Introduction
Lung cancer accounts for ~28% of cancer-associated
mortali-ties (1), the occurrence
of which is increasing worldwide. There are ~1.2 million novel
cases of lung cancer and ~1 million mortalities from lung cancer
each year (2). Lung cancer may be
subdivided into small cell lung carcinoma and non-small cell lung
carcinoma (NSCLC). The majority of lung cancer diagnoses are NSCLC
(3,4), which has a five-year survival rate of
~33% (5). At present, the standard
treatment for patients with resectable stage I to IIIA NSCLC is
surgical excision; however, the prognosis remains poor (6). In addition, chemotherapy with or
without surgery is not effective in the majority of cases;
therefore, it is essential to identify novel compounds, including
natural products, which may be employed for the treatment of lung
cancer.
Cantharidin (CTD) is a component of mylabris
(blister beetle), which has previously been used as a Traditional
Chinese Medicine (7). Previous
studies have reported that CTD induced cytotoxic effects in
leukemia stem cells (8) as well as
U937 (9), pancreatic cancer
(10), hepatocellular carcinoma
(11,12), colon cancer (13) and human lung cancer A549 (14) cells. In addition, CTD was found to
inhibit the activity of protein phosphatase 2A (PP2A) (9) and heat shock factor 1 (HSF1)
(15). Furthermore, it was shown
that CTD induced cell death in human colorectal cancer cells, which
was suggested to proceed through inhibiting the binding of heat
shock protein 70 (HSP70), B cell lymphoma 2-associated athanogene
domain 3 (BAG3) and HSF1 to promoters (15).
Genetic mutations in oncogenes and tumor suppressor
genes are present in cancer cells (16,17).
The development of cancer cells is well-known to be dependent on
oncogenes for tumor initiation and progression; this concept has
therefore been named oncogene addiction (18). Oncogenes are commonly used as
targets for drug-screening programs (19); however, other signaling pathways
have also been examined, such as the molecular chaperone pathway
(20). The present study aimed to
investigate the effect of CTD on the expression of key genes and
functional pathways of human H460 lung cancer cells using
complementary DNA microarray analysis. The results of the present
study showed that CTD affected DNA damage, the cell cycle and the
expression of apoptosis-associated genes in vitro.
Differentially expressed genes were then used to generate
interaction maps of signaling pathways. The epidermal growth factor
and vascular endothelial growth factor receptor pathways, provided
by the present study may be useful for the development of novel
molecular targeted therapies against lung cancer (21).
Materials and methods
Chemicals and reagents
Cantharidin (CTD), propidium iodide and dimethyl
sulfoxide (DMSO) were purchased from Sigma-Aldrich (St. Louis, MO,
USA). Minimum essential medium (MEM), fetal bovine serum (FBS),
L-glutamine and penicillin-streptomycin were purchased from
Gibco-BRL (Carlsbad, CA, USA). CTD was dissolved in DMSO and stored
at 20°C.
Lung cancer cell culture
The NCI-H460 human lung cancer cell line was
purchased from the Food Industry Research and Development Institute
(Hsinchu, Taiwan). Cells were grown in MEM containing 10% (v/v) FBS
as well as 100 U/ml penicillin and 100 μg/ml streptomycin in
a 37°C humidified incubator with 5% CO2. Cells were then
subcultured once they reached 80–90% confluence, as previously
described (22).
Complementary (c)DNA microarray
assay
H460 cells were placed on 12-well plates at a
density of 5×105 cells/well in 2 ml MEM with 10% (v/v)
FBS and 2 mM L-glutamine, as well as 100 U/ml penicillin and 100
μg/ml streptomycin for 24 h. Subsequently, cells were
treated with or without 10 μM CTD for a further 24 h. Cells
(3×106) were then harvested and washed twice with
phosphate-buffered saline (Gibco-BRL). Cells were lysed in
TRIzol® (Invitrogen Life Technologies, Carlsbad, CA,
USA) and total RNA was extracted using a Qiagen RNeasy Mini kit
(Qiagen, Valencia, CA, USA). RNA concentrations were determined
using a Qubit™ Fluorocytometer (Invitrogen Life Technologies).
Total RNA of CTD-treated and untreated H460 cells
was used for cDNA synthesis. Samples were hybridized using an
Affymetrix GeneChip Human Gene 1.0 ST array (Affymetrix, Santa
Clara, CA, USA) according to the manufacturer’s instructions.
Sample fluorescence was quantified by Asia BioInnovations Corp.
(Taipei, Taiwan), while data were analyzed using the Transcriptome
Analysis Console™ 2.0 Version 2.0.0.9. (Affymetrix) with default
robust multichip analysis parameters. A 2-fold change in gene
expression was used as the threshold to indicate an effect on
expression (7–10). An Oligo(dT) Maxime RT PreMix kit
(iNtRON Biotechnology, Gyeonggi-do, South Korea) was used to
reverse transcribe RNA into cDNA. The Affymetrix
GeneChip® Whole Transcript Sense Target (ST) Labeling
(cat. no. 900673; 30 Rxn; Affymetrix, Santa Clara, CA, USA) assay
is designed to generate amplified and biotinylated sense-strand DNA
targets from the entire expressed genome without bias. This assay
and associated reagents have been optimized specifically for use
with the GeneChip® ST arrays, and the probes on the
arrays have been selected to be distributed throughout the entire
length of each transcript. The gene list complete with Affymetrix
transcript identifiers, was uploaded from a spreadsheet onto
Metacore 5.0 software (GeneGo pathways analysis; http://www.genego.com). GeneGo recognizes the
Affymetrix identifiers and maps the gene to the MetaCore™ data
analysis suite, generating maps to describe common pathways or
molecular connections between genes in the list. Graphical
representations of the molecular associations between the genes
were generated using the GeneGo pathway analysis, based upon
processes exhibiting a significant association (P<0.05).
Gene ontology analysis
For detection of significantly over-represented GO
biological processes, the DAVID functional annotation clustering
tool (http://david.abcc.ncifcrf.gov) was
used (DAVID Bioinformatics Resources 6.7). Enrichment was
determined at the DAVID calculated Benjamini value <0.05. The
significance of the overexpression of individual genes was
determined using Student’s t-test.
Statistical analysis
Values are representative of three independent
experiments. Differences between control and CTD-experimental
groups are presented which >2-fold, where positive numbers
represent upregulation and negative numbers represent
downregulation.
Results
Upregulated and downregulated gene
expression in H460 cells exposed to CTD
H460 cells were incubated in the presence or absence
of 10 μM CTD in a 12-well plate for 24 h. Cells were then
harvested and following the extraction of total RNA, RNA
concentrations were determined and cDNA microarray analysis was
performed in order to determine the expression of genes. The
calculated upregulation and downregulation of gene expression, as
determined by the microarray, are shown in Tables I and II, respectively. As shown in Table I, the results indicated that in
CTD-treated H460 cells, 8 genes were upregulated >4-fold, 29
genes were upregulated >3–4-fold and 156 genes were upregulated
>2–3-fold compared with expression levels in the untreated
control cells. In addition, Table
II indicated that one gene was downregulated >4 fold, 14
genes were downregulated >3–4 fold and 150 genes were
downregulated >2–3 fold in H460 cells following exposure to CTD
compared with those in the untreated control cells. The results
presented in Table I demonstrated
that genes associated with DNA damage, including DN1T3 and GADD45A
were upregulated by 2.26-and 2.60-fold, respectively; in addition,
the expression of genes associated with the cell cycle progression
(check point proteins) were upregulated, including CCND2, CDKL3 and
RASA4, which were upregulated 2.72-, 2.19- and 2.72-fold,
respectively. Furthermore, the expression of apoptosis-associated
genes was upregulated, such as CARD6, which was upregulated
3.54-fold (Table I). By contrast,
the results presented in Table II
demonstrated that genes associated with DNA damage, cell cycle
progression and apoptosis were also downregulated, including DdiT4,
CDC42EP3 and STAT2, respectively. These genes were found to be
downregulated 3.14-, 2.16 and 2.04-fold, respectively (Table II). Overall, cDNA microarray
analysis of H460 cells following treatment with CTD demonstrated
that CTD induced the differential expression of numerous genes
associated with DNA damage, cell cycle progression and
apoptosis.
 | Table IUpregulation of gene expression in
catharidine-treated NCI-H460 cells. |
Table I
Upregulation of gene expression in
catharidine-treated NCI-H460 cells.
