Introduction
Cytochrome P450 (CYP)3A4 is the most abundant
hepatic and intestinal CYP450 enzyme in humans, contributing to the
metabolism of most drugs (1). It
shows large intra- and inter-individual variations, which
contribute to marked individual differences in drug responses in
terms of therapeutic effects as well as adverse effects (2). High CYP3A4 activity accelerates the
metabolic rate of drugs, shortens their half-life and reduces their
plasma concentration. Therefore, in individuals with high enzyme
activity, the anticipated therapeutic efficacy of certain drugs may
not be achieved at conventional doses. By contrast, individuals
with low CYP3A4 activity may show decreased drug clearance as well
as prolonged half-life and obvious accumulation of drugs.
Collectively, the variability of CYP3A4 activity or expression may
lead to the uncertainty of the therapeutic efficacy and studies on
the inter-individual variability of CYP3A4 may facilitate the
process of individualized pharmacotherapy and reduce adverse
effects of drugs (1,3).
Inter-individual variability in CYP3A4 expression is
thought to be largely heritable (4,5), and
>30 single nucleotide polymorphisms have been identified in the
CYP3A4 gene. However, the currently known genetic variants at the
CYP3A4 locus are unlikely to account for the proposed high
variability in CYP3A4-associated metabolic function (4,6–9).
CYP3A4 can be induced by a variety of structurally diverse
xenochemicals via xenobiotic receptors (10–19).
For instance, grapefruit juice is a potent inhibitor of
CYP3A4-mediated drug metabolism (20). Under specific pathophysiological
conditions, such as inflammatory stimulation, CYP3A4 can also be
induced, which is a key process involved in the toxic vs.
therapeutic effects of numerous drugs (21,22).
Furthermore, it has been shown that CYP3A4 activity is higher in
women than in men, suggesting enhanced pharmacokinetics in women
(23). In spite of this knowledge,
the detailed underlying mechanisms of the variation of CYP3A4
expression among individuals has remained elusive.
Vault particles are intracellular ribonucleoprotein
particles containing three different proteins and non-coding vault
RNAs (vRNAs), termed as vRNA1, vRNA2 and vRNA3 (24–26),
and are thought to be implicated in multi-drug resistance (10,25,27–29).
Studies on the biological roles of vaults have focused on their
protein components, while vRNAs have not been functionally
characterized. As vRNAs can theoretically fold into structures
resembling micro (mi)RNA precursors, Persson et al (30) sequenced a group of ~23
nucleotide-containing small RNAs matching the RNA components of the
vault particle and discovered the non-coding RNA svRNAb.
svRNAb, a ~23 nucleotide non-coding RNA, is encoded
by the 5′-arm of the stem-loop structure of non-coding vault RNA1
(vRNA1). pre-svRNAb, a ~32 nucleotide containing RNAs derived from
vRNA1 and vRNA2, ends precisely at the 3′ terminus of svRNAb.
svRNAb can associate with Argonaute proteins to guide
sequence-specific cleavage and regulate gene expression in the
pattern similar to that of miRNAs. CYP3A4 has been validated as the
target gene of svRNAb in the MCF7 cell line (30).
In the light of the importance of CYP3A4 in the
liver, the present study was the first to investigate the role of
svRNAb in the regulation of CYP3A4 expression in liver tissue
samples and the HepG2 cell line. The present study also quantified
svRNAb by reverse transcription-quantitative polymerase chain
reaction (RT-qPCR). The results revealed a significant correlation
between CYP3A4 and svRNAb in human liver tissue samples.
Furthermore, a luciferase assay validated that svRNAb directly
targets the 3′-untranslated region (3′-UTR) of CYP3A4 in the HepG2
cell line. Furthermore, the present study suggested a possible role
for svRNAb in drug metabolism via regulation of CYP3A4 expression
in multidrug-resistant cells.
Materials and methods
Sample collection
Human liver tissue samples were obtained from 19 Han
Chinese donors who underwent surgery at the First Affiliated
Hospital of Zhengzhou University (Zhengzhou, China) with the
donors' written informed consent. The present study was approved by
the Clinical Research Ethics Committee of the First Affiliated
Hospital of Zhengzhou University and complied with the Declaration
of Helsinki and its subsequent revisions. A total of 14 males and 5
females (age, 33–60 years; mean age, 50.3 years) were included in
the study. Among these donors, eight had mild cirrhosis and none
had Hepatitis B or C. All donors had normal liver functions. To
avoid any inhibitory or stimulatory effects on CYP3A4 activity,
none of the donors received any pre-operative medication affecting
CYP3A4, such as rifampicin, dexamethasone or propofol for two
weeks, or alcohol, grapefruit juice or caffeine within three days
prior to the surgery.
