Introduction
MicroRNAs (miRs) are a novel class of small,
non-coding, single-stranded RNA molecules (~22 nucleotides). They
mainly function via base pairing with the 3′-untranslated region
(3′-UTR) of their target mRNAs, thus inhibiting their translation
or repressing their levels (1–4). The
present study attempted to study the regulation of the 5′UTR and
3′-UTR of insulin-like growth factor-1 receptor (IGF-1R),
insulin-like growth factor binding protein-3 (IGFBP-3) and IGF-II,
which are members of the IGF axis and have a major role in
hepatocarcinogenesis, with the metastamiRs miR-96-5p and
miR-182-5p. miR-96 and miR-182 were previously shown to have a role
in hepatocellular carcinoma (HCC) (5–11).
They belong to the same family (miR-96/182/183 family) and are
members of a miRNA cluster that resides in the intergenic region
between two protein-coding genes in the chromosomal locus (7q31–34)
that is frequently amplified in HCC (5,6).
Gradual increase in miR-96 expression levels was linked to disease
progression from normal liver tissues to HCC (6–9).
miR-96 was identified as the most strongly associated HCC
recurrence-associated miR in non-tumor tissues and its expression
levels was found to be correlated with the invasive and metastatic
potential of HCC cells (7,10). Likewise, miR-182 was significantly
upregulated in metastatic HCC tissues compared with that in paired
normal tissues. This observed upregulation was significantly
associated with intrahepatic metastasis, early recurrence and
correlated with the HCC tumor grade (11). However, the expression status of
miR-96 and miR-182 in non-metastatic HCC as well as the underlying
mechanism of inducing carcinogenesis has not been investigated
previously, to the best of our knowledge. Therefore, the present
study aimed to explore their impact on the highly conserved IGF
tumorigenic pathway.
In HCC tissues, the expression of IGF-II ligand
protein was shown to be significantly higher than that in normal
liver tissues (12). Similarly,
the membrane-bound receptor IGF-1R is overexpressed in various HCC
cell lines (13). The expression
of IGFBP-3, which modulates the bioavailability of IGF-II ligand,
was found to be significantly downregulated in HCC tissues
(14). This reduced IGFBP-3
expression may increase the availability of biologically active
IGF-II potentiating the proliferative effects (15). Therefore, the present study aimed
to explore the impact of the single clustered miRNAs on their
downstream targets.
To achieve this aim, screening for miR-96 and
miR-182 in non-metastatic HCC tissues was performed followed by
manipulation of their expression in the HCC cell line HuH-7 in an
attempt to reveal their impact on the crucial members of the IGF
axis (IGF-1R, IGFBP-3 and IGF-II).
Materials and methods
Study patients
The present study included 22 HCC patients who
underwent liver transplant surgery at the Kasr El Einy Hospital,
Cairo University, Egypt. Ten liver biopsy specimens were obtained
from healthy donors during transplantation. Healthy donors were
non-diabetic, non-hypertensive and negative for hepatitis B and C
viruses. All participants gave their written informed consent.
The study was approved by the ethical review
committee of Cairo University (Cairo, Egypt). The institutional
ethics committees approving the protocol of the present study
comply with the principles set forth in the international reports
and guidelines as stated in the Helsinki Declaration and the
International Ethical Guidelines For Biomedical Research Involving
Human Subjects issued by the Council For International
Organizations of Medical Sciences (CIOMS).
All patients were non-metastatic with no
extrahepatic manifestations and no vascular invasion. Most of the
patients (68.2%) had more than one focal lesion as indicated in the
pathology report and were subjected to clinical assessment as shown
in Table I.
 | Table INumber/size of focal lesions according
to Milan criteria. |
Table I
Number/size of focal lesions according
to Milan criteria.
| Patient ID | Focal lesions
(n) | Size of focal lesions
(cm) |
|---|
| 1 | 3 | 1.5, 1, 1 |
| 2 | 1 | 2.5 |
| 3 | 3 | 2, 2.5, 3 |
| 4 | 3 | 2, 2, 3.5 |
| 5 | 1 | 1.5×2 |
| 6 | 3 | 3×4, 1, 1 |
| 7 | 1 | 4 |
| 8 | 3 | 4, 1, 1 |
| 9 | 3 | 1, 1, 1.5 |
| 10 | 1 | 2.5 |
| 11 | 2 | 1, 1.7 |
| 12 | 3 | 1, 1, 1 |
| 13 | 1 | 3 |
| 14 | 3 | 3, 1.5, 2 |
| 15 | 3 | 1, 1, 4 |
| 16 | 2 | 3, 1.5 |
| 17 | 2 | 1.5, 3 |
| 18 | 3 | 2.5, 2.5, 1.5 |
| 19 | 3 | 1.5, 1, 1 |
| 20 | 1 | 2 |
| 21 | 1 | 1.5 |
| 22 | 3 | 3, 2.5, 1 |
Cell culture
HuH-7 cells were maintained in Dulbecco’s modified
Eagle’s medium (DMEM) supplemented with 4.5 g/l glucose, 4 mmol/l
L-glutamine, 10% fetal bovine serum and mycozap at 1:500 dilution
(all Lonza, Basel, Switzerland) at 37˚C in a 5% CO2
atmosphere.