| Probe set ID | Fold change | Gene symbol | Gene description | mRNA accession
no. |
|---|
| 8108370 | 16.50 | EGR1 | Early growth response
1 | NM_001964 |
| 8012949 | 13.05 | CDRT1 | CMT1A duplicated
region transcript 1 | NM_006382 |
| 8012951 | 10.85 | CDRT1 | CMT1A duplicated
region transcript 1 | NM_006382 |
| 7916609 | 8.05 | JUN | Jun oncogene | NM_002228 |
| 7977075 | 6.45 | SNORA28 | Small nucleolar RNA,
H/ACA box 28 | NR_002964 |
| 8041168 | 5.54 | SNORD53 | Small nucleolar RNA,
C/D box 53 | NR_002741 |
| 7982084 | 5.06 | SNORD115-11 | Small nucleolar RNA,
C/D box 115-11 | NR_003303 |
| 8097991 | 4.27 | TDO2 | Tryptophan
2,3-dioxygenase | NM_005651 |
| 8114468 | 3.99 | SNORD63 | Small nucleolar RNA,
C/D box 63 | NR_002913 |
| 8158862 | 3.87 | SNORD62A | Small nucleolar
RNA, C/D box 62A | NR_002914 |
| 8158864 | 3.87 | SNORD62A | Small nucleolar
RNA, C/D box 62A | NR_002914 |
| 8007420 | 3.85 | AOC3 | Amine oxidase,
copper-containing 3 (vascular adhesion protein 1) | NM_003734 |
| 7975779 | 3.81 | FOS | FBJ murine
osteosarcoma viral oncogene homolog | NM_005252 |
| 8001746 | 3.74 | SNORA46 | Small nucleolar
RNA, H/ACA box 46 | NR_002978 |
| 7914322 | 3.70 | SNORD103A | Small nucleolar
RNA, C/D box 103A | NR_004054 |
| 7914324 | 3.70 | SNORD103A | Small nucleolar
RNA, C/D box 103A | NR_004054 |
| 793342 | 3.69 | PTPN20A | Protein tyrosine
phosphatase, non-receptor type 2 | NR_001042389 |
| 7918467 | 3.65 | Clorf103 | Chromosome 1 open
reading frame 103 | NM_018372 |
| 8053797 | 3.60 | LOC400986 | Protein
immuno-reactive with anti-parathyroid hormone polyclona |
ENST00000456556 |
| 8156848 | 3.59 | NR4A3 | Nuclear receptor
subfamily 4, group A, member 3 | NM_006981 |
| 8005483 | 3.56 | FBXW10 | F-box and WD repeat
domain-containing 10 | NM_031456 |
| 8105077 | 3.54 | CARD6 | Caspase recruitment
domain family, member 6 | NM_032587 |
| 8139840 | 3.48 | ERV3 | Endogenous
retroviral sequence 3 (includes zinc) | NM_001007253 |
| 8030831 | 3.45 | ZNF175 | Zinc finger protein
175 | NM_007147 |
| 7958200 | 3.45 | EID3 | EP300-interacting
inhibitor of differentiation 3 | NM_001008394 |
| 7936637 | 3.44 | SNORA19 | Small nucleolar
RNA, H/ACA box 19 | NR_002917 |
| 8107353 | 3.43 | ZRSR1 | Zinc finger (CCCH
type), RNA-binding motif and serine/arginine rich 1 | BC104811 |
| 8173600 | 3.26 | NAP1L2 | Nucleosome assembly
protein 1-like 2 | NM_021963 |
| 8126853 | 3.25 | C6orf138 | Chromosome 6 open
reading frame 138 | NM_001013732 |
| 8025301 | 3.20 | CD209 | CD209 molecule | NM_021155 |
| 7923119 | 3.17 | ZBTB41 | Zinc finger and BTB
domain-containing 41 | NM_194314 |
| 7985555 | 3.11 | EFTUD1 | Elongation factor
Tu guanine triphosphate binding domain-containing | NM_024580 |
| 7952986 | 3.09 | HSN2 | Hereditary sensory
neuropathy, type II | NM_213655 |
| 8114572 | 3.08 | HBEGF | Heparin-binding
epidermal growth factor-like growth factor | NM_001945 |
| 8049540 | 3.05 | LRRFIP1 | Leucine-rich repeat
(in FLII) interacting protein 1 | NM_001137550 |
| 8047926 | 3.03 | MAP2 |
Microtubule-associated protein 2 | NM_002374 |
| 7954382 | 3.02 | PYROXD1 | Pyridine
nucleotide-disulphide oxidoreductase domain 1 | NM_024854 |
| 7957260 | 2.99 | GLIPR1 | GLI
pathogenesis-related 1 | NM_006851 |
| 8054054 | 2.97 | ANKRD36B | Ankryin repeat
domain 36B | NM_025190 |
| 7907572 | 2.96 | PAPPA2 | Pappalysin 2 | NM_020318 |
| 8090688 | 2.93 | SNORA58 | Small nucleolar
RNA, H/ACA box 58 | NR_002985 |
| 8139935 | 2.89 | TYW1B | tRNA-yW
synthesizing protein 1 homolog B | NM_001145440 |
| 7982028 | 2.87 | SNORD115-11 | Small nucleolar
RNA, C/D box 115-11 | NR_003303 |
| 7982050 | 2.87 | SNORD115-11 | Small nucleolar
RNA, C/D box 115-11 | NR_003303 |
| 7982064 | 2.87 | SNORD115-11 | Small nucleolar
RNA, C/D box 115-11 | NR_003303 |
| 7982078 | 2.87 | SNORD115-11 | Small nucleolar
RNA, C/D box 115-11 | NR_003303 |
| 7982092 | 2.87 | SNORD115-11 | Small nucleolar
RNA, C/D box 115-11 | NR_003303 |
| 7905339 | 2.86 | GABPB2 | GA binding protein
transcription factor, β subunit | NM_144618 |
| 8140782 | 2.84 | ABCB1 | ATP-binding
cassette, sub-family B (multidrug resistance protein/transporter
associated with antigen processing) | NM_000927 |
| 8139482 | 2.83 | SNORA5A | Small nucleolar
RNA, H/ACA box 5A | NR_002919 |
| 8124756 | 2.83 | PPP1R10 | Protein phosphatase
1, regulatory (inhibitor) subunit | NM_002714 |
| 8124756 | 2.83 | PPP1R10 | Protein phosphatase
1, regulatory (inhibitor) subunit | NM_002714 |
| 8178358 | 2.83 | PPP1R10 | Protein phosphatase
1, regulatory (inhibitor) subunit | NM_002714 |
| 8179664 | 2.83 | PPP1R10 | Protein phosphatase
1, regulatory (inhibitor) subunit | NM_002714 |
| 8112731 | 2.77 | F2RL2 | Coagulation factor
II (thrombin) receptor-like 2 | NM_004101 |
| 8043687 | 2.74 | ANKRD36 | Ankryin repeat
domain 36 | NM_001164315 |
| 7977273 | 2.74 | ADSSL1 | Adenylosuccinate
synthase like 1 | NM_152328 |
| 7953200 | 2.72 | CCND2 | Cyclin D2 | NM_001759 |
| 8057954 | 2.72 | C2prf66 | Chromosome 2 open
reading frame 66 | AY358249 |
| 8049532 | 2.72 | LRRFIP1 | Leucine-rich repeat
