Introduction
Prostate cancer remains one of the most prevalent
types of cancer in males (1). In
the USA and Europe, prostate cancer is currently the second most
common cause of cancer mortality in males >40 years of age, and
the third most common cause of cancer-associated mortality in males
(2,3). A total of 238,590 novel cases and
29,720 mortalities due to prostate cancer were estimated for 2013
in the USA (2). Although the
etiology of this malignancy remains to be elucidated, several risk
factors have been identified that are considered to contribute to
prostate carcinogenesis, including hereditary and environmental
components; however, age, race and family history are the only
well-established risk factors (4).
Early-stage cancers are managed using various treatments, including
radical prostatectomy, radiation or hormone ablation therapy.
Prostate cancer is only temporarily manageable using hormone
deprivation, it will inevitably become resistant to hormonal
therapy, following which there is currently no effective treatment
(5). Therefore, understanding the
molecular basis of prostate cancer and the development of novel
treatments are crucial for improving the survival rates of prostate
cancer patients.
MicroRNAs (miRNAs) are a type of endogenously
expressed small, non-coding, single-stranded RNAs. miRNAs are able
to negatively regulate gene expression through binding to the 3′
untranslated region (3′UTR) of their target messenger RNAs (mRNAs)
(6). In mammalian cells, miRNAs
affect gene silencing via translational inhibition and mRNA
degradation; an individual miRNA is capable of regulating numerous
distinct mRNAs. miRNAs have been estimated to regulate ~30% of the
human genome (7). Hundreds of
evolutionarily conserved miRNAs have been identified in plants,
animals and viruses (7,8). These molecules have been reported to
control fundamental cell functions, including proliferation,
apoptosis and differentiation; this therefore indicated that they
may have a role in carcinogenesis (9–11).
Numerous studies have illustrated that miRNAs are aberrantly
expressed in human malignancies, such as prostate cancer (12–14).
Upregulated miRNAs in cancer may function as oncogenes through the
negative regulation of tumor suppressor genes (15). By contrast, downregulated miRNAs
have been reported to function as tumor suppressor genes and
inhibit cancer development through the regulation of oncogenes
(16). Therefore, the
identification of the target of miRNAs may be critical for
understanding the function of miRNAs in cancer development and
progression. It has also been suggested that miRNAs may be useful
therapeutic targets for novel cancer therapies.
miR-99a has been reported to be a frequently
downregulated miRNA in numerous types of human cancers, including
prostate, bladder, hepatocellular and ovarian carcinoma, squamous
cell carcinoma of the tongue, squamous cell lung carcinoma as well
as childhood adrenocortical tumors (17). A study by Sun et al
(18) reported that miR-99a
suppressed the expression of prostate-specific antigen and prostate
cancer cell proliferation by targeting switch/sucrose
nonfermentable-related matrix-associated actin-dependent regulator
of chromatin subfamily A member 5 and subfamily D member 1 as well
as the mechanistic target of rapamycin (mTOR). The aim of the
present study was to determine the effects of miR-99a on cell
proliferation, colony formation ability, migration and invasion in
prostate cancer cell lines. In addition, the effect of miR-99a
expression on fibroblast growth factor receptor 3 (FGFR3) was
examined.
Materials and methods
Cells and culture conditions
The human prostate cancer cell lines DU145 and PC-3
were obtained from the Shanghai Institute of Biochemistry and Cell
Biology (Shanghai, China). DU145 and PC-3 cells were cultured in
RPMI 1640 medium (Gibco-BRL, Grand Island, NY, USA) supplemented
with 10% heat-inactivated fetal bovine serum (FBS; Gibco-BRL) under
a humidified atmosphere of 5% CO2 at 37°C. To propagate
spheres in vitro, spheres were collected by gentle
centrifugation (200 × g), dissociated into single cells, and then
cultured to generate the next generation of spheres.
Transfection
Mature miR-99a mimics and miRNA mimics negative
control (NC) were designed and synthesized by GenePharma (Shanghai,
China). The sequence of miR-99a mimics was
5′-AACCCGUAGAUCCGAUCUUGUG-3′. The sequence of NC mimics was
5′-UUCUCCGAACGUGUCACGUTT-3′. Cells were transfected using
Lipofectamine 2000 (Invitrogen Life Technologies, Carlsbad, CA,
USA) according to the manufacturer’s instructions.
Quantitative polymerase chain reaction
(qPCR) for miR-99a
Total cellular RNA was extracted using TRIzol
reagent (Invitrogen Life Technologies). The RNA was stored in
diethylpyrocarbonate-treated water at −80°C and the quantity and
quality of the samples were evaluated prior to use with an ND-1000
NanoDrop spectrophotometer (NanoDrop Technologies, Wilmington, DE,
USA). qPCR for miR-99a was performed using a SYBRgreen microRNA
assay (Genepharm, Shanghai, China) according to the manufacturer’s
instructions. qPCR was performed using a 7300 Real-time PCR System
(Applied Biosystems, Foster City, CA, USA) with an miR-99a primer
set and double strand binding dye SYBRgreen. All primers were
obtained from the TaqMan miRNA assays. GAPDH (Genepharm, Shanghai,
China) was used as an internal control. Every sample was replicated
three times. Data were analyzed by comparing Ct values.