Transfection of oligonucleotides
For examining the effect of miR-96-5p and miR-182-5p
on IGF-1R, IGF-II and IGFBP-3 transcript expression, HuH-7 cells
were transfected with mimics and inhibitors of miR-96-5p and
miR-182-5p (Qiagen, Hilden, Germany). All transfection experiments
were performed in triplicate using HiPerfect Transfection Reagent
(Qiagen) according to the manufacturer’s instructions and
experiments were repeated three times. Cells that were only exposed
to transfection reagent were designated as mock cells, cells
transfected with miR-96-5p or miR-182-5p mimics were designated as
miR-96-5p cells or miR-182-5p cells, respectively, and cells
transfected with miR-96-5p or miR-182-5p inhibitors were designated
as anti-miR-96-5p or anti-miR-182-5p cells, respectively.
Total RNA extraction from liver biopsies
and HCC cells
mRNAs and miRs were extracted from liver biopsy
specimens and HCC cells. Fresh liver samples (HCC and healthy
tissues) were collected during surgery and were snap frozen in
liquid nitrogen. The specimens were manually pulverized in liquid
nitrogen and ~100 mg tissue powder was used for large and small RNA
extraction using the mirVana miRNA Isolation kit (Ambion Life
Technologies, Waltham, MA, USA) according to the manufacturer’s
instructions. HCC cells were harvested 48 h after transfection
according to the HiPerfect Transfection Reagent protocol; 150 ng
oligonucleotides were used for HuH-7-cell transfection in a
six-well plate.
miR and mRNA quantification
The extracted miRs were reverse transcribed into
single stranded complementary DNA (cDNA) using a TaqMan_MicroRNA
Reverse Transcription kit (Applied Biosystems Life Technologies)
using specific primers for Homo sapiens (hsa)-miR-96-5p,
hsa-miR-182-5p as well as RNU6B for normalization. IGF-1R, IGF-II
and IGFBP-3 mRNAs were reverse transcribed into cDNA using the
High-capacity cDNA Reverse Transcription kit (Applied Biosystems)
according to the manufacturer’s instructions. Relative expression
of miR-96-5p, miR-182-5p and RNU6B as well as IGF-1R, IGF-II,
IGFBP-3 and β-2-microglobulin (β2MG; for normalization) was
quantified using TaqMan Real-Time Q-PCR (assay IDs, 000186, 002334,
001093, Hs00609566_m1, Hs04188276_m1, Hs00365742_g1 and 4310886E,
respectively; Applied Biosystems) using a StepOne™ PCR system
(Applied Biosystems). Relative expression was calculated using the
2−ΔΔCt method. All PCR reactions including controls were
run in duplicate.
Bioinformatics
Putative downstream targets for miR-96-5p and
miR-182-5p were predicted using bioinformatics algorithms. 3′-UTR
target regions were predicted using microrna.org
(www.microrna.org), Diana Lab (www.diana.cslab.ece.ntua.gr/) and TargetScan
(www.targetscan.org/), while 5′-UTR
target regions were predicted using bibiserv software version 2
(Bielefeld University Bioinformatics Server; http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/submission.html).
Statistical analysis
The Mann-Whitney U test (non-parametric and
two-tailed) was performed to compare gene expression between two
groups. Pearson’s method of parametric statistical correlation was
used for correlation analysis, with a Pearson r>0.3 indicating a
significantly positive correlation and a Pearson r<-0.3
indicating a significantly negative correlation. A P-value <0.05
was considered to indicate a statistically significant difference
between values. P-values are indicated as ***P<0.001,
**P<0.01, *P<0.05 and ns, statistically
not significant. All the data were statistically analyzed using
GraphPad Prism 5.00 software (GraphPad, La Jolla, CA, USA).