CYP3A4 enzymatic activity
Liver microsomes were extracted from tissues using
the CaCl2 (Sigma-Aldrich, St. Louis, MO, USA) method.
Liver microsomes were incubated with midazolam (0.25–75 mmol/l;
Sigma-Aldrich) at 37°C for 10 min. The 1-OH midazolam concentration
was measured using an Odyssil C18 HPLC system (Agela Technologies,
Wilmington, DE, USA) with a mobile phase of 20 mM/l ammonium
acetate-acetonitrile, a column temperature of 40°C and a flow rate
of 1 ml/min. Michaelis constants (Km, mM) were estimated to
quantify enzymatic activity with five replicates (31).
RT-qPCR analysis
Approximately 0.1 g of liver was cut into small
pieces and ground with a grinding rod (Shanghai Ding Jie
Technology, Ltd., Shanghai, China) while total RNA was extracted
using 1 ml TRIzol (Life Technologies; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) according to the manufacturer's
instructions, followed by gel electrophoresis for quality control.
Total RNAs were reverse-transcribed to cDNA using a Revert Aid
First Strand cDNA Synthesis kit (Thermo Fisher Scientific, Inc.).
cDNA was then amplified by PCR using FastStart Universal SYBR Green
Master Mix (Roche, Basel, Switzerland) with the following primers
(Invitrogen; Thermo Fisher Scientific, Inc.): CYP3A4 forward,
5′-CCAAGCTATGCTCTTCACCG-3′ and reverse, 5′-TCAGGCTCCACTTACGGTGC-3′;
GAPDH forward, 5′-ATCACCATCTTCCAGGAGCGA-3′ and reverse,
5′-GCTTCACCACCTTCTTGATGT-3′) The thermocycling conditions included
1 cycle at 95°C for 10 min, and 40 cycles of 95°C for 15 sec and
60°C for 1 min. Relative quantities were normalized to GAPDH as the
endogenous control and were calculated using the 2−ΔΔCq
method (32). For svRNAb
quantification, RT-qPCR was performed using TaqMan®
Small RNA Assays (Applied Biosystems; Thermo Fisher Scientific,
Inc.). U6 small nuclear RNA (Invitrogen; Thermo Fisher Scientific,
Inc.) was used as the endogenous control. The cycling conditions
included 1 cycle at 95°C for 10 min, and 40 cycles of 95°C for 15
sec and 60°C for 1 min. All qPCRs were run on a 7900 HT instrument
(Applied Biosystems) in three replicates.
Plasmid construction and sequencing
The target PCR products were recovered and ligated
into the pGMT vector (Tiangen Biotech Co., Beijing, China). Next,
PCR products were further separated using electrophoresis on 2%
agarose gels (Tiangen Biotech Co.). A Zymoclean™ Gel DNA Recovery
kit (Zymo Research Corp., Irvine, CA, USA) was used to recover pure
DNA from agarose gels. Through TA cloning with EcoV (New
England BioLabs, Inc., Ipswich, MA, USA), the PCR products of
svRNAb and pre-svRNAb were inserted into the PGMT vector. The
universal forward primer (Invitrogen; Thermo Fisher Scientific,
Inc.; 3′-TGTAATACGACTCACTATAGGG-5′) was used for sequencing, which
was conducted by Major BioShanghai Technologies Co., Ltd.
(Shanghai, China).