Cell viability assay
Cell proliferation was determined using the MTT
assay. The transfected cells (miR-99a mimics and NC) were seeded in
a 96-well plate at a density of 3,000 cells per well. Cell
viability assays were performed every 24 h for 5 days. In brief, 20
μl MTT solution was added to each well and incubated at 37°C for 4
h. Plates were then centrifuged (200 × g, 10 min), and the purple
colored precipitates of formazan were dissolved in 200 μl dimethyl
sulfoxide. Absorbance was measured at 490 nm in an ELISA reader
(Model 550; Bio-Rad, Richmond, CA, USA). The suppression rate was
calculated using the formula: Suppression
rate=(1-ODmiR-145/ODmiR-NC)x100%. Proliferation curves were drawn
on the basis of mean absorbance at each time-point. All the
experiments were performed in triplicate.
Colony formation assay
The colony-forming ability of DU145 and PC-3 cells
transfected with miR-99a was assessed using a colony formation
assay. In brief, the transfected cells (miR-99a mimics and NC)
growing in log phase were trypsinized and seeded into six-well
plates with a density of 2,000 cells per well. The cells were kept
in an incubator at 37°C for seven days. On day eight, the colonies
were washed with phosphate-buffered saline (PBS), fixed with
formalin (10%; Beyotime Institute of Biotechnology, Haimen, China)
and stained with methyl violet. Finally, the methyl violet dye was
washed off with PBS. The number of colonies was counted using a
microscope (Olympus IX53; Olympus Corporation, Tokyo, Japan).
Colony-inhibition rate=[1−(number of colonies in experimental
groups/control group)]x100%; and colony-forming
efficiency=(1−colony-inhibition rate), were calculated.
Cell migration and invasion assay
Cell migration and invasion were assayed using
Transwell chambers (8 μm; Costar, Cambridge, MA, USA). For the
migration assay, the transfected cells (miR-99a mimics and NC)
growing in the log phase were trypsinized and resuspended as single
cell solutions. A total of 1×105 cells per well were
placed into the upper chamber cultured in medium with 2% FBS, while
500 μl medium containing 20% FBS was added to the lower chamber.
For the invasion assay, a Transwell chamber coated with
Matrigel® (BD Biosciences, San Jose, CA, USA) and a
total of 1×105 cells were seeded into the upper chamber,
while the bottom chamber was incubated with 500 μl medium
containing 20% serum. Cells were incubated for 12 h for the
migration assay and 24 h for the invasion assay. At the end of the
experiments, the cells on the upper surface of the membrane were
removed, and the migrated cells were fixed with 100% methanol
(Beyotime Institute of Biotechnology) for two min, stained in 0.5%
crystal violet (Beyotime Institute of Biotechnology) for two min,
rinsed in PBS and then subjected to microscopic inspection
(magnification, ×200). Five visual fields of each insert were
randomly selected and counted under a light microscope (Olympus
IX53). Each condition was assayed in triplicate and each experiment
repeated at least three times.
Western blot analysis
Primary antibodies used in the present study,
including FGFR3 (rabbit, polyclonal) and β-actin (rabbit,
monoclonal), were purchased from Bioworld Technology (Louis Park,
MN, USA). Transfected cells were washed with ice cold PBS and lysed
with 1% radioimmunoprecipitation assay lysis buffer (Beyotime
Institute of Biotechnology) 72 h following transfection. The
protein concentration was determined using the bicinchoninic acid
assay kit (Beyotime Institute of Biotechnology). Equal amounts of
the proteins were separated by 10% SDS PAGE (Beyotime Institute of
Biotechnology) and transferred to polyvinylidene difluoride
membranes (Beyotime Institute of Biotechnology). The membrane was
blocked with 5% skimmed milk, followed by an overnight incubation
at 4°C with a primary antibody at dilutions specified by the
manufacturer’s instructions. Following washing, the membrane was
incubated with the corresponding horseradish peroxidase-conjugated
secondary antibody (goat anti-rabbit; Bioworld Technology) in
tris-buffered saline with Tween (Beyotime Institute of
Biotechnology, China). The blot was then developed using an
enhanced chemilluminescence solution (Pierce, Rockford, IL, USA)
and images were captured using a FluorChem imaging system (Model
number: 92-13779-00 FC2; Alpha Innotech, San Leandro, CA, USA).
Luciferase assay
The DU145 and PC-3 cells were transfected with 0.5
μg reporter plasmid, 40 nmol miR-99a mimics or NC in a 12-well
plate using Lipofectamine 2000 according to the manufacturer’s
instructions. The primers used for cloning FGFR3 mRNA 3′UTR were:
Forward, GGGCTCGAGGGCCACTGGTCCCCAACAATGTG and reverse,
GGGCGGCCGCCCAGTAACAGTACAGAACGA ACCAAC. Assays were performed using
the Dual-Luciferase Reporter Assay system (Promega, Manheim,
Germany) following 48 h of transfection. The firefly and renilla
luciferase activities were measured using a TECAN Infinite 200
luminometer (Tecan, Männedorf, Switzerland). The firefly luciferase
activity was normalized to the renilla luciferase activity for each
transfected well. To determine whether miR-99a targets the FGFR3
3′-UTR, TARGETSCAN 5.2 (www.targetscan.org) and PICTAR (pictar.mdc-berlin.de)
were used to assess the complementarity of miR-99a to the FGFR3
3′-UTR. Each reporter plasmid was transfected at least three times
(on different days) and each sample was assayed in triplicate.
Statistical analysis
Values are presented as the mean ± standard
deviation. Statistical differences were analyzed using the
Student’s t test. Stata 10.0 software (StataCorp LP, College
Station, TX, USA) was used for statistical analysis. P<0.05 was
considered to indicate a statistically significant difference
between values.