Results
miR-96-5p and miR-182-5p screening in
liver tissues and HCC cell lines
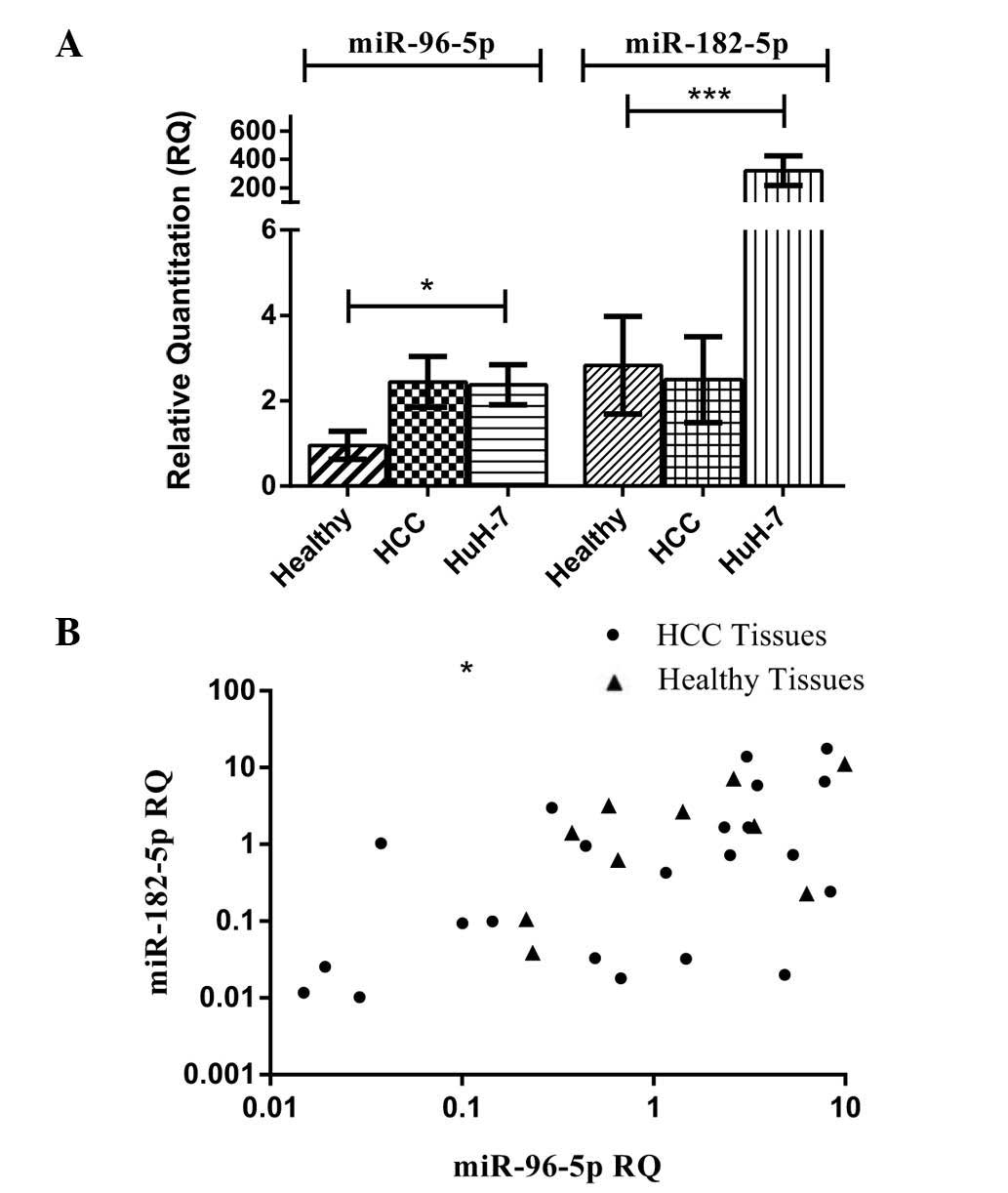
The relative expression of miR-96 and miR-182 was
assessed in the HCC and healthy liver tissues groups, as well as in
a HCC cell line (HuH-7), and its expression was normalized to
RNU6B. Expression levels of miR-96 and miR-182 in all HCC biopsy
specimens (n=22) were similar to those in healthy tissues (P=0.5555
and P=0.1864, respectively). However, expression of miR-96 in HuH-7
cells was significantly higher than that in healthy tissues
(P=0.0225) (Fig. 1A). In a similar
manner, the expression of miR-182-5p in HuH-7 cells showed a
significantly higher expression compared to that in healthy tissues
and HCC tissues (P=0.0002) (Fig.
1A).
Correlation analysis between miR-96-5p
and miR-182-5p mRNA expression in HCC tissues and healthy
controls
miR-96-5p expression was quantified in all healthy
and HCC tissues and correlated to miR-182-5p expression in the same
patients and healthy controls. Using Pearson’s statistical method
of correlation, miR-96-5p expression was found to be directly
correlated with miR-182-5p expression in all HCC tissues studied,
as indicated by a Pearson correlation coefficient of r=0.5119
(P=0.0149, Fig. 1B). Similarly, in
healthy controls, the same direct correlation was observed with a
Pearson correlation coefficient of r=0.6708 (P=0.0337) (Fig. 1B).
Bioinformatics analysis
miR-96-5p and miR-182-5p accession numbers and
mature sequences were retrieved using miRBase database (http://www.mirbase.org/). The mature sequences of
miR-96-5p and miR-182-5p showed an identical seed sequence
(ACGGUU). Using different in silico algorithms for
predicting binding to the downstream targets at the 3′-UTR and
5′-UTR positions, miR-96 and miR-182 were shown to target IGF-1R,
IGFBP-3 and IGF-II transcripts.
According to microrna.org,
Diana Lab and TargetScan, it was shown that miR-96 and miR-182
targeted the 3′-UTR of IGF-1R at two binding sites by the same seed
sequence. In silico algorithms further predicted targeting
of the IGFBP-3 3′-UTR at one binding site by miR-96 and miR-182.