Dual-luciferase assay
HepG2 cells (Fudan University IBS Cell Bank,
Shanghai, China) were cultured in Dulbecco's modified Eagle's
medium (Invitrogen; Thermo Fisher Scientific, Inc.) containing 10%
fetal bovine serum (Invitrogen; Thermo Fisher Scientific, Inc.) at
37°C in a humidified atmosphere of 5% CO2. For
validation of CYP3A4 as a direct target of svRNAb, a luciferase
reporter assay was performed using the pGL3 Luciferase Reporter
Vector containing wild-type (WT) CYP3A4 3′-UTR and mutant (MT)
CYP3A4 3′-UTR. Total RNA was reverse transcribed using a RevertAid
First Strand cDNA Synthesis kit (Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions. A 931 base-pair
fragment of the CYP3A4 3′-UTR covering all three svRNAb seed
matches was amplified from cDNA of the human liver tissue using
specific primers obtained from Invitrogen (Thermo Fisher
Scientific, Inc.; forward, 5′-GAATTCTCTAGATGTGCCTGAGAACACCAGAG-3′
and reverse, 5′-GAATTCTCTAGAACGTGCTTCAAAAAGGCATA-3′) which have
XbaI sites. The PCR products were purified using DNA Clean
& Concentrator™-5 (Zymo Research Corp.) according to the
manufacturer′s instructions. Following the XbaI enzyme
digestion (New England BioLabs, Inc.) of the purified product and
the pGL3 Renilla luciferase vector (Promega Corp. Madison,
WI, USA), the plasmids were ligated with T4 DNA ligase (New England
BioLabs, Inc.). Deletions of seed matches were sequentially
introduced using the KOD-Plus-Mutagenesis kit (Toyobo Co. Ltd.,
Osaka, Japan). The following primers (Invitrogen; Thermo Fisher
Scientific, Inc.) were used: Forward,
5′-ATGCATGTACAGAATCCCCGGTTA-3′ and reverse,
5′-CTCTCATTGTCTGTGTAGAGTGTTATAC-3′ (site 1); forward,
5′-GAGGAGTTAATGGTGCTAACTGG-3′ and reverse,
5′-CTGATAAGAGAATCAACATTTCTCAATAAT-3′ (site 2); and forward,
5′-TTCAACATCCGCCTCCCAGGTT-3′ and reverse
5′-TGAGATTGCACCACTGCACTCC-3′ (site 3). Co-transfection experiments
were performed in 96-well plates. A total of 100 ng WT or MT
reporter constructs and 50 nM svRNAb mimics or negative control
mimics (JIMA, Shanghai, China) were co-transfected into HepG2 cells
using Lipofectamine 2000 transfection reagent (Invitrogen)
according to the manufacturer's instructions. After 48 h,
luciferase activity was measured with the dual luciferase reporter
assay system (Promega Corp.). The relative luciferase activity was
normalized to that of firefly luciferase.
Statistical analysis
Each experiment was performed at least three times.
All values are expressed as the mean ± standard deviation. The
establishment of the standard curve of svRNAb and Spearman's rank
correlation were performed using SPSS for Windows, version 11.0
(International Business Machines, Inc., Armonk, NY, USA). All tests
were two-tailed and P<0.05 was considered to indicate a
statistically significant difference.
Results
Inter-individual variability of hepatic
CYP3A4
CYP3A4 varied considerably at the mRNA expression
level as well as in terms of enzyme activity in the 19 liver tissue
samples assessed in the present study (Table I). The variation was 55-fold and
17-fold for mRNA and enzyme activity, respectively. The coefficient
of variation as a normalized measure of variability was then
calculated to be 89.05 for mRNA expression and 69.61 for enzyme
activity. These values are comparable to those of a previous study
on CYP3A4 variation (7).
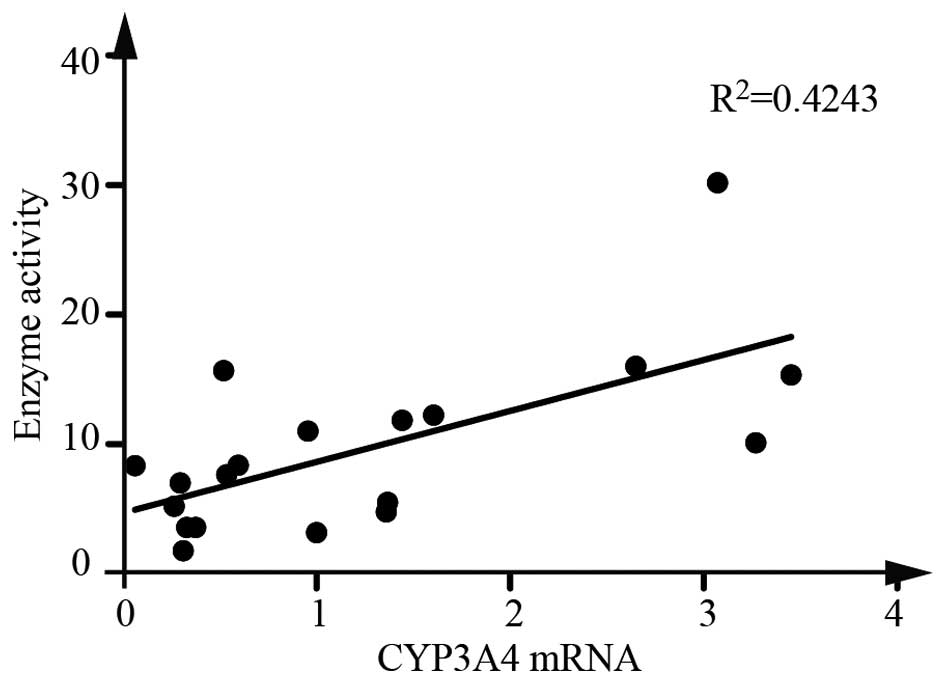
Furthermore, a Spearman correlation analysis revealed that CYP3A4
mRNA expression was significantly correlated with the enzyme
activity (Fig. 1).