Results
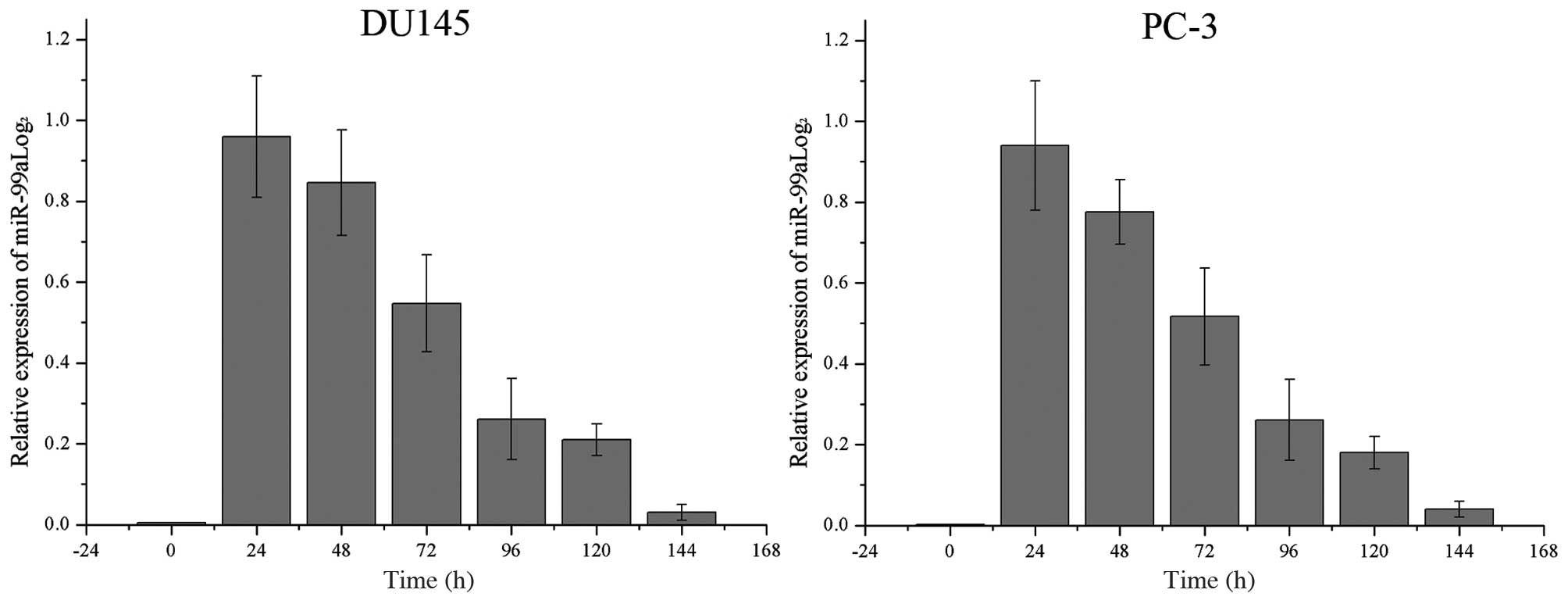
Relative miR-99a levels increase
significantly following transfection into DU145 and PC-3 cells
The endogenous levels of miR-99a in DU145 and PC-3
cells as well as its levels following transfection of miR-99a were
recorded every 24 h. As shown in Fig.
1, miR-99a levels were significantly increased until ~120 h
following transfection in the two cell lines. The levels of miR-99a
declined gradually following a peak at 24 h.
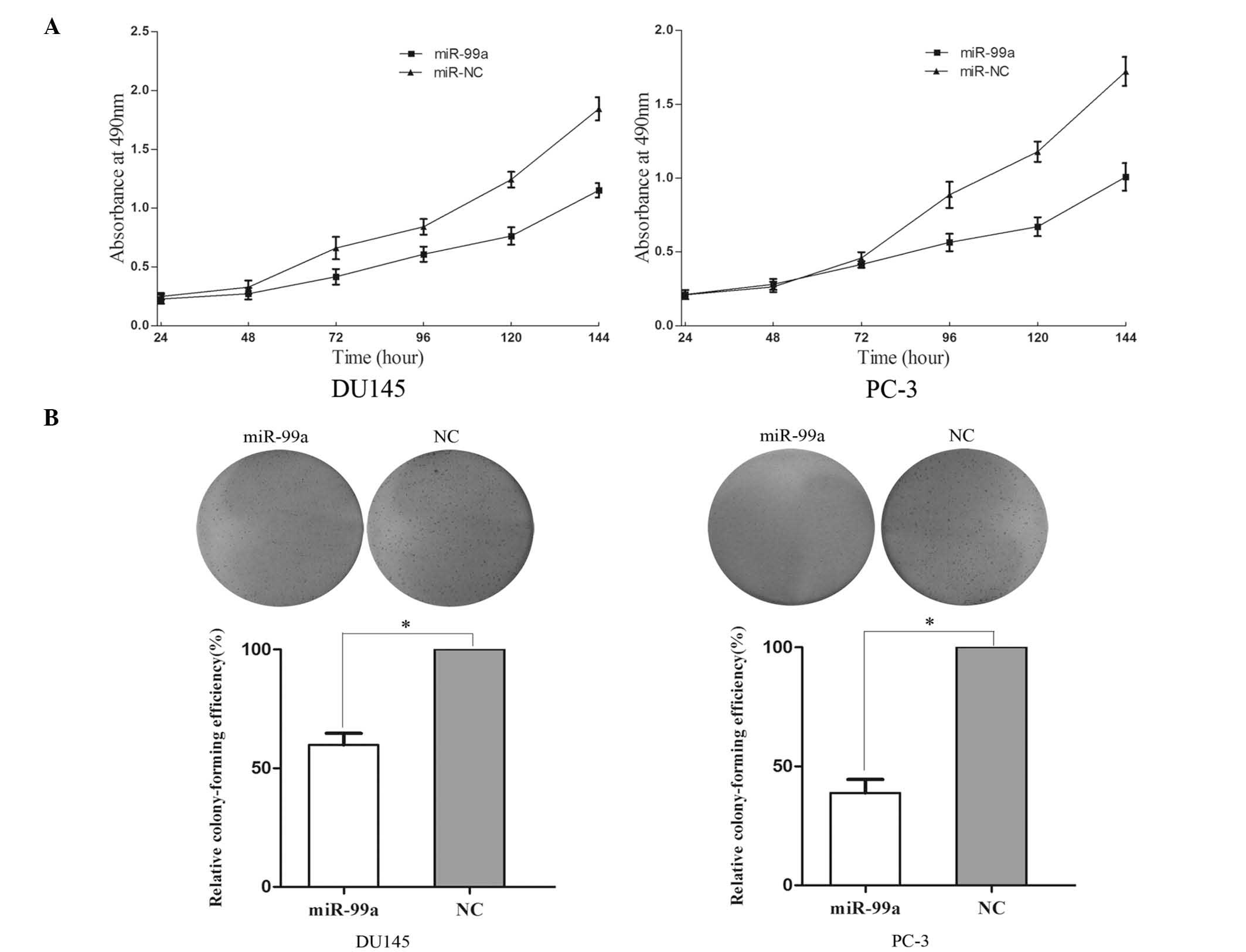
miR-99a reduces cell proliferation and
colony formation ability of DU145 and PC-3 cells
An MTT assay was used in order to investigate the
influence of miR-99a on cell proliferation. Upregulation of miR-99a
significantly inhibited cell proliferation (Fig. 2A). MTT assays revealed that
following 144 h of treatment, the growth suppression rate of
miR-99a reached 33.42±2.5% in DU145 cells and 29.13±2.8% in PC-3
cells.
A colony formation assay was used to determine the