However, the two miRNAs were not predicted to target the 3′-UTR of
IGF-II (Table II).
 | Table IIPositions of binding sites of
hsa-miR-96-5p and hsa-miR-182-5p to UTRs of IGF-1R, IGFBP-3 and
IGF-II (binding sites’ positions: 5′-UTR as predicted by bibiserv
and 3′-UTR as predicted by microrna.org). |
Table II
Positions of binding sites of
hsa-miR-96-5p and hsa-miR-182-5p to UTRs of IGF-1R, IGFBP-3 and
IGF-II (binding sites’ positions: 5′-UTR as predicted by bibiserv
and 3′-UTR as predicted by microrna.org).
| miR | Position of 5′-UTR
target regions
| Position of 3′-UTR
target regions
|
|---|
| IGF-1R | IGFBP-3 | IGF-II | IGF-1R | IGFBP-3 | IGF-II |
|---|
| miR-96-5p | 17–29 | 65–88 | 630–652 | 564–592
5799–5827 | 41–63 | N/A |
| miR-182-5p | 14–21 | 65–78 | 600–627 | 564–592
5799–5827 | 40–62 | N/A |
However, miR-96 and miR-182 were shown to target the
5′-UTR of IGF-1R, IGFBP-3 and IGF-II according to bibiserv software
(Table II).
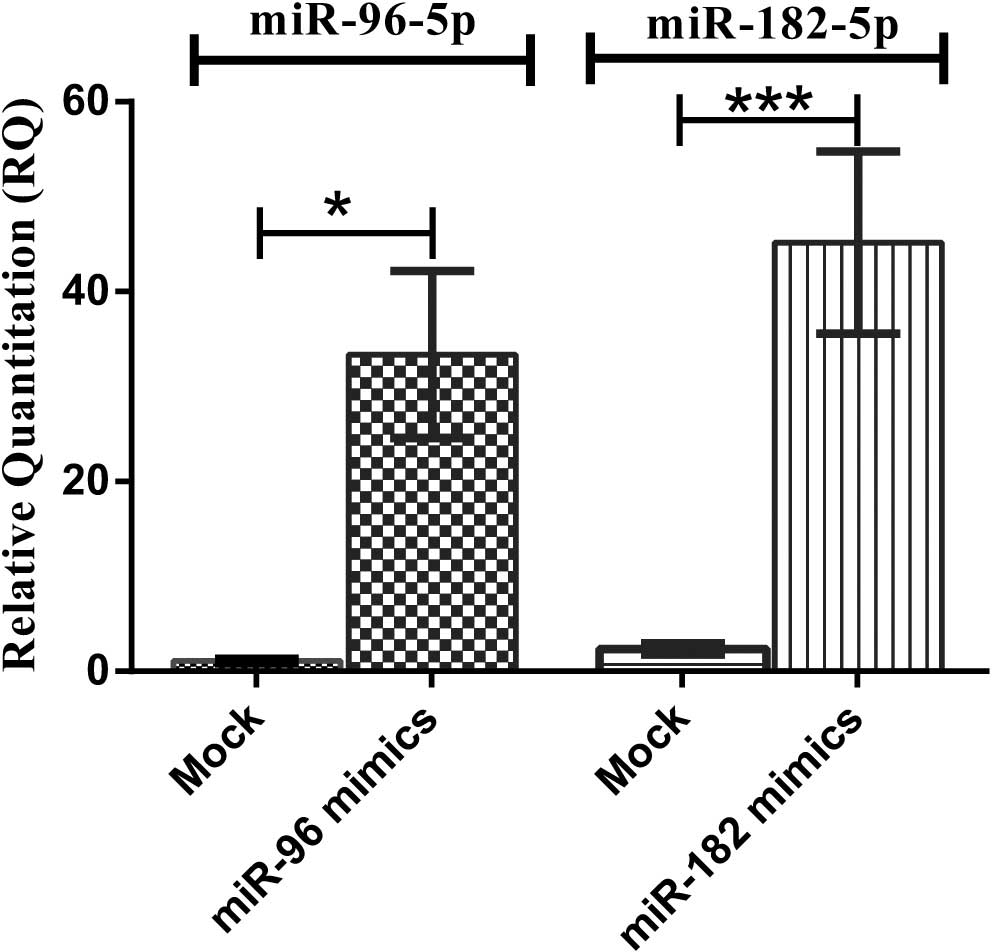
Transfection efficiency of miR-96-5p and
miR-182-5p oligonucleotides
HuH-7 cells transfected with miR-96 and miR-182
mimics showed upregulation of miR-96 and miR-182 by up to 47.18-and
98.59-fold, respectively (P=0.0294 and P<0.0001, respectively)
compared to mock-transfected cells (Fig. 2).
Impact of miR-96-5p and miR-182-5p on
IGF-1R
Transfection of HuH-7 cells with miR-96 mimics
resulted in a significant upregulation of IGF-1R mRNA levels
(P<0.0001) compared to those in the mock controls. Of note,
inhibitors of miR-96 in HuH-7 caused a significant decrease in
IGF-1R mRNA levels compared to those in mimic-transfected cells
(P<0.0001) (Fig. 3A). Of note,
transfection of HuH-7 cells with miR-182 mimics showed a
significant downregulation of IGF-1R mRNA levels compared to those
in mock controls (P=0.0267). However, inhibitors of miR-182 in
HuH-7 did not cause any significant changes in IGF-1R mRNA levels
compared to those in mock controls (P=0.1266) (Fig. 3A).