(in FLII) interacting protein 1 | NM_001137550 |
| 8141843 | 2.72 | RASA4 | RAS p21 protein
activator 4 | NM_006989 |
| 8098752 | 2.72 | ABCA11P | Adenosine
triphosphate-binding cassette, sub-family A, member 11,
pseudogene | NR_002451 |
| 7982046 | 2.70 | SNORD115-20 | Small nucleolar
RNA, C/D box 115-20 | NR_003312 |
| 7982016 | 2.70 | SNORD115-12 | Small nucleolar
RNA, C/D box 115-12 | NR_003304 |
| 7982024 | 2.70 | SNORD115-12 | Small nucleolar
RNA, C/D box 115-12 | NR_003304 |
| 7982030 | 2.70 | SNORD115-12 | Small nucleolar
RNA, C/D box 115-12 | NR_003304 |
| 8054064 | 2.69 | ANKRD36B | Ankyrin repeat
domain 36B | NM_025190 |
| 7910047 | 2.68 | DNAH14 | Dynein, axonemal,
heavy chain 14 | NM_001373 |
| 8016239 | 2.68 | PLEKHM1 | Pleckstrin homology
domain-containing, family M (with RUN domain) member 1 | NR_027774 |
| 8060949 | 2.67 | ANKRD5 | Ankyrin repeat
domain 5 | NM_022096 |
| 8064375 | 2.62 | SRXN1 | Sulfiredoxin 1
homolog (S. Cerevisiae) | NM_080725 |
| 8077612 | 2.61 | TTLL3 | Tubulin tyrosine
ligase-like family, member 3 | NM_001025930 |
| 8151559 | 2.60 | SLC10A5 | Solute carrier
family 10 (sodium/bile acid cotransporter family), member 5 | NM_001010893 |
| 7902227 | 2.60 | GADD45A | Growth arrest and
DNA-damage-inducible, α | NM_001924 |
| 7982058 | 2.59 | SNORD115-26 | Small nucleolar
RNA, C/D box 115-26 | NR_003343 |
| 8118023 | 2.55 | GTF2H4 | General
transcription factor II human, polypeptide 4 | NM_001517 |
| 8006336 | 2.54 | LRRC37B | Leucine-rich
repeat-containing 37B | NM_052888 |
| 8108006 | 2.51 | LEAP2 | Liver-expressed
antimicrobial peptide 2 | NM_052971 |
| 7971388 | 2.50 | SLC25A30 | Solute carrier
family 25, member 30 | NM_001010875 |
| 7987163 | 2.48 | FMN1 | Formin 1 |
ENST00000414268 |
| 7938293 | 2.47 | SNORA45 | Small nucleolar
RNA, H/ACA box 45 | NR_002977 |
| 8113651 | 2.45 | ATG12 | ATG12
autophagy-related 12 homolog (S. Cerevisiae) | NR_033362 |
| 8049542 | 2.45 | LRRFIP1 | Leucine-rich repeat
(in FLII) interacting protein 1 | NM_001137550 |
| 8097435 | 2.45 | C4orf33 | Chromosome 4 open
reading frame 33 | NM_173487 |
| 8168345 | 2.45 | ACRC | Acidic
repeat-containing | NM_052957 |
| 7980828 | 2.42 | CCDC88C | Coiled-coil
domain-containing 88C | NM_001080414 |
| 8045423 | 2.42 | SNORA40 | Small nucleolar
RNA, H/ACA box 40 | NR_002973 |
| 8112331 | 2.42 | ISCA1 | Iron-sulfur cluster
assembly 1 homolog | NM_030940 |
| 8043697 | 2.41 | ANKRD36B | Ankyrin repeat
domain 36B | NM_025190 |
| 8033667 | 2.40 | ZNF558 | Zinc finger protein
558 | NM_144693 |
| 8142232 | 2.39 | LAMB4 | Laminin, β4 | NM_007356 |
| 7960052 | 2.39 | SNORA49 | Small nuclear RNA,
H/ACA box 49 | NR_002979 |
| 8031837 | 2.39 | ZNF587 | Zinc finger protein
587 | AF294842 |
| 8174715 | 2.38 | SNORA69 | Small nuclear RNA,
H/ACA box 69 | NR_002584 |
| 8001748 | 2.38 | SNORA50 | Small nuclear RNA,
H/ACA box 50 | NR_002980 |
| 8042503 | 2.38 | MXD1 | MAX dimerization
protein 1 | NM_002357 |
| 8072678 | 2.36 | HMOX1 | Heme oxygenase
(decycling) 1 | NM_002133 |
| 7997904 | 2.35 | ZNF778 | Zinc finger protein
778 | AK295122 |
| 8053648 | 2.35 | KRCC1 | Lysine-rich
coiled-coil 1 | NM_016618 |
| 8035793 | 2.35 | ZNF737 | Zinc finger protein
737 | NM_001159293 |
| 7977732 | 2.34 | SNORD8 | Small nuclear RNA,
C/D box 8 | NR_002916 |
| 8153457 | 2.31 | EEF1D | Eukaryotic
translation elongation factor 1δ | AY358690 |
| 8069574 | 2.31 | C21orf91 | Chromosome 21 open
reading frame 91 | NM_001100420 |
| 8112841 | 2.30 | HOMER1 | Homer homolog 1
(Drosophila) | NM_004272 |
| 8038919 | 2.29 | ZNF350 | Zinc finger protein
50 | NM_021632 |
| 9175288 | 2.29 | MOSPD1 | Motile sperm
domain-containing 1 | NM_019556 |
| 8160912 | 2.28 | C9orf131 | Chromosome 9 open
reading frame 131 | NM_203299 |
| 7934334 | 2.28 | TTC18 | Tetratricopeptide
repeat domain 18 | NM_145170 |
| 8056572 | 2.27 | SPC25 | SPC25, NDC80
kinetochore complex component | NM_020675 |
| 8161919 | 2.27 | TLE1 | Transducin-like
enhancer of split 1 (Drosophila) | NM_005077 |
| 7964460 | 2.26 | DDIT3 |
DNA-damage-inducible transcript 3 | NM_004083 |
| 8019857 | 2.26 | NDC80 | NDC80 Homolog,
kinetochore complex component (S. cerevisiae) | NM_006101 |
| 8045587 | 2.24 | ACVR2A | Activin A receptor,
type IIA | NM_001616 |
| 8002660 | 2.24 | TXNL4B | Thioredoxin-like
4B | NM_017853 |
| 7911329 | 2.24 | 14-Sep | Septin 14 | NM_207366 |
| 8080980 | 2.24 | FLJ10213 | Endogenous
Borna-like N element-1 | NM_018029 |
| 8014115 | 2.22 | MYOID | Myosin ID | NM_015194 |
| 7949916 | 2.20 | CHKA | Choline kinase
α | NM_001277 |
| 7938295 | 2.20 | RPL27A | Ribosomal protein
L27a | NM_000990 |
| 8168146 | 2.20 | KIF4A | Kinesin family
member 4A | NM_012310 |
| 8114171 | 2.19 | CDKL3 | Cycline-dependent
kinase-like 3 | NM_001113575 |
| 8008700 | 2.19 | FLJ11710 | Hypothetical
protein FLJ11710 | AK021772 |
| 8108321 | 2.18 | FAM53C | Family with
sequence similarity 53, member C | NM_001135647 |
| 8047161 | 2.18 | OBFC2A |
Oligonucleotide/oligosaccharide-binding
fold-containing 2a | NM_001031716 |
| 8053576 | 2.17 | RNF103 | Ring finger protein