effect of miR-99a upregulation on colony formation ability. As
shown in Fig. 2B, the relative
colony-formation efficiency was significantly reduced to 59.8±5.6%
in DU145 cells and 38.7±6.1% in PC-3 cells. These results therefore
indicated that miR-99a may have an important role in the regulation
of prostate cancer DU145 and PC-3 cell lines.
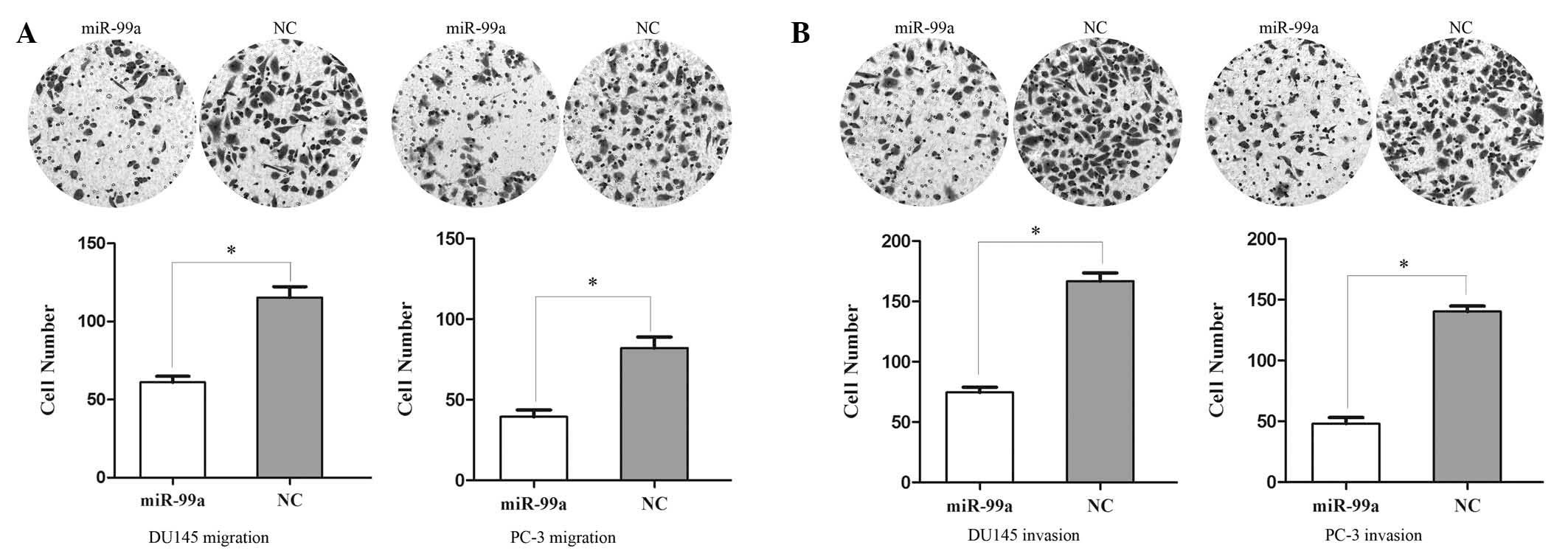
miR-99a suppresses cell migration and
invasion in prostate cancer DU145 and PC-3 cells
A Transwell assay was performed in order to measure
the effect of miR-99a on tumor cell migration and invasion.
Migration of miRNA-99a-transfected cells was significantly
decreased to 47.14±5.38% in DU145 cells and 51.95±6.18% in PC-3
cells (Fig. 3A). In the invasion
assay (Fig. 3B), the miR-99a
groups were found to have decreased cell invasion of 55.28±6.42% in
DU145 cells and 65.76±7.37% in PC-3 cells. These results indicated
that miR-99a reduced migration and invasion in prostate cancer
DU145 and PC-3 cell lines.