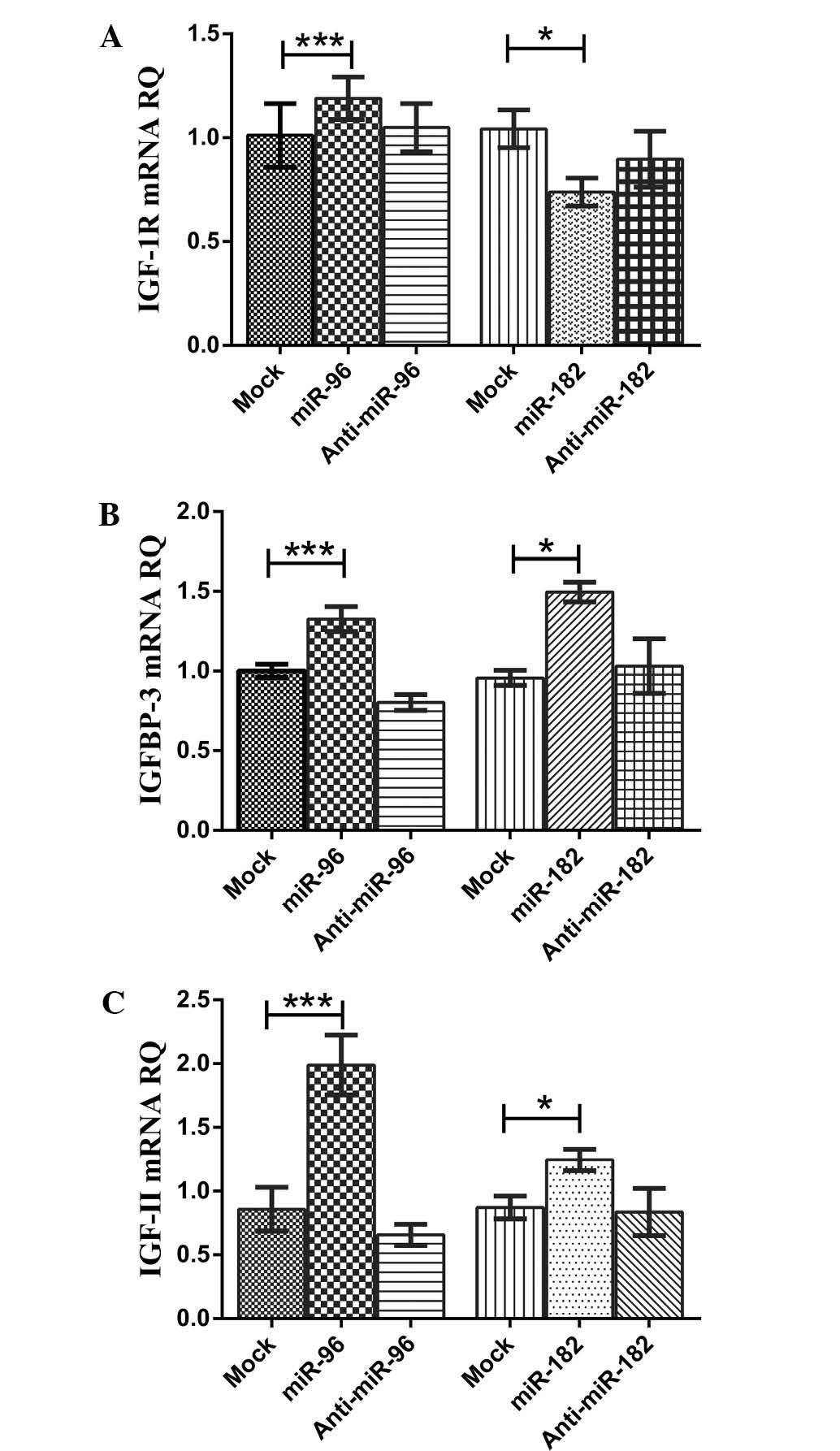 | Figure 3Impact of miR-96-5p and miR-182-5p on
IGF-1R, IGFBP-3 and IGF-II transcript expression. (A) Transfection
with mimics of miR-96-5p in HuH-7 cells resulted in a significant
upregulation of IGF-1R mRNA levels compared to those in
mock-transfected controls, while inhibitors of miR-96-5p caused a
significant downregulation in the expression of IGF-1R transcripts
compared to that in mimic-transfected cells. Mimicking of
miR-182-5p in HuH-7 cells resulted in a significant downregulation
of IGF-1R mRNA levels compared to those in mock-transfected
controls, while inhibitors of miR-182-5p did not significantly
change the expression of IGF-1R transcript levels compared to those
in mock-transfected cells. (B) Transfection of HuH-7 cells with
miR-96-5p mimics resulted in a significant upregulation of IGFBP-3
mRNA levels compared to those in mock-transfected controls.