 | Table IPopulation variability of hepatic
CYP3A4 expression phenotypes (n=19). |
Table I
Population variability of hepatic
CYP3A4 expression phenotypes (n=19).
| Parameter | CYP3A4/GAPDH
mRNA
Relative units | Enzyme activity |
|---|
| Median | 0.96 | 8.36 |
| Minimum | 0.06 | 1.78 |
| Maximum | 3.45 | 30.17 |
| Ratio max/min | 55.18 | 16.91 |
| Normal
distribution | No | No |
| Coefficient of
variation (%) | 89.05 | 69.61 |
PCR amplification of svRNAb and
pre-svRNAb
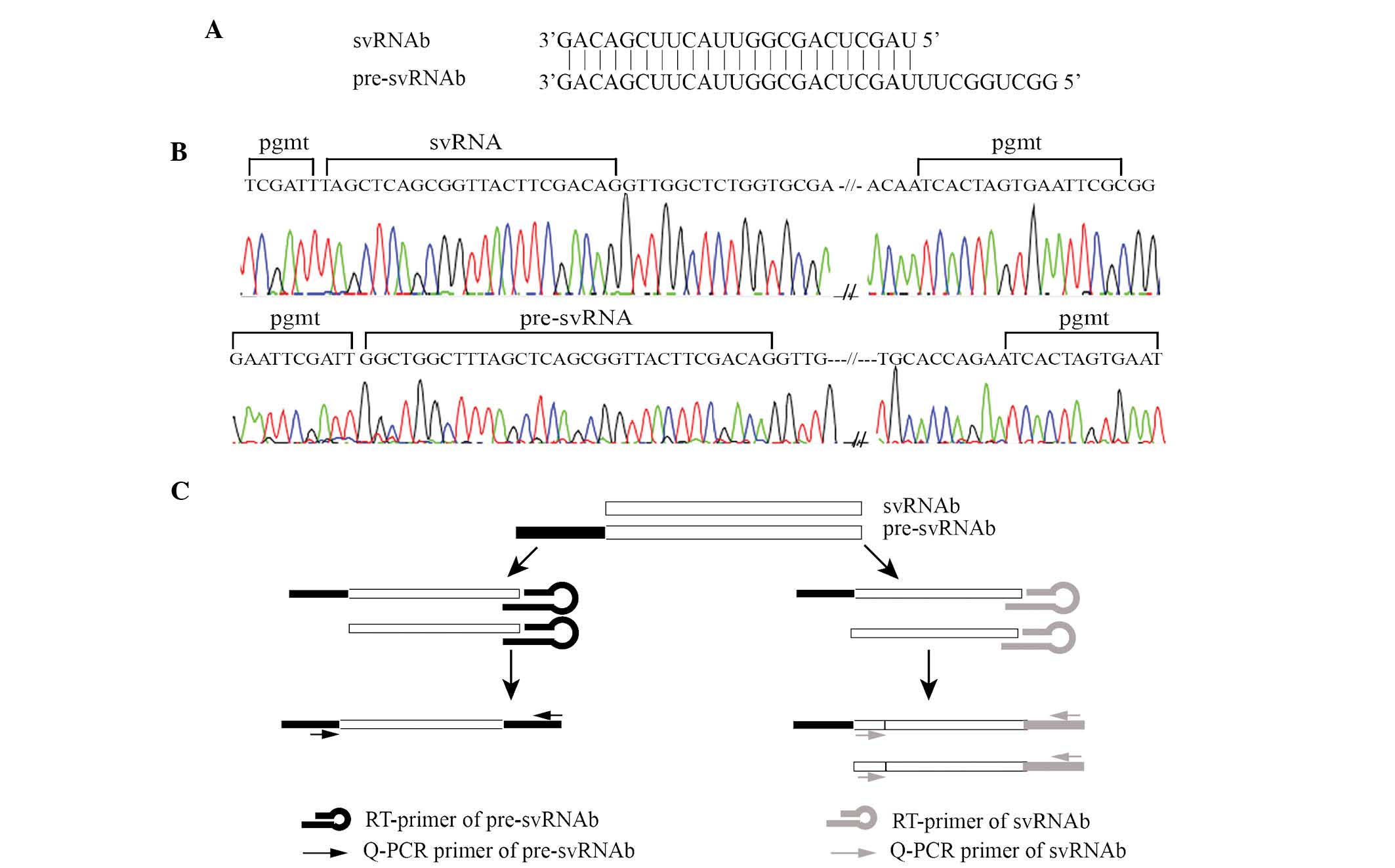
As shown in Fig.
2A, pre-svRNAb has an identical 3′ end to that of svRNAb, while
the former is nucleotides longer at the 5′ end. As the abundance of
svRNAb was expected to be low in liver tissues, the present study
tested whether the PCR primers for svRNAb and pre-svRNAb were able
to produce specific amplification products. Direct sequencing of