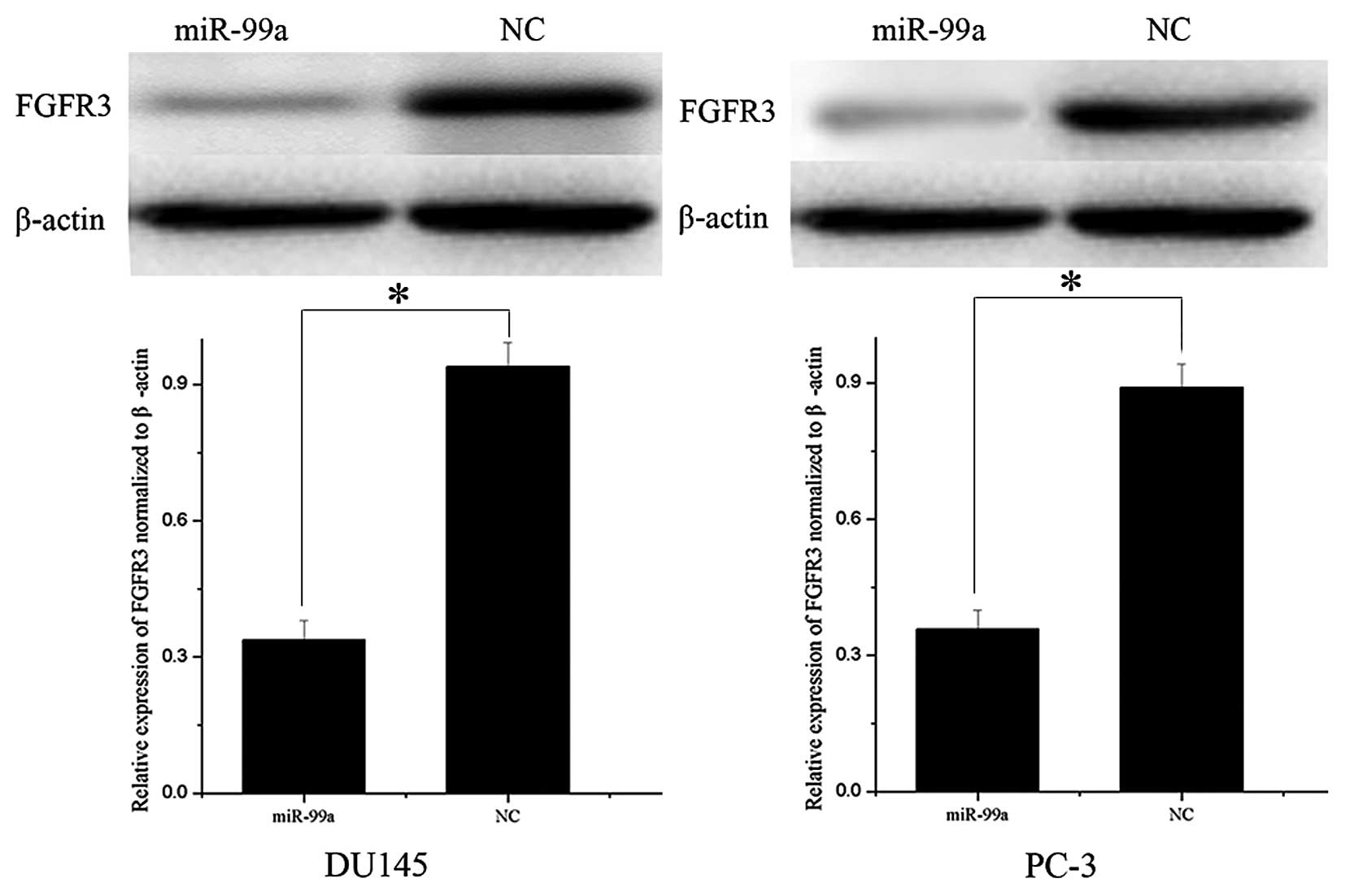
FGFR3 is downregulated following
overexpression of miR-99a in DU145 and PC-3 cells
Western blot analysis was used to determine whether
FGFR3 expression was altered following miR-99a transfection into
prostate cancer DU145 and PC-3 cells. As shown in Fig. 4, the relative expression of FGFR3
was significantly downregulated in DU145 and PC-3 cell lines
following overexpression of miR-99a (P<0.05). These results
indicated that miR-99a reduced protein levels of FGFR3 in prostate
cancer cells.