103 | NM_005667 |
| 8006237 | 2.17 | LOC400590 | Hypothetical
LOC400590 |
ENST00000433145 |
| 8136341 | 2.17 | BPGM |
2,3-bisphosphoglycerate mutase | NM_199186 |
| 8146225 | 2.16 | C8orf40 | Chromosome 8 open
reading frame 40 | NM_001135674 |
| 8147057 | 2.16 | CHMP4C | Chromatin modifying
protein 4C | NM_152284 |
| 7921228 | 2.15 | ETV3 | E26
transforming-specific variant 3 | NM_001145312 |
| 8065607 | 2.15 | PLAGL2 | Pleiomorphic
adenoma gene-like 2 | NM_002657 |
| 8096511 | 2.14 | BMPR1B | Bone morphogenetic
protein receptor, type 1B | NM_001203 |
| 7927389 | 2.14 | MAPK8 | Mitogen-activated
protein kinase 8 | NM_002750 |
| 8098958 | 2.14 | POLN | Polymerase (DNA
directed) nu | NM_181808 |
| 8038989 | 2.14 | ZNF600 | Zinc finger protein
600 | NM_198457 |
| 7951654 | 2.14 | FDXACB1 | Ferredoxin-fold
anticodon binding domain-containing 1 | NM_138378 |
| 8036341 | 2.13 | ZNF461 | Zinc finger protein
461 | NM_153257 |
| 7981998 | 2.13 | SNORD116-25 | Small nucleolar
RNA, C/D box 116-25 | NM_003339 |
| 8041179 | 2.13 | CLIP4 |
Cytoskeleton-associated
protein-glycine-rich domain-containing linker protein family member
4 | NM_024692 |
| 7969096 | 2.13 | CDADC1 | Cytidine and
deoxycytidine monophosphate deaminase domain-containing 1 | NM_030911 |
| 7969243 | 2.13 | CKAP2 |
Cytoskeleton-associated protein 2 | NM_018204 |
| 7986350 | 2.12 | ARRDC4 | Arrestin
domain-containing 4 | NM_183376 |
| 8063382 | 2.12 | SNAI1 | Snail homolog 1
(Drosophila) | NM_005985 |
| 8053801 | 2.12 | ANKRD36 | Ankyrin repeat
domain 36 | NM_001164315 |
| 7999588 | 2.12 | PLA2G10 | Phospholipase A2,
group X | NM_003561 |
| 8008795 | 2.11 | C17orf71 | Chromosome 17 open
reading frame 71 | NM_018149 |
| 8029340 | 2.11 | ZNF155 | Zinc finger protein
155 | NM_003445 |
| 8166104 | 2.11 | OFD1 | Oral-facial-digital
syndrome 1 | NM_003611 |
| 8123825 | 2.11 | SLC35B3 | Solute carrier
family 35 member B3 | NM_015948 |
| 7901052 | 2.11 | SNORD38B | Small nucleolar
RNA, C/D box 38B | NM_001457 |
| 8084880 | 2.10 | HES1 | Hairy and enhancer
of split 1 | NM_005524 |
| 7925672 | 2.10 | ZNF670 | Zinc finger protein
670 | NM_033213 |
| 7982294 | 2.10 | OTUD7A | OTU
domain-containing 7A | NM_130901 |
| 7962112 | 2.09 | CAPRIN2 | Caprin family
member 2 | NM_001002259 |
| 7973948 | 2.09 | BRMSIL | Breast cancer
metastasis-suppressor 1-like | NM_032352 |
| 8117685 | 2.09 | ZKSCAN3 | Zinc finger with
KRAB and SCAN domains 3 | NM_024493 |
| 8010082 | 2.08 | SNORD1A | Small nucleolar
RNA, C/D box 1A | NR_004395 |
| 8041982 | 2.08 | ACYP2 | Acylphosphatase 2,
muscle type | NM_138448 |
| 8137693 | 2.08 | COX19 | Cytochrome c
oxidase assembly homolog 19 | NM_001031617 |
| 7917779 | 2.08 | GCLM | Glutamate-cysteine
ligase, modifier subunit | NM_002061 |
| 7938364 | 2.08 | WEE1 | WEE1 homolog 1
(S. Pombe) | BX641032 |
| 8007414 | 2.08 | AOC2 | Amine oxidase,
copper-containing 2 (retina-specific) | NM_009590 |
| 8139656 | 2.08 | GRB10 | Growth factor
receptor-bound protein 10 | NM_001001555 |
| 8059852 | 2.08 | MSL3L2/MSL3-like
2 | Male-specific
lethal 3-like 2 (Drosophila) | NM_001166217 |
| 8109484 | 2.07 | KIF4B | Kinesin family
member 4B | NM_001099293 |
| 8022559 | 2.07 | ANKRD29 | Ankyrin repeat
domain 29 | NM_173505 |
| 7910030 | 2.07 | DNAH14 | Dynein, axonemal,
heavy chain 14 | NM_00145154 |
| 8052143 | 2.07 | GPR75 | G protein-coupled
receptor 75 | NM_006794 |
| 7931643 | 2.07 | CYP2E1 | Cytochrome P450,
family 2, subfamily E, polypeptide 1 | NM_000773 |
| 8102789 | 2.06 | TERF1 | Telomeric repeat
binding factor (NIMA-interacting) 1 | NM_003218 |
| 7953603 | 2.06 | C1S | Complement
component 1, s subcomponent | NM_201442 |
| 8104930 | 2.05 | SLC1A3 | Solute carrier
family 1 (glial high affinity glutamate transporter), member 3 | NM_004172 |
| 7953211 | 2.05 | C12orf5 | Chromosome 12 open
reading frame 5 | NM_020375 |
| 8114326 | 2.04 | FAM13B | Family with
sequence similarity 13, member B | NM_0166603 |
| 7936826 | 2.04 | IKZF5 | IKAROS family zinc
finger (Pegasus) | NM_022466 |
| 8013567 | 2.04 | C17orf108 | Chromosome 17 open
reading frame 108 | NM_001076680 |
| 7975066 | 2.04 | AKAP5 | A-kinase anchor
protein 5 | NM_004857 |
| 8142524 | 2.04 | TSPAN12 | Tetraspanin 12 | NM_012338 |
| 7952673 | 2.04 | FLJ45950 | FLJ45950
protein | AK127847 |
| 8081128 | 2.04 | NSUN3 | Nucleolar protein 2
homolog/Sun domain family, member 3 | NM_022072 |
| 7922846 | 2.04 | FAM129A | Family with
sequence similarity 129, member A | NM_052966 |
| 8013305 | 2.04 | ZNF286B | Zinc finger protein
286B | NM_001145045 |
| 8153935 | 2.03 | ZNF252 | Zinc finger protein
252 | NM_023392 |
| 8162490 | 2.03 | HIATL1 | Hippocampus
abundant transcript-like 1 | NM_032558 |
| 8128698 | 2.02 | SESN1 | Sestrin 1 | NM_014454 |
| 8010778 | 2.02 | CSNK1D | Casein kinase 1,
δ | NM_001893 |
| 8141311 | 2.02 | FAM200A | Family with
sequence similarity 200, member A | NM_145111 |
| 7944867 | 2.02 | SIAE | Sialic acid
acetylesterase | NM_170601 |
| 7961829 | 2.02 | BCAT1 | Branched-chain
amino-acid transaminase 1, cytosolic | NM_005504 |
| 7994161 | 2.02 | RBBP6 | Etinoblastoma
