:
Association between survivin gene promoter −31 C/G polymorphism and
urothelial carcinoma risk in Taiwanese population. Urology.
73:670–674. 2009.
|
|
39
|
Yang X, Xiong G, Chen X, et al:
Polymorphisms of survivin promoter are associated with risk of
esophageal squamous cell carcinoma. J Cancer Res Clin Oncol.
135:1341–1349. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Borbély AA, Murvai M, Szarka K, et al:
Survivin promoter polymorphism and cervical carcinogenesis. J Clin
Pathol. 60:303–306. 2007.
|
|
41
|
Wierstra I: Sp1: emerging roles - beyond
constitutive activation of TATA-less housekeeping genes. Biochem
Biophys Res Commun. 372:1–13. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Grant SF, Reid DM, Blake G, Herd R,
Fogelman I and Ralston SH: Reduced bone density and osteoporosis
associated with a polymorphic Sp1 binding site in the collagen type
I alpha 1 gene. Nat Genet. 14:203–205. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Liu W, Innocenti F, Wu MH, et al: A
functional common polymorphism in a Sp1 recognition site of the
epidermal growth factor receptor gene promoter. Cancer Res.
65:46–53. 2005.PubMed/NCBI
|
|
44
|
Miao X, Yu C, Tan W, et al: A functional
polymorphism in the matrix metalloproteinase-2 gene promoter
(−1306C/T) is associated with risk of development but not
metastasis of gastric cardia adenocarcinoma. Cancer Res.
63:3987–3990. 2003.
|
|
45
|
Yu C, Pan K, Xing D, et al: Correlation
between a single nucleotide polymorphism in the matrix
metalloproteinase-2 promoter and risk of lung cancer. Cancer Res.
62:6430–6433. 2002.PubMed/NCBI
|
|
46
|
Cheng Q, Ling X, Haller A, et al:
Suppression of survivin promoter activity by YM155 involves
disruption of Sp1-DNA interaction in the survivin core promoter.
Int J Biochem Mol Biol. 3:179–197. 2012.PubMed/NCBI
|
|
47
|
Estève PO, Chin HG and Pradhan S:
Molecular mechanisms of transactivation and doxorubicin-mediated
repression of survivin gene in cancer cells. J Biol Chem.
282:2615–2625. 2007.PubMed/NCBI
|