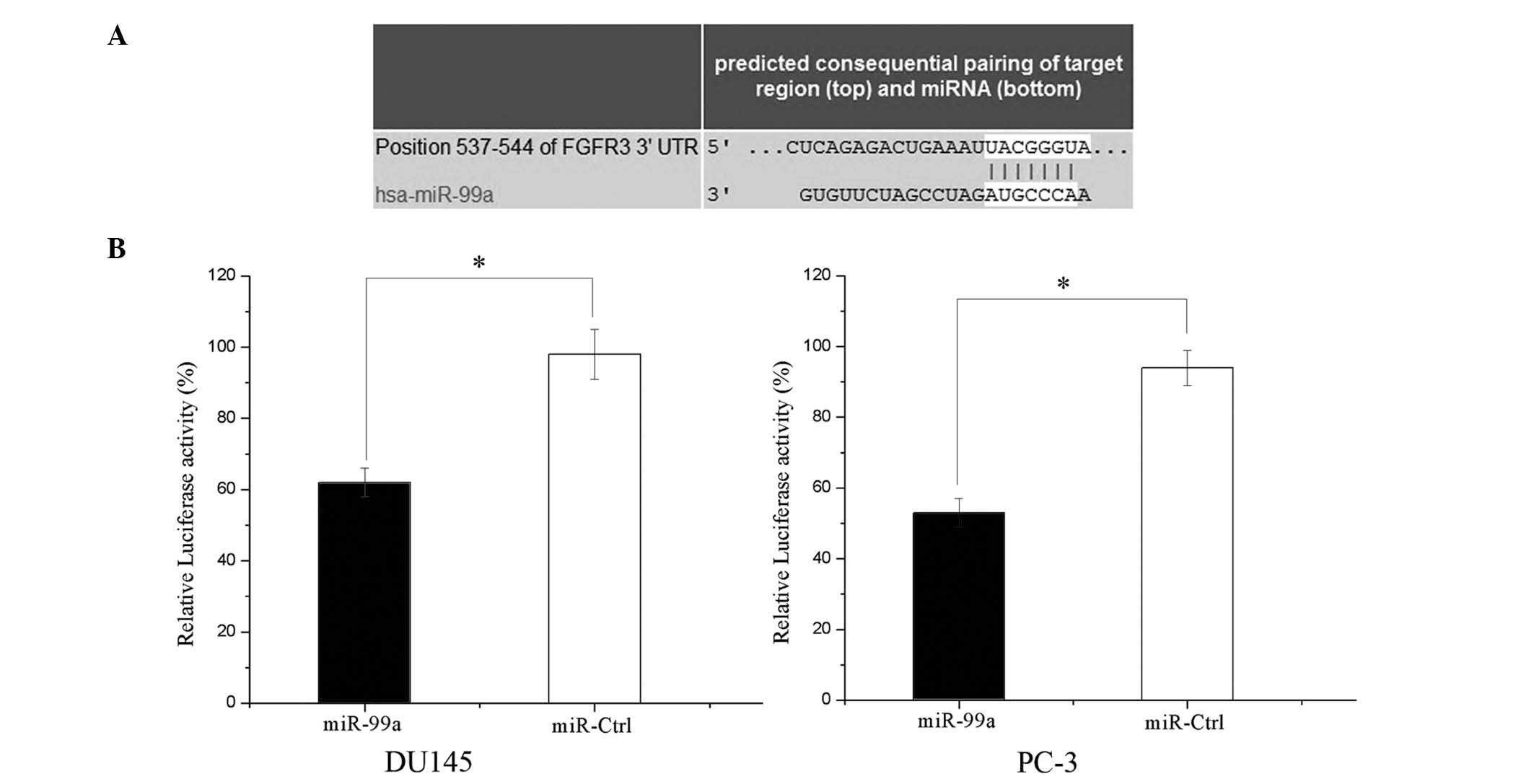
FGFR3 is a direct target gene of miR-99a
in prostate cancer
In order to determine whether miR-99a targets the
FGFR3 3′-UTR, TARGETSCAN 5.2 and PICTAR were used to assess the
complementarity of miR-99a to the FGFR3 3′-UTR. The results
revealed that FGFR3 mRNA contained a miR-99a seven-nucleotide seed
match at position 537-544 of the FGFR3 3′-UTR (Fig. 5A).
Luciferase reporter assays were performed to
investigate whether FGFR3 is a target of miR-99a. As shown in
Fig. 5B, overexpression of miR-99a
suppressed FGFR3 3′UTR-luciferase activity by 36% in DU145 cells
and by 43% in PC-3 cells (P<0.05). This therefore implied that
FGFR3 may be a direct target of miR-99a in vitro.
Discussion
miRNAs are major regulatory molecules that govern
numerous essential cellular functions, including proliferation,
differentiation, cell cycle control and apoptosis (19).
Dysregulation of miRNAs has been reported to result
in dysregulated gene expression of oncogenes and tumor suppressor
genes, subsequently leading to human diseases such as cancer
(15). The miR-99 family of miRNAs
is comprised of three members, miR-99a, miR-99b and miR-100; the
mature sequences of these three members are located on different
chromosomes (20). miR-99a is
transcribed from 21q21, a region that has been reported to be
commonly deleted in human lung cancers (21). The upregulation of miR-99a has also
been demonstrated in childhood acute myeloid leukemia (22). However, the role of miR-99a in
various malignant diseases remains to be elucidated. Previous
studies have reported that the downregulation of miR-99a
contributed to tumorigenesis of several types of cancer, including
prostate, bladder, hepatocellular and ovarian carcinoma, squamous
cell carcinoma of the tongue, childhood adrenocortical tumors as
well as lung cancer (17). The
controversial effects of miR-99a can in part be explained by miRNAs
binding to numerous 3′UTRs with complementary sites and therefore
having multiple downstream target proteins. The role of microRNAs
in cancer may be due to target proteins that have a tumor-promoting
or tumor-inhibiting function.
miR-99a expression was reported to correlate with
the survival rates of hepatocellular carcinoma patients; the
upregulation of miR-99a was found to suppress growth of
hepatocellular carcinoma cells via targeting the downstream
proteins insulin-like growth factor 1 and mTOR (23). In renal cell carcinoma, it was
demonstrated that downregulation of miR-99a resulted in increased
tumorigenicity through targeting of the mTOR pathway; conversely,
following the upregulation of miR-99a, tumorigenicity of renal cell
carcinoma cell lines was suppressed in vitro and in
vivo (17). In prostate
cancer, miR-99a suppressed the expression of prostate-specific
antigen and may be used as an indicator of the progression of
prostate cancer (18). This
therefore implied that the upregulation of miRNA-99a or the
production of a synthetic analogue may provide effective
therapeutic targets for the treatment of tumors that result from
specific oncogene activation or overexpression.