binding protein 6 | NM_006910 |
| 7981273 | 2.02 | CCDC85C | Coiled-coil
domain-containing 85C | NM_001144995 |
| 8110649 | 2.02 | TRIM41 | Tripartite
motif-containing 41 | NM_033549 |
| 8101839 | 2.01 | EIF4E | Eukaryotic
translation initiation factor 4E | NM_001968 |
| 8103226 | 2.01 | TMEM154 | Transmembrane
protein 154 | NM_152680 |
 | Table IIDownregulation of gene expressions in
catharidine-treated NCI-H460 cells. |
Table II
Downregulation of gene expressions in
catharidine-treated NCI-H460 cells.
| Probe set ID | Fold change | Gene symbol | Gene
description | mRNA accession
no. |
|---|
| 8175016 | −3.08 | APLN | Apelin | NM_017413 |
| 8124413 | −3.06 | HIST1H4D | Histone cluster 1,
H4d | NM_003539 |
| 8105302 | −3.05 | FST | Follistatin | NM_006350 |
| 7953665 | −3.04 | DPPA3 | Developmental
pluripotency-associated 3 | NM_199286 |
| 8117426 | −2.99 | HIST1H2BH | Histone cluster 1,
H2bh | NM_003524 |
| 8117898 | −2.97 | HIST1H4J | Histone cluster 1,
H4j | NM_021968 |
| 8117337 | −2.95 | HIST1H1E | Histone cluster 1,
H1e | NM_005321 |
| 7911241 | −2.93 | OR2L8 | Olfactory receptor,
family 2, subfamily L, member 8 | NM_001001963 |
| 8048749 | −2.88 | KCNE4 | Potassium
voltage-gated channel, IsK-related family, member 4 | NM_080671 |
| 8124437 | −2.87 | HIST1H3F | Histone cluster 1,
H3f | NM_021018 |
| 8117395 | −2.83 | HIST1H2BF | Histone cluster 1,
H2bf | NM_003522 |
| 8015798 | −2.79 | LOC100130581 | Hypothetical
LOC100130581 | NR_027413 |
| 7919642 | −2.78 | HIST2H2AB | Histone cluster 2,
H2ab | NM_175065 |
| 8059470 | −2.76 | IRS1 | Insulin receptor
substrate 1 | NM_005544 |
| 8077270 | −2.75 | CHL1 | Cell adhesion
molecule with homology to L1 cell adhesion molecule | NM_006614 |
| 7915592 | −2.74 | RNU5D | RNA, U5D small
nuclear | NR_002755 |
| 7906767 | −2.74 | FCGR2C | Fc fragment of
immunoglobulin G, low affinity IIc, receptor for (CD32)
(gene/pseudogene) | NM_201563 |
| 8117594 | −2.74 | HIST1H2BM | Histone cluster 1,
H2bm | NM_003521 |
| 8117589 | −2.72 | HIST1H3H | Histone cluster 1,
H3h | NM_003536 |
| 7981728 | −2.71 | LOC100293211 | Similar to
hCG2042717 |
ENST00000390601 |
| 8124397 | −2.71 | HIST1H1C | Histone cluster 1,
H1c | NM_005319 |
| 8135734 | −2.71 | C7orf58 | Chromosome 7 open
reading frame 58 | NM_024913 |
| 8138988 | −2.70 | DPY19L2P1 | Dpy-19-like 2
pseudogene 1 (C. elegans) | NR_002833 |
| 8117583 | −2.65 | HIST1H2AI | Histone cluster 1,
H2ai | NM_003509 |
| 8153258 | −2.65 | SLC45A4 | Solute carrier
family 45, member 4 | BC033223 |
| 7960865 | −2.61 | SLC2A3 | Solute carrier
family 2 (facilitated glucose transporter), member 3 | NM_006931 |
| 8116921 | −2.59 | EDN1 | Endothelin 1 | NM_001955 |
| 8101757 | −2.58 | GPRIN3 | G protein-regulated
inducer of neurite outgrowth family member 3 | NM_198281 |
| 7971723 | −2.58 | FLJ37307 | Hypothetical
LOC283521 | NR_027047 |
| 8117580 | −2.56 | HIST1H2AI | Histone cluster 1,
H2ai | NM_003509 |
| 8167573 | −2.56 | GAGE1 | G antigen 1 | NM_001468 |
| 8165295 | −2.56 | LCN8 | Lipocalin 8 |
ENST00000371686 |
| 7927876 | −2.53 | TET1 | Ten-eleven
translocation oncogene 1 | NM_030625 |
| 7963054 | −2.52 | TUBA1A | Tubulin, α1a | NM_006009 |
| 7957386 | −2.51 | ACSS3 | Acyl-CoA synthetase
short-chain family member 3 | NM_024560 |
| 7915919 | −2.49 | TAL1 | T cell acute
lymphocytic leukemia 1 | NM_003189 |
| 8174985 | −2.48 | SMARCA1 | Switch/sucrose
non-fermentable-related, matrix-associated, actin-dependent
regulator of chromatin, subfamily a, member 1 | NM_003069 |
| 8105495 | −2.47 | PART1 | Prostate
androgen-regulated transcript 1 (non-protein coding) | NR_028508 |
| 8146967 | −2.46 | CRISPLD1 | Cysteine-rich
secretory protein LCCL domain-containing 1 | NM_031461 |
| 8144786 | −2.46 | SLC7A2 | Solute carrier
family 7 (cationic amino acid transporter, y+ system), member
2 | NM_003046 |
| 8029280 | −2.45 | CD177 | CD177 molecule | NM_020406 |
| 7984524 | −2.45 | PAQR5 | Progestin and
adipoQ receptor family member V | NM_001104554 |
| 7973182 | −2.44 | LOC554207 | Hypothetical
LOC554207 |
ENST00000320322 |
| 8145795 | −2.44 | LOC100293539 | Similar to
ribosomal protein 10 | XM_002346094 |
| 8095697 | −2.44 | CXCL1 | Chemokine (C-X-C
motif) ligand 1 (melanoma growth stimulating activity, α) | NM_001511 |
| 8124406 | −2.42 | HIST1H2BC | Histone cluster 1,
H2bc | NM_003526 |
| 7978260 | −2.42 | DHRS1 |
Dehydrogenase/reductase family member
1 | NM_001136050 |
| 7920271 | −2.42 | S100A4 | S100 calcium
binding protein A4 | NM_019544 |
| 8166469 | −2.42 | SAT1 | Spermidine/spermine
N1-acetyltransferase 1 | NR_027783 |
| 8124527 | −2.40 | HIST1H1B | Histone cluster 1,
H1b | NM_005322 |
| 8008321 | −2.40 | ACSF2 | Acyl-CoA synthetase
family member 2 | NM_025149 |
| 8124385 | −2.39 | HIST1H4B | Histone cluster 1,
H4b | NM_003544 |
| 800731 | −2.38 | TUBG2 | Tubulin, γ2 | NM_016437 |
| 8122365 | −2.36 | GPR126 | G protein-coupled
receptor 126 | NM_020455 |
| 8015273 | −2.34 | KRT31 | Keratin 31 | NM_002277 |
| 7904465 | −2.34 | HIST2H2BA | Histone cluster 2,
H2ba | NR_027337 |