However, inhibitors of miR-96-5p caused a significant
downregulation in the expression of IGFBP-3 transcript levels
compared with those in mock-and mimic-transfected cells.
Transfection of HuH-7 cells with miR-182-5p mimics resulted in a
significant upregulation of IGFBP-3 mRNA levels compared to those
in mock-transfected controls. However, inhibitors of miR-182-5p did
not cause any significant changes in the expression of IGFBP-3
transcript levels compared to those in mock-transfected cells. (C)
Transfection of HuH-7 cells with miR-96-5p mimics resulted in a
significant upregulation of IGF-II mRNA levels compared to those in
mock-transfected controls. Of note, inhibitors of miR-96-5p caused
a significant downregulation in the expression of IGF-II transcript
levels compared to those in mock-and mimic-transfected cells.
miR-182-5p mimics caused a significant upregulation of IGF-II mRNA
levels in HuH-7 cells compared to those in mock-transfected
controls. However, inhibitors of miR-182-5p did not cause any
significant change in the expression of IGF-II transcript levels
compared to those in mock-transfected cells. The Mann-Whitney U
test was performed. ***P<0.001,
*P<0.05. RQ, relative quantitation, IGFBP,
insulin-like growth factor binding protein; IGF-1R, insulin-like
growth factor-1 receptor; miR, microRNA. |
Impact of miR-96-5p and miR-182-5p on
IGFBP-3
Transfection of HuH-7 cells with miR-96 mimics
resulted in a significant upregulation of IGFBP-3 mRNA levels
(P<0.0001) compared to those in mock-transfected controls. While
inhibitors of miR-96 caused a significant downregulation of IGFBP-3
mRNA levels in HuH-7 cells compared to those in mock-transfected
controls (P<0.0001) and mimic-transfected cells (P<0.0001)
(Fig. 3B). Transfection of HuH-7
cells with miR-182 caused a significant upregulation of IGFBP-3
mRNA levels compared to those in mock-transfected controls
(P=0.0357). However, inhibitors of miR-182 did not cause any
significant change in IGFBP-3 mRNA levels in HuH-7 cells compared
to those in mock cells (P=0.7937) (Fig. 3B).
Impact of miR-96-5p and miR-182-5p on
IGF-II
Transfection of HuH-7 cells with miR-96 mimics
resulted in a significant upregulation of IGF-II mRNA levels
(P<0.0001) compared to those in mock-transfected controls. Of
note, inhibitors of miR-96 caused a significant downregulation of
IGF-II mRNA levels in HuH-7 cells compared to those in the
mock-transfected controls (P<0.0001) and mimic-transfected cells
(P<0.0001) (Fig. 3C).
Transfection of HuH-7 cells with miR-182 mimics caused a
significant upregulation of IGF-II mRNA levels compared to those in
mock-transfected controls (P=0.0317). Inhibitors of miR-182 did not
cause any significant changes in IGF-II mRNA levels in HuH-7 cells
compared with those in mock-transfected controls (P=0.4762)
(Fig. 3C).
Discussion
miR-96-5p and miR-182-5p, which belong to the same
family of single clustered miRs, were recently shown to be oncomiRs
that promote oncogenesis with an essential role in HCC progression
and metastasis (5,10,11,16).
To the best of our knowledge, the present study was
the first to perform expression profiling of miR-96 and miR-182 in
non-metastatic HCC tissues. The two miRNAs were found to have
similar expression levels in non-metastatic HCC tissues as compared
to that in healthy liver tissues. However, when screened in the
HuH-7 cell line, expression levels of the two miRNAs were
significantly upregulated. Alongside these results, miR-96 and
miR-182 were previously reported to be upregulated in metastatic
tissues (10,11).
Furthermore, there was a significant direct
correlation between the expression of miR-96 and miR-182 in 22 HCC
biopsy samples and in 10 healthy controls. This similar expression
pattern may be explained by the fact that they belong to the same
family (miR-96/182/183 family), which consorts with the findings of
a previous study where a positive correlation was observed between
miR-96, miR-182 and miR-183 expression in prostate cancer tissues
(17).
By performing bioinformatics analyses, miR-96-5p and
miR-182-5p were found to target the 5′-UTR and 3′-UTR of three
crucial IGF axis genes in HCC, namely IGF-1R, IGFBP-3 and IGF-II
transcripts. Therefore, the present study aimed at exploring the
impact of miR-96 and miR-182 on IGF axis members to understand
their role in HCC cancer progression.
The aberrant expression pattern of IGF axis members
in human HCC tissues with overexpression of IGF-1R and IGF-II and
downregulation of IGFBP-3 was reported to promote HCC progression
and metastasis (14,18,19).