The results of the present study indicated that
miR-99a may function as a tumor suppressor via the direct targeting
of FGFR3 in prostate cancer. Transfection of miR-99a into prostate
cancer cell lines resulted in decreased cell viability and colony
formation ability as well as reduced cell migration and invasion.
This therefore suggested that miR-99a may have a potential
therapeutic role through regulation of the oncogenic FGFR3 in
prostate cancer patients.
The fibroblast growth factor (FGF) gene family
consists of >19 genes encoding associated polypeptide mitogens.
FGFs interact with a family of four distinct, high-affinity
tyrosine kinase receptors, designated FGFR1-4 (24). FGFRs are composed of an
extracellular domain that consists of three immunoglobulin-like
domains and an intracellular tyrosine kinase domain. The affinity
of FGF binding varies dependent on the FGFR (25); activation of FGFRs triggers
different responses in different cell types, and these responses
include proliferation, migration and differentiation as well as the
inhibition of proliferation and/or cell death (26). Therefore, the role of deregulated
FGF signaling in carcinogenesis has been the focus of numerous
studies.
The family of FGFs and FGFRs are important in
prostate organogenesis as well as the pathogenesis of prostate
cancer (27). FGFR3 has been
reported to have a major role in numerous types of cancer. The
extracellular portion of FGFR3 interacts with FGF3, inducing trans
autophosphorylation at the tyrosine residues of the cytoplasmic
domain and subsequently stimulating intrinsic catalytic activity
and activation of downstream signaling pathways (28). Several of these FGFR3-stimulated
signal transduction pathways have been implicated in oncogenesis,
including the Ras/extracellular-signal-regulated
kinase/mitogen-activated protein kinase, the phospholipase
Cc/protein kinase C, phosphatidylinositol 3-kinase, and the signal
transducers and activators of transcription pathways (29). FGFR signaling has also been
reported to be involved in the activation of nuclear factor
kappa-light-chain-enhancer of activated B cells (30,31),
the dysregulated activation of which is prevalent in human cancer
(32,33) and closely correlates with cancer
hallmarks (34).
The overexpression of FGFR3 typically occurs through
mutations within its extracellular and transmembrane domains as
well as through overexpression of the wild type receptor (35). Hernández et al (36) reported that the highest recorded
number of FGFR3 mutations was in patients with additional tumors as
well as prostate cancer, i.e. bladder, skin, and colon tumors. In
bladder cancers, mutations occurred predominantly in non-muscle
invasive disease and significantly less frequently in
muscle-invasive lesions. This therefore indicated that these
mutations may be associated with a favorable course of the disease
in non-invasive papillary bladder cancer (37). In prostate cancer, FGFR3 is
expressed in the majority of benign prostatic hyperplasia (BPH) and
prostate cancer. The expression pattern was reported to be
predominantly epithelial with significant nuclear signals in BPH as
well as the malignant prostate dysplasia (38). Hernández et al (36) reported an association between FGFR3
mutation frequency in low-grade prostate cancer and prostate cancer
in patients with other coinciding malignancies. These results
therefore indicated that FGFR3 may be a potential target for
inhibition in prostate cancer.
The results of the present study revealed that
miR-99a suppressed prostate cancer cell proliferation, colony
formation ability, migration and invasion via the downregulation of
FGFR3, therefore suggesting miR-99a and FGFR3 as targets of
therapeutic drugs for prostate cancer.
In conclusion, to the best of our knowledge, the
present study was the first to report that miR-99a reduced cell
proliferation, colony formation ability, migration and invasion
through regulation of FGFR3 in prostate cancer. The identification
of candidate target genes of miR-99a may help to elucidate the
carcinogenic mechanisms involved in prostate cancer. The results of
the present study have promising therapeutic implications and
therefore provide the basis for further studies for the treatment
of prostate cancer. Future studies are required to address whether
the potential of miR-99a may be fully realized in cancer treatment.
If so, these may provide beneficial results for treatment of
prostate cancer.
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
McGoldrick CA, Jiang YL, Paromov V,
Brannon M, Krishnan K and Stone WL: Identification of oxidized
protein hydrolase as a potential prodrug target in prostate cancer.
BMC Cancer. 14:772014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Damber JE and Aus G: Prostate cancer.
Lancet. 371:1710–1721. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shavers VL, Underwood W and Moser RP:
Race/ethnicity and the perception of the risk of developing
prostate cancer. Am J Prev Med. 37:64–67. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ma S, Chan YP, Kwan PS, et al:
MicroRNA-616 induces androgen-independent growth of prostate cancer
cells by suppressing expression of tissue factor pathway inhibitor
TFPI-2. Cancer Res. 71:583–592. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Filipowicz W, Bhattacharyya SN and
Sonenberg N: Mechanisms of post-transcriptional regulation by
microRNAs: are the answers in sight? Nat Rev Genet. 9:102–114.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bartel DP: MicroRNAs: target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Esquela-Kerscher A and Slack FJ: Oncomirs
- microRNAs with a role in cancer. Nature Rev Cancer. 6:259–269.
2006. View
Article : Google Scholar
|
|
11
|
Lu J, Getz G, Miska EA, et al: MicroRNA
expression profiles classify human cancers. Nature. 435:834–838.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Schaefer A, Stephan C, Busch J, Yousef GM
and Jung K: Diagnostic, prognostic and therapeutic implications of
microRNAs in urologic tumors. Nat Rev Urol. 7:286–297. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fendler A, Jung M, Stephan C, et al:
miRNAs can predict prostate cancer biochemical relapse and are
involved in tumor progression. Int J Oncol. 39:1183–1192.