| 8124416 | −2.33 | HIST1H3D | Histone cluster 1,
H3d | NM_003530 |
| 8074458 | −2.33 | C22orf39 | Chromosome 22 open
reading frame 39 | NM_173793 |
| 8046048 | −2.33 | CSRNP3 |
Cysteine-serine-rich nuclear protein
3 | NM_001172173 |
| 8007493 | −2.32 | ARL4D | Adenosine
diphosphate-ribosylation factor-like 4D | NM_001661 |
| 8157804 | −2.31 | OLFML2A | Olfactomedin-like
2A | NM_182487 |
| 7948836 | −2.31 | TMEM223 | Transmembrane
protein 223 | NM_001080501 |
| 8033458 | −2.31 | LYPLA2 | Lysophospholipase
II | NM_007260 |
| 7919619 | −2.30 | HIST2H2AA3 | histone cluster 2,
H2aa3 | NM_003516 |
| 7905079 | −2.30 | HIST2H2AA3 | Histone cluster 2,
H2aa3 | NM_003516 |
| 7927631 | −2.29 | DKK1 | Dickkopf homolog 1
(Xenopus laevis) | NM_012242 |
| 7975598 | −2.28 | ACOT1 | Acyl-CoA
thioesterase 1 | NM_001037161 |
| 8071801 | −2.27 | GSTTP1 | Glutathione
S-transferase θ pseudogene 1 | NR_003081 |
| 7928429 | −2.27 | PLAU | Plasminogen
activator, urokinase | NM_002658 |
| 8068898 | −2.27 | HIST1H2BK | Histone cluster 1,
H2bk | NM_080593 |
| 8041467 | −2.26 | VIT | Vitrin | NM_053276 |
| 8077160 | −2.26 | ARSA | Arylsulfatase
A | NM_000487 |
| 7991754 | −2.25 | HBZ | Hemoglobin, ζ | NM_005332 |
| 8049534 | −2.24 | LRRFIP1 | Leucine-rich repeat
(in FLII) interacting protein 1 | NM_001137550 |
| 7909789 | −2.23 | TGFB2 | Transforming growth
factor β2 | NM_001135599 |
| 7919612 | −2.23 | HIST2H3D | Histone cluster 2,
H3d | NM_001123375 |
| 8100578 | −2.22 | EPHA5 | Ephrin receptor
A5 | NM_004439 |
| 8169541 | −2.22 | DOCK11 | Dedicator of
cytokinesis 11 | NM_144658 |
| 8124430 | −2.21 | HIST1H1D | Histone cluster 1,
H1d | NM_005320 |
| 8124524 | −2.21 | HIST1H2AK | Histone cluster 1,
H2ak | NM_003510 |
| 8124524 | −2.21 | HIST1H2AK | Histone cluster 1,
H2ak | NM_003510 |
| 7906775 | −2.20 | HSPA7 | Heat shock 70kDa
protein 7 (HSP70B) | NR_024151 |
| 7953291 | −2.20 | CD9 | CD9 molecule | NM_001769 |
| 8033319 | −2.19 | SH2D3A | Src Homology 2
domain-containing 3A | NM_005490 |
| 7905088 | −2.19 | HIST2H2AC | Histone cluster 2,
H2ac | NM_003517 |
| 7975602 | −2.19 | ACOT2 | Acyl-CoA
thioesterase 2 | NM_006821 |
| 7982854 | −2.19 | DLL4 | δ-like 4
(Drosophila) | NM_019074 |
| 8019778 | −2.19 | PCYT2 | Phosphate
cytidylyltransferase 2, ethanolamine | NM_002861 |
| 8046048 | −2.19 | HIST1H4C | Histone cluster 1,
H4c | NM_003542 |
| 8007493 | −2.18 | VWA5A | Von Willebrand
factor A domain-containing 5A | NM_001130142 |
| 8157804 | −2.18 | PLG | Plasminogen | NM_000301 |
| 7948836 | −2.18 | CD24 | CD24 molecule | NM_013230 |
| 8033458 | −2.17 | FSTL4 | Follistatin-like
4 | NM_015082 |
| 7919619 | −2.16 | CA2 | Carbonic anhydrase
II | NM_000067 |
| 7905079 | −2.16 | CDH19 | Cadherin 19, type
2 | NM_021153 |
| 7927631 | −2.16 | CDC42EP3 | Cell division cycle
42 effector protein (Rho guanosine triphosphatase binding) 3 | NM_006449 |
| 7975598 | −2.16 | ACCN2 | Amiloride-sensitive
cation channel 2, neuronal | NM_020039 |
| 8071801 | −2.15 | HIST2H3D | Histone cluster 2,
H3d | NM_001123375 |
| 7928429 | −2.15 | RFX2 | Regulatory factor
X, 2 (influences human leukocyte antigen class II expression) | NM_000635 |
| 8068898 | −2.15 | NES | Nestin | NM_006617 |
| 8041467 | −2.15 | LOC25845 | Hypothetical
LOC25845 | NR_024158 |
| 8077160 | −2.15 | THSD7A | Thrombospondin,
type I, domain-containing 7A | NM_015204 |
| 7991754 | −2.14 | LOC147727 | Hypothetical
LOC147727 | NR_024333 |
| 8049534 | −2.14 | CALML6 | Calmodulin-like
6 | NM_138705 |
| 7909789 | −2.14 | DEFB109P1B | Defensin, β 109,
pseudogene 1B | NR_003668 |
| 7919612 | −2.13 | EPOR | Erythropoietin
receptor | NM_000121 |
| 8100578 | −2.13 | EEF2K | Eukaryotic
elongation factor-2 kinase | NM_013302 |
| 8169541 | −2.13 | EMP3 | Epithelial membrane
protein 3 | NM_001425 |
| 8124430 | −2.13 | TMEM84 | Transmembrane
protein 84 | NR_026949 |
| 8124524 | −2.13 | CXXC5 | CXXC finger 5 | NM_016463 |
| 7906775 | −2.12 | PCYT2 | Phosphate
cytidylyltransferase 2, ethanolamine | NM_002861 |
| 7953291 | −2.12 | LYPD1 | Ly6/plasminogen
activator, urokinase 1 receptor domain-containing | NM_144586 |
| 8033319 | −2.12 | PHLDB2 | Pleckstrin
homology-like domain, family B, member 2 | NM_001134439 |
| 7905088 | −2.11 | LRFN2 | Leucine-rich repeat
and fibronectin type III domain-containing 2 | NM_020737 |
| 7975602 | −2.11 | C9orf23 | Chromosome 9 open
reading frame 23 | NM_148179 |
| 7982854 | −2.11 | FLJ13744 | Hypothetical
FLJ13744 | BC070061 |
| 8018445 | −2.11 | UNK | Unkempt homolog
(Drosophila) | NM_001080419 |
| 8038407 | −2.10 | RRAS | Related RAS viral
(r-ras) oncogene homolog | NM_006270 |
| 7987230 | −2.10 | LPCAT4 |
Lysophosphatidylcholine acyltransferase
4 | NM_153613 |
| 8031514 | −2.10 | LOC100133142 | Hypothetical
LOC100133142 | XM_001718400 |
| 8130374 | −2.10 | FBXO5 | F-box protein
5 | NM_012177 |
| 7908409 | −2.09 | RGS2 | Regulator of
G-protein signaling 2 | NM_002923 |
| 8111255 | −2.09 | CDH10 | Cadherin 10, type 2
(T2-cadherin) | NM_006727 |
| 7965335 | −2.09 | DUSP6 | Dual specificity
phosphatase 6 | NM_001946 |
| 8065537 | −2.09 | LOC100134868 | Hypothetical