amplification products confirmed that the PCR primers efficiently
amplified the specific products (Fig.
2B). Due to the partial structural identify of pre-svRNAb and
svRNAb, the stem loop RT primer can reversely transcribe pre-svRNAb
as well as svRNAb in the RT reactions. The PCR primer for
pre-svRNAb amplified pre-svRNAb only, while the PCR primer for
svRNAb amplified svRNAb and pre-svRNAb (Fig. 2C).
CYP3A4 is correlated with svRNAb in liver
tissue samples
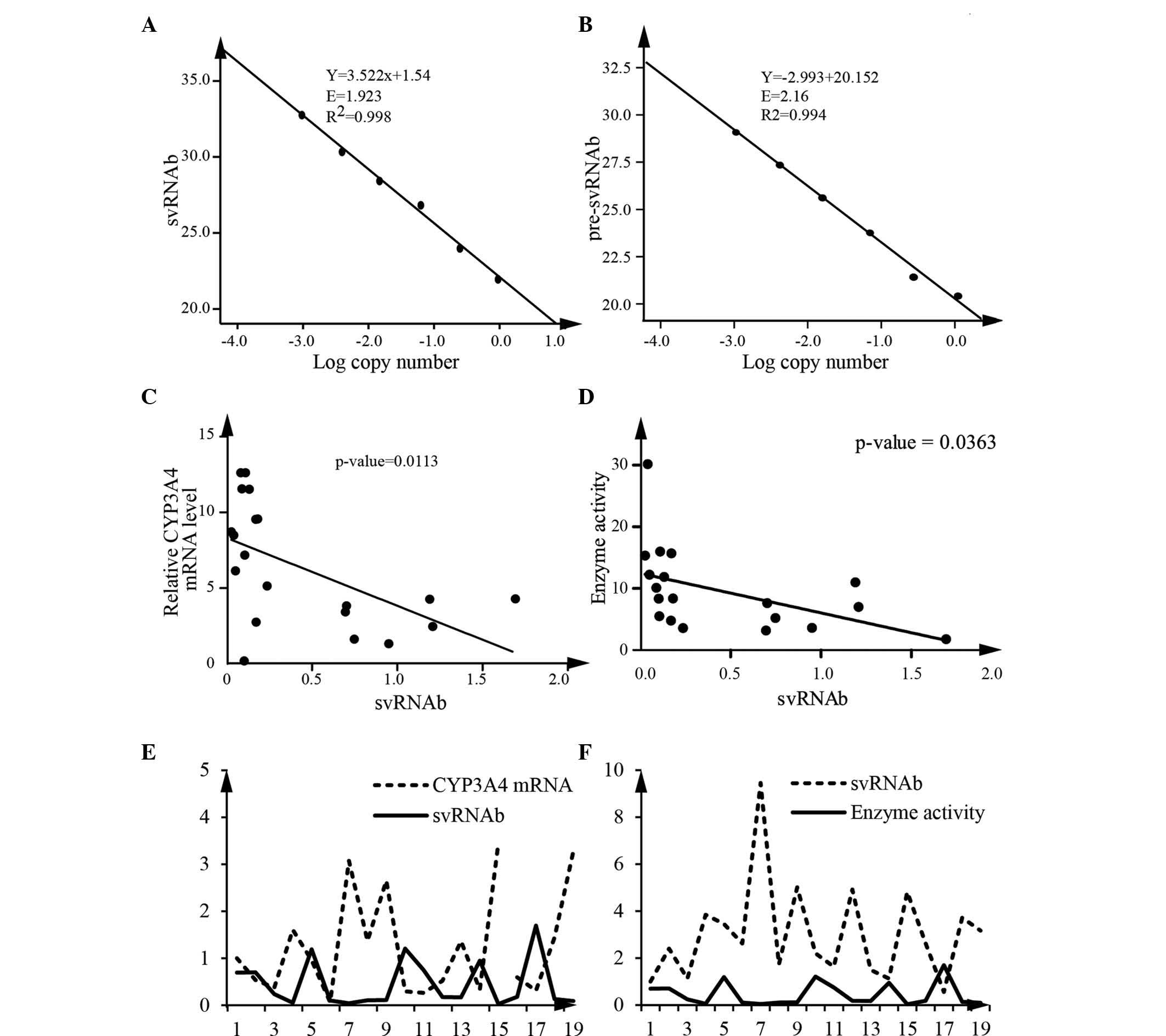
For accurate quantification, qPCR was used to
quantify pre-svRNAb and the sum of svRNAb and pre-svRNAb, with the
primer sets designed for pre-svRNAb and svRNAb, respectively.
First, standard curves were generated. The amplification efficiency
of svRNAb was 1.92 and that of the pre-svRNAb was 2.16 (Fig. 3A and B). The calculated difference