2011.PubMed/NCBI
|
|
14
|
Schaefer A, Jung M, Kristiansen G, et al:
MicroRNAs and cancer: current state and future perspectives in
urologic oncology. Urol Oncol. 28:4–13. 2010. View Article : Google Scholar
|
|
15
|
Zhou Y, Wu D, Tao J, Qu P, Zhou Z and Hou
J: MicroRNA-133 inhibits cell proliferation, migration and invasion
by targeting epidermal growth factor receptor and its downstream
effector proteins in bladder cancer. Scand J Urol. 47:423–432.
2013. View Article : Google Scholar
|
|
16
|
Ventura A and Jacks T: MicroRNAs and
cancer: short RNAs go a long way. Cell. 136:586–591. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cui L, Zhou H, Zhao H, et al: MicroRNA-99a
induces G1-phase cell cycle arrest and suppresses tumorigenicity in
renal cell carcinoma. BMC Cancer. 12:5462012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sun D, Lee YS, Malhotra A, et al: miR-99
family of MicroRNAs suppresses the expression of prostate-specific
antigen and prostate cancer cell proliferation. Cancer Res.
71:1313–1324. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lerman G, Avivi C, Mardoukh C, et al:
MiRNA expression in psoriatic skin: reciprocal regulation of
hsa-miR-99a and IGF-1R. PloS One. 6:e209162011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sun J, Chen Z, Tan X, et al:
MicroRNA-99a/100 promotes apoptosis by targeting mTOR in human
esophageal squamous cell carcinoma. Med Oncol. 30:4112013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nagayama K, Kohno T, Sato M, Arai Y, Minna
JD and Yokota J: Homozygous deletion scanning of the lung cancer
genome at a 100-kb resolution. Genes Chromosomes Cancer.
46:1000–1010. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li XJ, Luo XQ, Han BW, Duan FT, Wei PP and
Chen YQ: MicroRNA-100/99a, deregulated in acute lymphoblastic
leukaemia, suppress proliferation and promote apoptosis by
regulating the FKBP51 and IGF1R/mTOR signalling pathways. Br J
Cancer. 109:2189–2198. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li D, Liu X, Lin L, et al: MicroRNA-99a
inhibits hepatocellular carcinoma growth and correlates with
prognosis of patients with hepatocellular carcinoma. J Biol Chem.
286:36677–36685. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Johnson DE and Williams LT: Structural and
functional diversity in the FGF receptor multigene family. Adv
Cancer Res. 60:1–41. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kwabi-Addo B, Ropiquet F, Giri D and
Ittmann M: Alternative splicing of fibroblast growth factor
receptors in human prostate cancer. Prostate. 46:163–172. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lafitte M, Moranvillier I, Garcia S, et
al: FGFR3 has tumor suppressor properties in cells with epithelial
phenotype. Mol Cancer. 12:832013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Dorkin TJ, Robinson MC, Marsh C, Neal DE
and Leung HY: aFGF immunoreactivity in prostate cancer and its
co-localization with bFGF and FGF8. J Pathol. 189:564–569. 1999.
View Article : Google Scholar
|
|
28
|
Azab AK, Azab F, Quang P, et al: FGFR3 is
overexpressed waldenstrom macroglobulinemia and its inhibition by
Dovitinib induces apoptosis and overcomes stroma-induced
proliferation. Clin Cancer Res. 17:4389–4399. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wesche J, Haglund K and Haugsten EM:
Fibroblast growth factors and their receptors in cancer. BiochemJ.
437:199–213. 2011. View Article : Google Scholar
|
|
30
|
Ettelaie C, Fountain D, Collier ME, Elkeeb
AM, Xiao YP and Maraveyas A: Low molecular weight heparin
downregulates tissue factor expression and activity by modulating
growth factor receptor-mediated induction of nuclear factor-κB.
Biochim Biophys Acta. 1812:1591–1600. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lungu G, Covaleda L, Mendes O,
Martini-Stoica H and Stoica G: FGF-1-induced matrix
metalloproteinase-9 expression in breast cancer cells is mediated
by increased activities of NF-kappaB and activating protein-1. Mol
Carcinog. 47:424–435. 2008. View
Article : Google Scholar
|
|
32
|
Chaturvedi MM, Sung B, Yadav VR, Kannappan
R and Aggarwal BB: NF-κB addiction and its role in cancer: ‘one
size does not fit all’. Oncogene. 30:1615–1630. 2011. View Article : Google Scholar :
|
|
33
|
Perkins ND: The diverse and complex roles
of NF-κB subunits in cancer. Nat Rev Cancer. 12:121–132.
2012.PubMed/NCBI
|
|
34
|
Hanahan D and Weinberg RA: The hallmarks
of cancer. Cell. 100:57–70. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tomlinson DC, Hurst CD and Knowles MA:
Knockdown by shRNA identifies S249C mutant FGFR3 as a potential
therapeutic target in bladder cancer. Oncogene. 26:5889–5899. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hernández S, de Muga S, Agell L, et al:
FGFR3 mutations in prostate cancer: association with low-grade
tumors. Mod Pathol. 22:848–856. 2009.PubMed/NCBI
|
|
37
|
Hernández S, López-Knowles E, Lloreta J,
et al: Prospective study of FGFR3 mutations as a prognostic factor
in nonmuscle invasive urothelial bladder carcinomas. J Clin Oncol.
24:3664–3671. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Gowardhan B, Douglas DA, Mathers ME, McKie
AB, McCracken SR, Robson CN and Leung HY: Evaluation of the
fibroblast growth factor system as a potential target for therapy
in human prostate cancer. Br J Cancer. 92:320–327. 2005.PubMed/NCBI
|