LOC100134868 | NR_004846 |
| 8138466 | −2.08 | MACC1 | Metastasis
associated in colon cancer 1 | NM_182762 |
| 7902687 | −2.08 | CYR61 | Cysteine-rich,
angiogenic inducer, 61 | NM_001554 |
| 8036136 | −2.08 | TMEM149 | Transmembrane
protein 149 | NM_024660 |
| 8098916 | −2.08 | TMEM129 | Transmembrane
protein 129 | NM_001127266 |
| 7955663 | −2.07 | TENC1 | Tensin-like C1
domain-containing phosphatase (tensin 2) | NM_170754 |
| 7939897 | −2.07 | FOLH1 | Folate hydrolase
(prostate-specific membrane antigen) 1 | NM_004476 |
| 7920191 | −2.07 | LCE3A | Late cornified
envelope 3A | NM_178431 |
| 7951437 | −2.06 | GUCY1A2 | Guanylate cyclase
1, soluble, α2 | NM_000855 |
| 8022653 | −2.06 | LOC728606 | Hypothetical
LOC728606 | NR_024259 |
| 7929816 | −2.06 | SCD | Stearoyl-CoA
desaturase (δ-9-desaturase) | NM_005063 |
| 7940565 | −2.06 | FADS2 | Fatty acid
desaturase 2 | NM_004265 |
| 7951157 | −2.06 | CCDC82 | Coiled-coil
domain-containing 82 | AK313893 |
| 7936100 | −2.06 | CALHM2 | Calcium homeostasis
modulator 2 | NM_015916 |
| 7954090 | −2.06 | EMP1 | Epithelial membrane
protein 1 | NM_001423 |
| 8005951 | −2.05 | SNORD42B | Small nucleolar
RNA, C/D box 42B | NR_000013 |
| 8148917 | −2.05 | MFSD3 | Major facilitator
superfamily domain-containing 3 | NM_138431 |
| 7937990 | −2.04 | HBG1 | Hemoglobin, γA | NM_000559 |
| 7937993 | −2.04 | HBG2 | Hemoglobin, γG | NM_000184 |
| 8033233 | −2.04 | TUBB4 | Tubulin, β4 | NM_006087 |
| 8048350 | −2.04 | PLCD4 | Phospholipase C,
δ4 | NM_032726 |
| 8037408 | −2.04 | KCNN4 | Potassium
intermediate/small conductance calcium-activated channel, subfamily
N, member 4 | NM_002250 |
| 7964119 | −2.04 | STAT2 | Signal transducer
and activator of transcription 2 | NM_005419 |
| 8016841 | −2.03 | TMEM100 | Transmembrane
protein 100 | NM_001099640 |
| 7958948 | −2.03 | DDX54 | DEAD box
polypeptide 54 | NM_0011111322 |
| 8151512 | −2.02 | PAG1 | Phosphoprotein
associated with glycosphingolipid microdomains 1 | NM_018440 |
| 8005549 | −2.02 | GRAPL | Growth factor
receptor-bound protein 2-related adaptor protein-like | NM_001129778 |
| 8033159 | −2.02 | PSPN | Persephin | NM_004158 |
| 7986639 | −2.02 | VSIG6 | V-set and
immunoglobulin domain-containing 6 |
ENST00000338567 |
| 7938741 | −2.01 | MRGPRX3 | MAS-related G
protein coupled receptor, member X3 | NM_054031 |
| 8047174 | −2.01 | SLC39A10 | Solute carrier
family 39 (zinc transporter), member 10 | NM_001127257 |
GeneGo analysis
A GeneGo analysis program was used to analyze the
CTD-treated NCI-H460 cells in order to determine the top scoring
genes which were differentially expressed, as determined by the
number of pathway networks involved. The results of the GeneGo
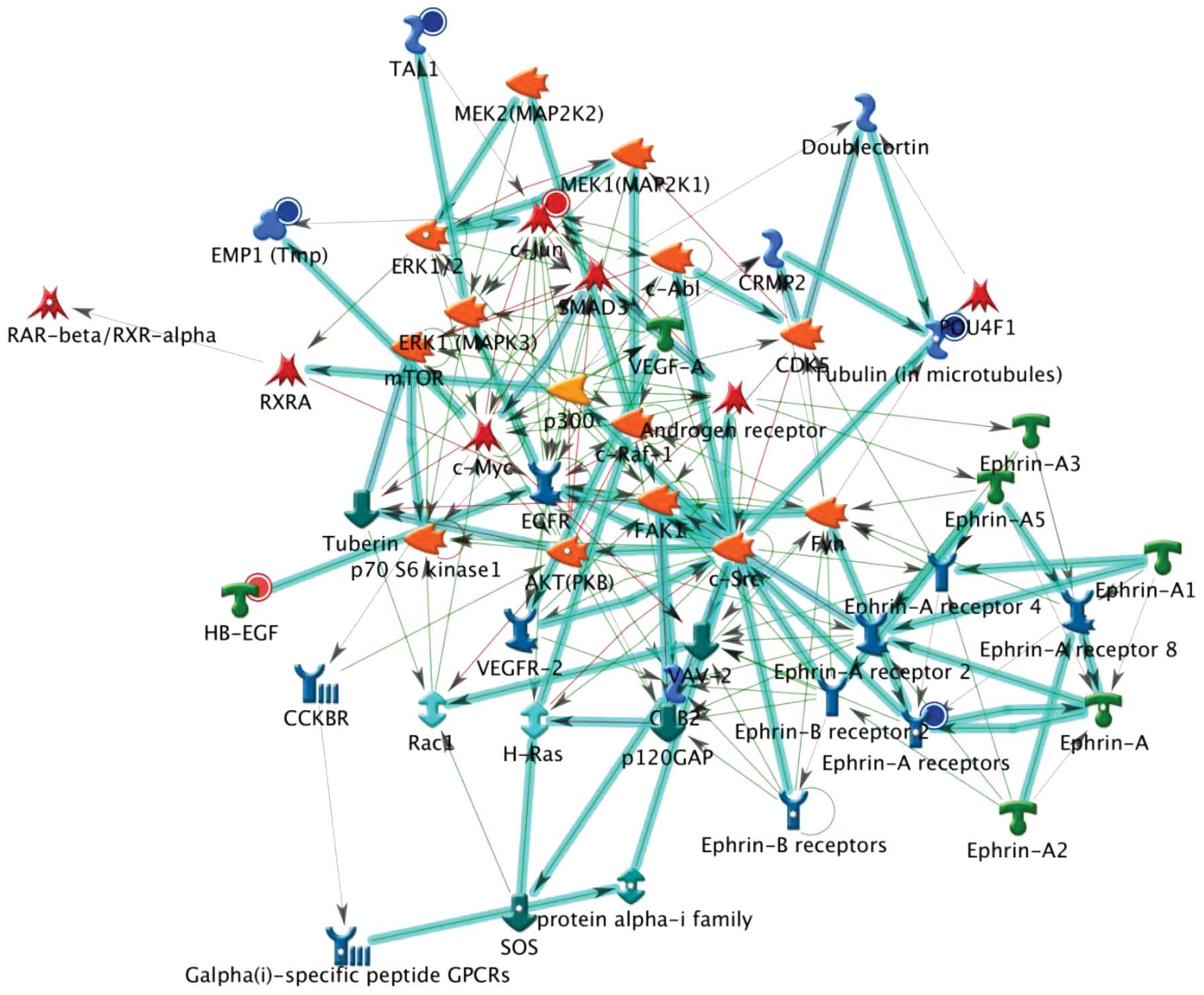
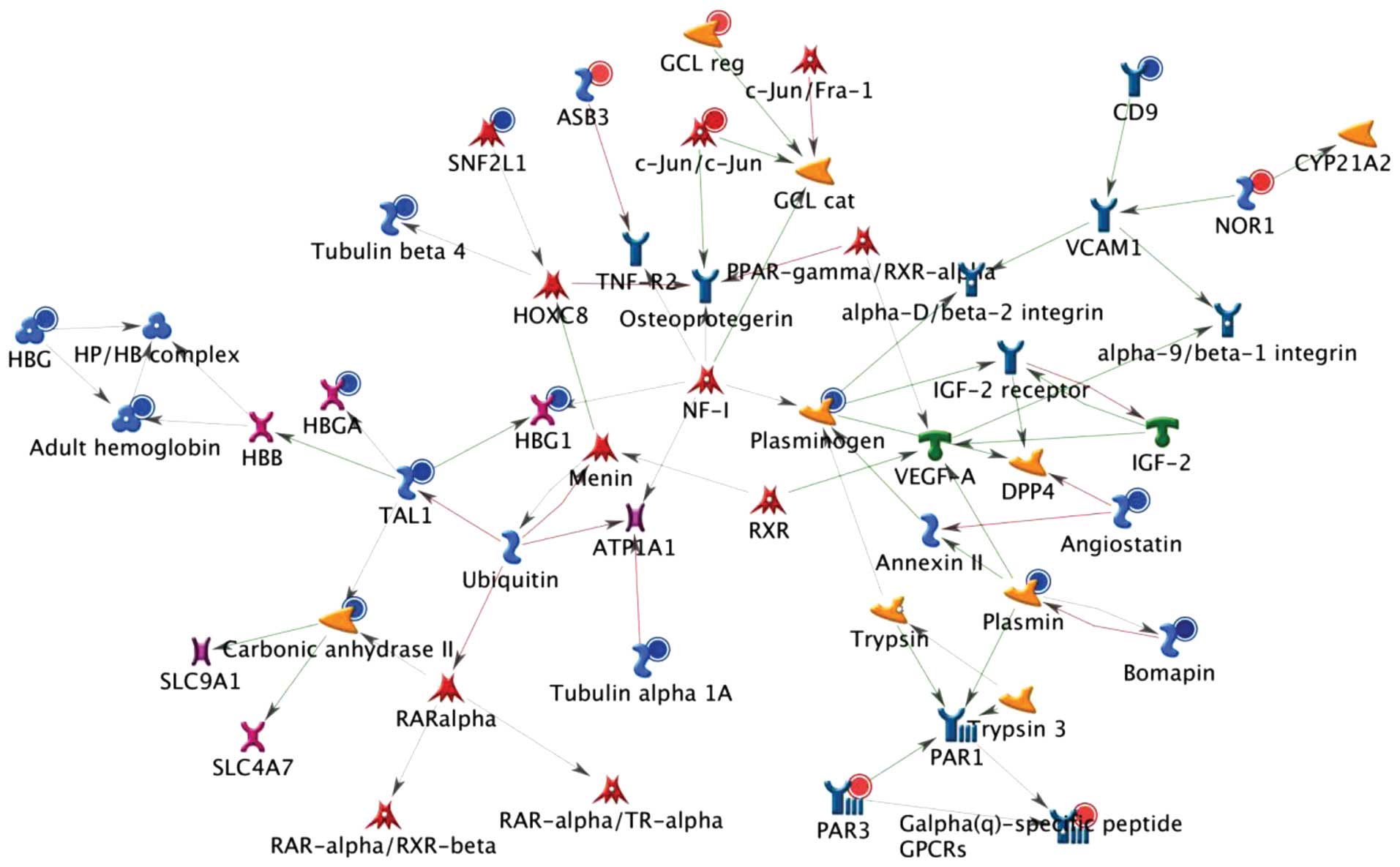
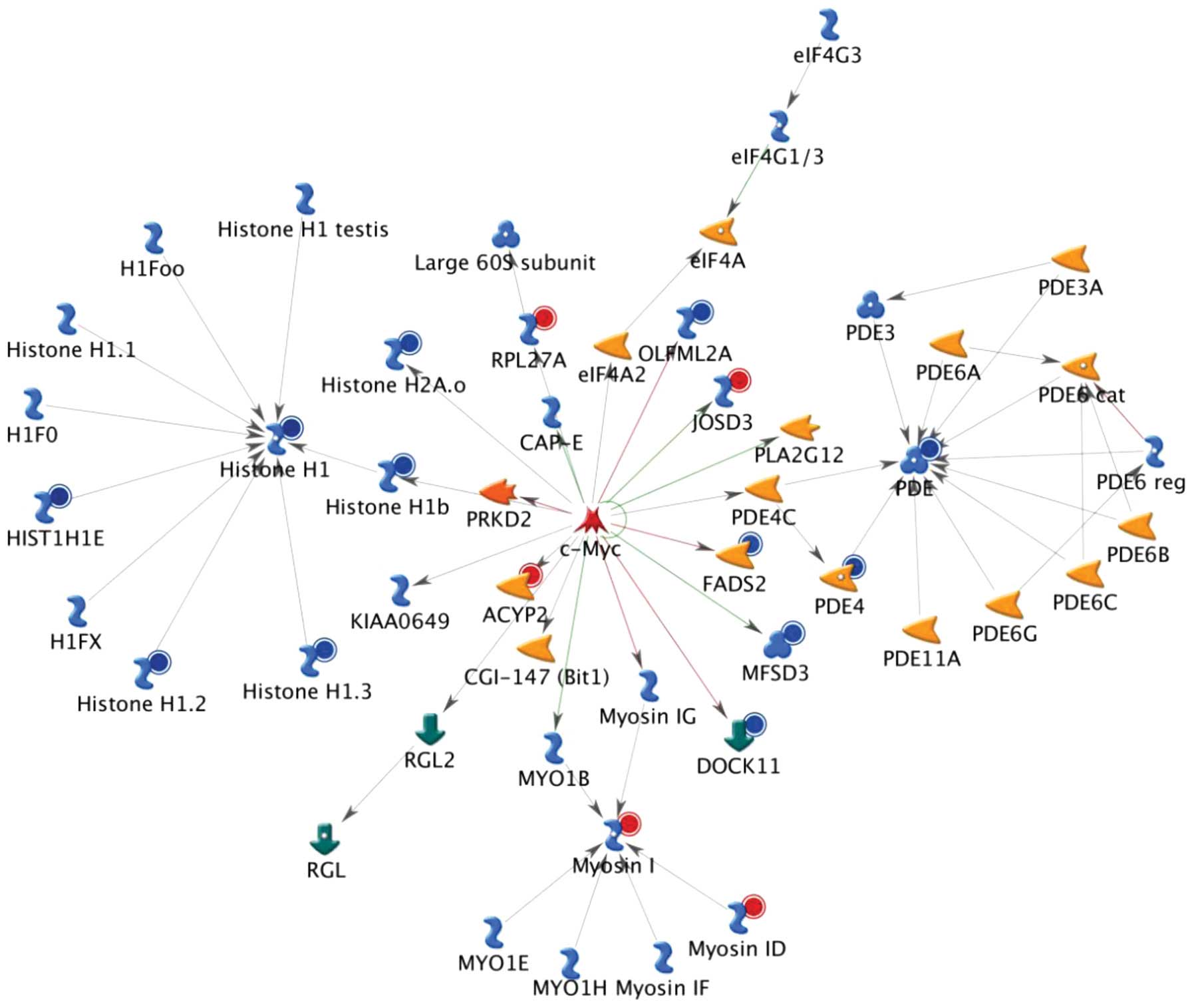
analyses are shown in Figs.
1Figure 23, which reveal the top, second and third
scored genes by the number of pathways, respectively. Experimental
data were used to generate maps of the pathway interactions and
genes which were upregulated (indicated by red circles) and
down-regulated (indicated by blue circles) in H460 cells following
treatment with CTD. It was indicated that these genes may also be
involved in DNA damage, cell cycle arrest and apop-tosis-associated
responses in CTD-treated H460 cells.
Discussion
CTD has been reported to have cytotoxic effects in
numerous different types of cancer cell (8–15).
The results of previous studies have also demonstrated that
CTD-induced cell death occurred due to the induction of apoptosis
in human lung cancer cells (data not shown) (23). However, the effects of CTD on gene
expression in cancer cells have remained to be elucdiated. To the
best of our knowledge, the present study was the first to report on
the effects of CTD on gene expression in H460 cells. Therefore, the
present study not only advanced the understanding of the
differential gene expression following treatment with CTD in lung
cancer cells, but may additionally provide several potential
biomarkers for use as future therapeutic clinical targets for the
treatment of lung cancer.
It has been well documented that the tumor
microenvironment, which contains matrix proteins, stromal cells and
associated secreted molecules, including cytokines and associated
genes, which may be used as targets of cancer therapeutic drugs
(24–26). Therefore, an increasing number of
studies focus on elucidating the tumor microenvironment and
associated gene expression in order to determine potential novel
therapeutic agents for treating cancer patients (27). Over the past decade, there have
been numerous clinical trials of treatments for lung cancer
patients, including adjuvant chemotherapy trials and neo-adjuvant
chemotherapy trials (28–30); however, the results of these trials
have not yet provided a successful, effective treatment for lung
cancer. Numerous studies have demonstrated that chemotherapeutics
may result in cell death through DNA damage, cell cycle arrest and
the induction of apoptosis (31,32).