of copy numbers between these two sets of primers was equal to the
copy number of svRNAb. Using this method, the expression levels of
svRNAb in 19 human liver tissue samples were measured. The results
revealed a statistically significant correlation between CYP3A4
mRNA and svRNAb (Fig. 3C). Enzyme
activity of CYP3A4 was also negatively correlated with the svRNAb
(Spearman, P<0.05) (Fig. 3D).
Analysis of 19 liver tissue samples implied that svRNAb had a
negative correlation with CYP3A4 mRNA and enzyme activity,
respectively (Fig. 3E and F). All
of these results indicated that svRNAb regulates CYP3A4 expression
and contributes to the variability of the expression of CYP3A4.
svRNAb directly regulates CYP3A4
expression in HepG2 cells
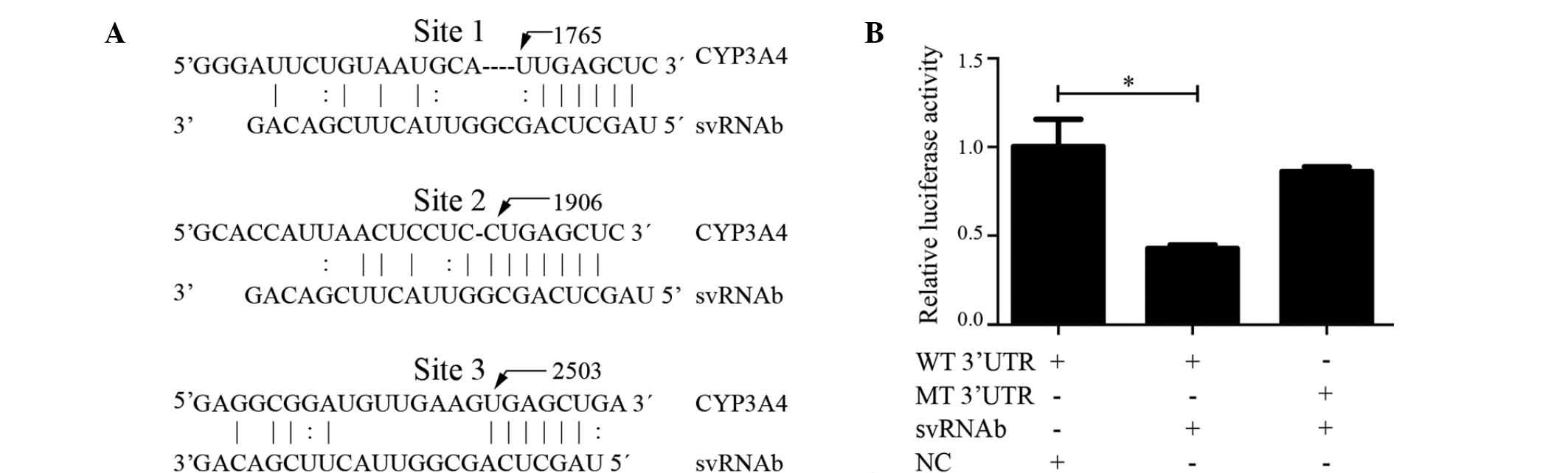
Three binding sites for svRNAb are present in the
3′-UTR of the CYP3A4 transcript (30) (Fig.