Bioinformatics tools showed that miR-96-5p and
miR-182-5p have the same potential binding sites in two regions of
the 3′-UTR of IGF-1R mRNA. The two miRNAs were also predicted to
target two different regions in the 5′-UTR of IGF-1R mRNA. In spite
of the fact that miR-96 and miR-182 share an identical seed region
(UUGGCA, nucleotides 2–7), the IGF-1R expression levels obtained
upon transfection of HuH-7 cells with miR-96 and miR-182 mimics
were not the same: In HuH-7 cells, IGF-1R mRNA was upregulated by
miR-96 mimics and downregulated by miR-182 mimics compared to that
in their respective mock-transfected cells. These findings are in
agreement with those of another study, in which the glypican-3
(GPC3) 3′-UTR was predicted as a common target for the two miRs due
to exhibiting one miR-96/182 binding site. miR-96 downregulated
GPC3 expression by targeting its mRNA 3′-UTR; however, miR-182
failed to regulate it despite the seed similarity between the two
miRNAs (20).
The second potential target, IGFBP-3 mRNA, was
predicted to be targeted by the two miRs at the 5′-and 3′-UTRs.
IGFBP-3 mRNA was found to be significantly upregulated upon ectopic
expression of miR-96 in HuH-7 cells compared to that in
mock-transfected cells, and the same results were observed upon
transfection of HuH-7 cells with miR-182 mimics. This
overexpression of the downstream target was restored to normal
levels upon antagonizing miR-96 and miR-182 using specific
antagomirs in HuH-7 cells. The finding that the two miRNAs have the
same effect on their target IGFBP-3 mRNA consorts with another
study where the 3′-UTR of chloride intracellular channel 5, which
contains a single miR-96/-182 binding site, was found to be
directly targeted; however, in that case it was downregulated by
the two miRs at the mRNA and protein level (21). The observed induction of IGFBP-3
mRNA by miR-96 and miR-182 may be due to 5′-UTR binding leading to
stabilization of the mRNA, thus activating gene expression, which
is considered a novel mechanism of action for miRs (22). This inducing impact was previously
reported when miR-122 directly targeted two sites in the 5′-UTR of
Hepatitis C virus RNA stimulating its translation and increasing
its abundance (23–25).
Following the exploration of the effect of miR-96
and miR-182 on IGF-1R and IGFBP-3 mRNAs, the present study examined
the effect of the two miR mimics on IGF-II mRNA, which was
predicted to be targeted by both miRs at the 5′-UTR but not the
3′-UTR. Unexpectedly, it was found that IGF-II mRNA levels were
significantly upregulated following transfection of HuH-7 cells
with each of the miR mimics compared with those in their respective
mock-transfected cells. The same enhancing effect on target mRNA by
a miR was reported by a previous study on the receptor-interacting
protein 140 (RIP140) mRNA, where miR-346 targeted the 5′-UTR of the
RIP 140 transcript, leading to an upregulation of its protein
levels (26).
This expression variability of IGF axis components
upon miR-96 mimic-transfection raises a question about the
simultaneous increase of IGF-1R and IGF-II, which are mitogenic, as
well as IGFBP-3, which is anti-tumorigenic. This may be explained
as a compensatory mechanism where the upregulation of IGF-1R and
IGF-II may be neutralized by the upregulation of IGFBP-3,
counteracting the tumorigenic effect of the mitogens and mediating
their effect. This mediation by upregulated IGFBP-3 is realized
through its binding to the overexpressed IGF-II, which inhibits the
binding and consequent activation of IGF-1R and subsequent
downstream signaling. By contrast, miR-182 has a dual role where it
upregulates the mitogenic IGF-II and, as a compensatory mechanism,
upregulates IGFBP-3 and downregulates IGF-1R to regulate the
anti-mitogenic effect.
In conclusion, the present study showed for the
first time, to the best of our knowledge, that miR-96 and miR-182
have similar expression levels in non-metastatic HCC tissues and in
healthy controls, contradicting previous studies reporting their
upregulation in metastatic tissues (10,11).
This highly implies that miR-96-5p and miR-182-5p are metastatic
miRs in HCC. The present study also shows for the first time, to
the best of our knowledge, the pleiotropic effect of miR-96-5p and
miR-182-5p in targeting various components of the oncogenic IGF
signaling pathway in HCC. Thus, these data provide the rationale
for potential targeting of miR-96-5p and miR-182-5p in HCC
therapy.
Acknowledgments
The authors acknowledge the patients and healthy
volunteers for their valuable participation in this study.
Furthermore, the authors would like to thank the Science and
Technology Development Fund (STDF; grant no. 4242) for funding this
work.
References
|
1
|
Baskerville S and Bartel DP: Microarray
profiling of microRNAs reveals frequent coexpression with
neighboring miRNAs and host genes. RNA. 11:241–247. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Guo H, Ingolia NT, Weissman JS and Bartel
DP: Mammalian microRNAs predominantly act to decrease target mRNA
levels. Nature. 466:835–840. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Selbach M, Schwanhausser B, Thierfelder N,
Fang Z, Khanin R and Rajewsky N: Widespread changes in protein
synthesis induced by microRNAs. Nature. 455:58–63. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gottwein E and Cullen BR: Viral and
cellular microRNAs as determinants of viral pathogenesis and
immunity. Cell Host Microbe. 3:375–387. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Segura MF, Hanniford D, Menendez S, et al:
Aberrant miR-182 expression promotes melanoma metastasis by
repressing FOXO3 and microphthalmia-associated transcription
factor. Proc Natl Acad Sci USA. 106:1814–1819. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang Y, Lee AT, Ma JZ, et al: Profiling
microRNA expression in hepatocellular carcinoma reveals
microRNA-224 up-regulation and apoptosis inhibitor-5 as a
microRNA-224-specific target. J Biol Chem. 283:13205–13215. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sato F, Hatano E, Kitamura K, et al:
MicroRNA profile predicts recurrence after resection in patients
with hepatocellular carcinoma within the milan criteria. PLoS One.