In the present study, H460 cells were treated with CTD and
incubated in 12-well plates, and their RNA was then isolated in
order to determine which genes exhibited altered expression
following treatment with CTD. The results revealed that CTD
effected the upregulation and downregulation, respectively, of the
expression of certain genes which are known to be associated with
DNA damage, cell cycle progression and apoptosis in H460 cells.
In order to further elucidate the molecular
signaling pathways associated with altered gene expression in H460
cells following exposure to CTD, GeneGo Process Networks were used
in the present study in order to analyze the altered gene
expression results of the microarray, in order to determine the
possible signaling pathways involved. Based on GeneGo pathway and
canonical pathway maps, which represent a set of ~650 signaling and
metabolic maps covering human biology (signaling and metabolism) in
a comprehensive way. A preset network of protein interaction
characteristics for the process was used for each process, and the
experimental data were mapped regarding the specific process. The
obtained hypothetical molecular signaling pathways indicated that
CTD affects numerous associated signaling pathways, indicated by
the involvement of the differentially expressed genes in the
network of the respective the signaling pathways. The gene content
of the uploaded files was used as the input list for the generation
of biological networks using the Analyze Networks algorithm with
default settings. This is a variant of the shortest paths
algorithm, with main parameters of relative enrichment with the
uploaded data, and relative saturation of the networks with
canonical pathways. The network provides data listing interacting
proteins. In this workflow the network is prioritized based on the
number of fragments of canonical pathways on the network.
In conclusion, the results of the present study
revealed that treatment with CTD induced the upregulation and
downregulation of numerous genes in H460 cells. In addition, these
differentially expressed genes were associated with DNA damage,
cell cycle progression and apoptotic cell death in human lung
cancer H460 cells. The present study also revealed possible
signaling pathways, which may provide more information on the
possible mechanism of CTD in H460 cells; however, further studies
are required.
Acknowledgments
The present study was supported by a grant from
China Medical University [grant no. MU 101-AWARD-03(1/2); Taichung,
Taiwan].
References
|
1
|
Bilello KS, Murin S and Matthay RA:
Epidemiology, etiology, and prevention of lung cancer. Clin Chest
Med. 23:1–25. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang P, Gao WY, Turner S and Ducatman BS:
Gleevec (STI-571) inhibits lung cancer cell growth (A549) and
potentiates the cisplatin effect in vitro. Mol Cancer. 2:12003.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Midthun DE and Jett JR: Chemotherapy for
advanced lung cancer. When to expect a response. Postgrad Med.
101:187–194. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Stewart DJ: Tumor and host factors that
may limit efficacy of chemotherapy in non-small cell and small cell
lung cancer. Crit Rev Oncol Hematol. 75:173–234. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chansky K, Sculier JP, Crowley JJ, et al:
International Staging Committee and Participating Institutions: The
International Association for the Study of Lung Cancer Staging
Project: prognostic factors and pathologic TNM stage in surgically
managed non-small cell lung cancer. J Thorac Oncol. 4:792–801.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang W, Ma YZ, Song L, Wang CH, Qi TG and
Shao GR: Effect of cantharidins in chemotherapy for hepatoma: A
retrospective cohort study. Am J Chin Med. 42:561–567. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dorn DC, Kou CA, Png KJ and Moore MA: The
effect of cantharidins on leukemic stem cells. Int J Cancer.
124:2186–2199. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Huh JE, Kang KS, Chae C, Kim HM, Ahn KS
and Kim SH: Roles of p38 and JNK mitogen-activated protein kinase
pathways during cantharidin-induced apoptosis in U937 cells.
Biochem Pharmacol. 67:1811–1818. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li W, Xie L, Chen Z, et al: Cantharidin, a
potent and selective PP2A inhibitor, induces an oxidative
stress-independent growth inhibition of pancreatic cancer cells
through G2/M cell-cycle arrest and apoptosis. Cancer Sci.
101:1226–1233. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yeh CB, Su CJ, Hwang JM and Chou MC:
Therapeutic effects of cantharidin analogues without bridging ether
oxygen on human hepatocellular carcinoma cells. Eur J Med Chem.
45:3981–3985. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chang C, Zhu YQ, Mei JJ, Liu SQ and Luo J:
Involvement of mitochondrial pathway in NCTD-induced cytotoxicity
in human hepG2 cells. J Exp Clin Cancer Res. 29:1452010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Huang WW, Ko SW, Tsai HY, et al:
Cantharidin induces G2/M phase arrest and apoptosis in human
colorectal cancer colo 205 cells through inhibition of CDK1
activity and caspase-dependent signaling pathways. Int J Oncol.
38:1067–1073. 2011.PubMed/NCBI
|
|
14
|
Zhang WD, Zhao HR, Yan Y, Wang XH, Zong ZH
and Liu Y: Apoptosis induced by cantharidin in human pulmonary
carcinoma cells A549 and its molecular mechanisms. Zhonghua Zhong
Liu Za Zhi. 27:330–334. 2005.In Chinese. PubMed/NCBI
|
|
15
|
Kim JA, Kim Y, Kwon BM and Han DC: The
natural compound cantharidin induces cancer cell death through
inhibition of heat shock protein 70 (HSP70) and Bcl-2-associated
athanogene domain 3 (BAG3) expression by blocking heat shock factor
1 (HSF1) binding to promoters. J Biol Chem. 288:28713–28726. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Osborne C, Wilson P and Tripathy D:
Oncogenes and tumor suppressor genes in breast cancer: potential
diagnostic and therapeutic applications. Oncologist. 9:361–377.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Heneghan HM, Miller N and Kerin MJ: MiRNAs
as biomarkers and therapeutic targets in cancer. Curr Opin
Pharmacol. 10:543–550. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Weinstein IB: Cancer. Addiction to
oncogenes – the Achilles heal of cancer. Science. 297:63–64. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Luo J, Solimini NL and Elledge SJ:
Principles of cancer therapy: oncogene and non-oncogene addiction.
Cell. 136:823–837. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Solimini NL, Luo J and Elledge SJ:
Non-oncogene addiction and the stress phenotype of cancer cells.
Cell. 130:986–988. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Méndez M, Custodio A and Provencio M: New
molecular targeted therapies for advanced non-small cell lung
cancer. J Thorac Dis. 3:30–56. 2011.
|
|
22
|
Chang YM, Velmurugan BK, Kuo WW, et al:
Inhibitory effect of alpinate Oxyphyllae fructus extracts on Ang
II-induced cardiac pathological remodeling-related pathways in H9c2
cardio-myoblast cells. BioMedicine. 3:148–152. 2013. View Article : Google Scholar
|
|
23
|
Hsia TC, Yu CC, Hsu SC, et al: Cantharidin
induces apoptosis of H460 human lung cancer cells through
mitochondria-dependent pathways. Int J Oncol. 45:245–254.
2014.PubMed/NCBI
|
|
24
|
Ayala F, Dewar R, Kieran M and Kalluri R:
Contribution of bone microenvironment to leukemogenesis and
leukemia progression. Leukemia. 23:2233–2241. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Straussman R, Morikawa T, Shee K, et al:
Tumour micro-environment elicits innate resistance to RAF
inhibitors through HGF secretion. Nature. 487:500–504. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wilson TR, Fridlyand J, Yan Y, et al:
Widespread potential for growth-factor-driven resistance to
anticancer kinase inhibitors. Nature. 487:505–509. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Moniri MR, Dai LJ and Warnock GL: The
challenge of pancreatic cancer therapy and novel treatment strategy
using engineered mesenchynmal stem cells. Cancer Gene Ther.
21:12–23. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Pignon JP, Tribodet H, Sagliotti GV, et
al: LACE Collaborative Group: Lung adjuvant cisplatin evaluation: A
pooled analysis by the LACE Collaborative Group. J Clin Oncol.
26:3552–3559. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Pepe C, Hasan B, Winton TL, et al National
Cancer Institute of Canada and Intergroup Study FBR.10: Adjuvant
vinorelbine and cisplatin in elderly patients: National Cancer
Institute of Canada and Intergroup Study JBR.10. J Clin Oncol.
25:1553–1561. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Glynne-Jones R and Hoskin P: Neoadjuvant
cisplatin chemotherapy before chemoradiation: A flawed paradigm? J
Clin Oncol. 25:5281–5286. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ricci MS and Zong WX: Chemotherapeutic
approaches for targeting cell death pathways. Oncologist.
11:342–357. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tan TT and White E: Therapeutic targeting
of death pathways in cancer: Mechanisms for activating cell death
in cancer cells. Adv Exp Med Biol. 615:81–104. 2008.PubMed/NCBI
|