4A). To identify whether svRNAb targeted CYP3A4 in HepG2 cells
via these three binding sites, a dual-luciferase reporter gene
assay was performed. Luciferase reporter plasmids containing the
wild-type 3′-UTR (Luc-CYP3A4-wt) or mutant 3′-UTR (Luc-CYP3A4-mt)
of CYP3A4 were constructed to verify the targeted region. The three
validated binding sites in the 3′-UTR were mutated simultaneously
and every corresponding svRNAb binding site had one deletion
mutation. The results showed that svRNAb significantly decreased
the firefly luciferase activity of the reporter vector containing
the wild-type 3′UTR (P<0.05); however, the activity of the
mutant 3′-UTR vector remained unaffected (P>0.05) (Fig. 4B). These results indicated that
svRNAb targeted the CYP3A4 gene through interacting with the three
binding sites in its 3′-UTR in HepG2 cells, which was consistent
with the result of a previous study (30). However, the expression of CYP3A4 in
HepG2 cells was so low that the mRNA and protein was almost
undetectable. Further study is also required to validate the
expression of mRNA and protein after the transfection of svRNAb
mimics into human primary hepatocytes.
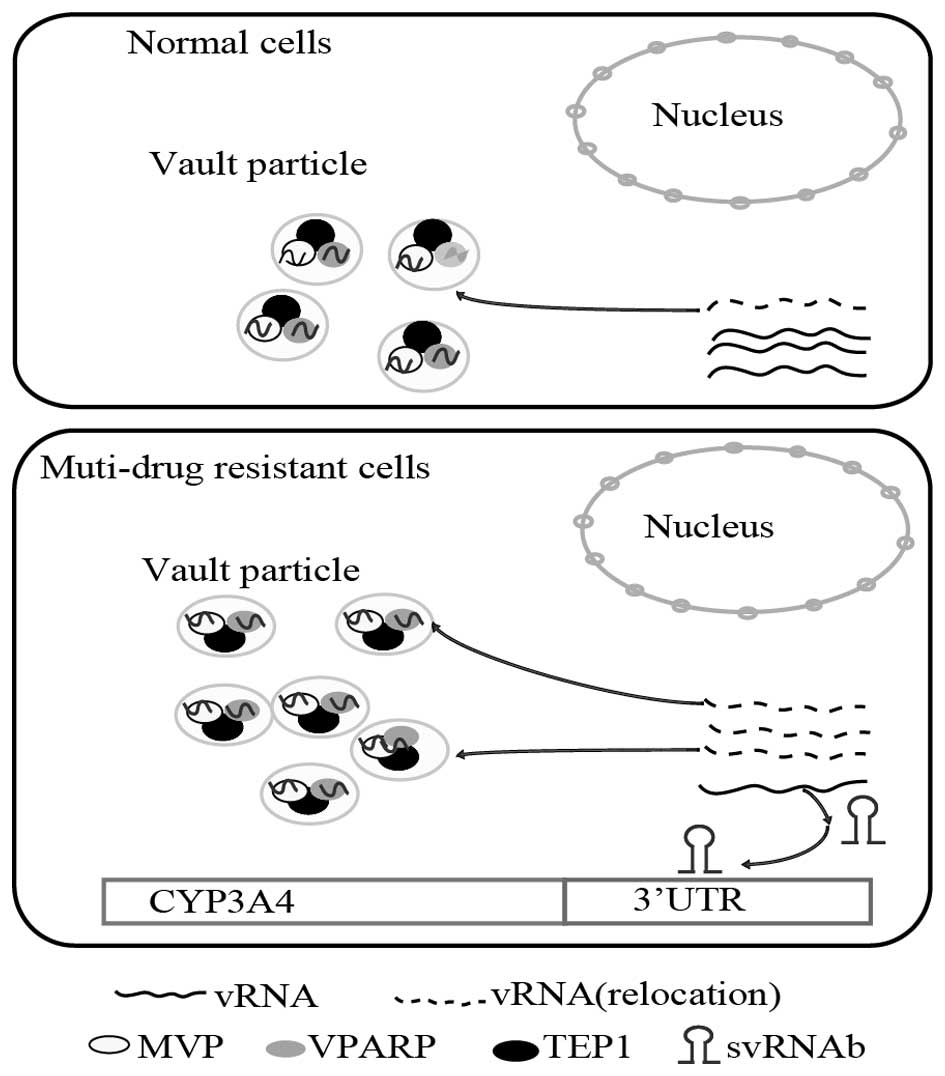
svRNAb may be involved in drug metabolism
by regulating CYP3A4 expression in multidrug resistant cells
Vilalta et al (24) reported a dynamic association
between the vRNA and vault particles, and that there was a pool of
vRNAs, from which a certain fraction was re-located to constitute
the vault particle at any given time (30). The present study demonstrated that
in multi-drug resistant cells, vRNAs were shown to re-locate from
the cytoplasm and form excess vault particles. vRNAs were located
in vault particles as well as being freely abundant in the
cytoplasm. As svRNAb was processed from vRNA1, it was reasonable to
assume that excess formation of vault particles in multi-drug
resistant cell lines would decrease the amount of free vRNA1 in the
cytoplasm (10,26). Thus, the present study speculated
that svRNAb expression was reduced, while CYP3A4 mRNA was augmented
in multi-drug resistant cell lines. The suggested mechanism is
illustrated in the scheme shown in Fig. 5. From this viewpoint, is may be
assumed that svRNAb is involved in drug metabolic mechanisms by
regulating CYP3A4 expression in drug-resistant cells.