6:e164352011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ladeiro Y, Couchy G, Balabaud C, et al:
MicroRNA profiling in hepatocellular tumors is associated with
clinical features and oncogene/tumor suppressor gene mutations.
Hepatology. 47:1955–1963. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pineau P, Volinia S, McJunkin K, et al:
miR-221 overexpression contributes to liver tumorigenesis. Proc
Natl Acad Sci USA. 107:264–269. 2010. View Article : Google Scholar :
|
|
10
|
Chen RX, Xia YH, Xue TC and Ye SL:
Suppression of microRNA-96 expression inhibits the invasion of
hepatocellular carcinoma cells. Mol Med Rep. 5:800–804. 2012.
|
|
11
|
Wang J, Li J, Shen J, Wang C, Yang L and
Zhang X: MicroRNA-182 downregulates metastasis suppressor 1 and
contributes to metastasis of hepatocellular carcinoma. BMC Cancer.
12:2272012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dong ZZ, Yao DF, Wu W, et al: Correlation
between epigenetic alterations in the insulin growth factor-II gene
and hepatocellular carcinoma. Chin J Hepatol. 20:593–597. 2012.In
Chinese.
|
|
13
|
Scharf JG, Schmidt-Sandte W, Pahernik SA,
Ramadori G, Braulke T and Hartmann H: Characterization of the
insulin-like growth factor axis in a human hepatoma cell line
(PLC). Carcinogenesis. 19:2121–2128. 1998. View Article : Google Scholar
|
|
14
|
Gong Y, Cui L and Minuk GY: The expression
of insulin-like growth factor binding proteins in human
hepatocellular carcinoma. Mol Cell Biochem. 207:101–104. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gray SG, Eriksson T, Ekstrom C, et al:
Altered expression of members of the IGF-axis in hepatoblastomas.
Br J Cancer. 82:1561–1567. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xu D, He X, Chang Y, et al: Inhibition of
miR-96 expression reduces cell proliferation and clonogenicity of
HepG2 hepatoma cells. Oncol Rep. 29:653–661. 2013.
|
|
17
|
Yin Y, Li M, Li H, et al: Expressions of 6
microRNAs in prostate cancer. Natl J Androl. 16:599–605. 2010.In
Chinese.
|
|
18
|
Nussbaum T, Samarin J, Ehemann V, et al:
Autocrine insulin-like growth factor-II stimulation of tumor cell
migration is a progression step in human hepatocarcinogenesis.
Hepatology. 48:146–156. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Aleem E, Nehrbass D, Klimek F, Mayer D and
Bannasch P: Upregulation of the insulin receptor and type I
insulin-like growth factor receptor are early events in
hepatocarcinogenesis. Toxicol Pathol. 39:524–543. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jalvy-Delvaille S, Maurel M, Majo V, et
al: Molecular basis of differential target regulation by miR-96 and
miR-182: the Glypican-3 as a model. Nucleic Acids Res.
40:1356–1365. 2012. View Article : Google Scholar :
|
|
21
|
Gu C, Li X, Tan Q, Wang Z, Chen L and Liu
Y: MiR-183 family regulates chloride intracellular channel 5
expression in inner ear hair cells. Toxicol In Vitro. 27:486–491.
2013. View Article : Google Scholar
|
|
22
|
Henke JI, Goergen D, Zheng J, et al:
microRNA-122 stimulates translation of hepatitis C virus RNA. EMBO
J. 27:3300–3310. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Villanueva RA, Jangra RK, Yi M, Pyles R,
Bourne N and Lemon SM: miR-122 does not modulate the elongation
phase of hepatitis C virus RNA synthesis in isolated replicase
complexes. Antiviral Res. 88:119–123. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Norman KL and Sarnow P: Modulation of
hepatitis C virus RNA abundance and the isoprenoid biosynthesis
pathway by microRNA miR-122 involves distinct mechanisms. J Virol.
84:666–670. 2010. View Article : Google Scholar :
|
|
25
|
Jopling CL: Regulation of hepatitis C
virus by microRNA-122. Biochem Soc Trans. 36:1220–1223. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tsai NP, Lin YL and Wei LN: MicroRNA
mir-346 targets the 5′-untranslated region of receptor-interacting
protein 140 (RIP140) mRNA and up-regulates its protein expression.
Biochem J. 424:411–418. 2009. View Article : Google Scholar : PubMed/NCBI
|