Discussion
CYP3A4 is the most abundant hepatic CYP450 enzyme in
humans, contributing to the metabolism of most drugs in current
clinical use. However, the detailed underlying mechanisms of
inter-individual variability with regard to CYP3A4 levels and the
resulting drug responses and metabolism have largely remained
elusive. svRNAb, a newly identified non-coding RNA, has been found
to target the CYP3A4 gene in the MCF7 cell line (30). The present study validated the
association between svRNAb and CYP3A4 in human liver tissue samples
and the HepG2 cell line.
A significant negative correlation was observed
between svRNAb and CYP3A4 expression in human liver tissue samples.
This hypothesis was further confirmed by a luciferase activity
assay, which demonstrated that svRNAb was able to target the 3′UTR
of CYP3A4, and the binding site was consistent with the seed
regions previously reported (30).
These data demonstrated that svRNAb is able to regulate CYP3A4
expression in the liver, which provided insight into the underlying
mechanisms of the inter-individual variability of hepatic CYP3A4
expression.
It is worth pointing out a few limitations and
drawbacks of the present study. Although Persson et al
(30) detected the expression of
svRNAb using RNase protection assays, the expression of svRNA in
liver tissues remained undetectable. Furthermore, the PCR primer
for svRNAb allowed for distinguishing svRNAb from pre-svRNAb. The
present study was the first to quantitatively detect svRNAb
expression using RT-qPCR. However, whether this method can quantify
svRNAb accurately and effectively requires further evaluation. In
addition, the mechanism provided by Persson et al (30) and the present study, suggesting
that svRNAb is involved in drug metabolism by regulating CYP3A4
expression in multi-drug resistant cells, requires additional
verification.
In conclusion, the present study reported that
svRNAb is able to regulate the CYP3A4 expression in the liver,
thereby providing insight into the mechanisms of inter-individual
variability in the therapeutic and toxic effects of drugs.
Acknowledgments
The authors are grateful to all of the participants
of this study. This work was supported by grants from the Shanghai
Municipal Commission of Science and Technology Program (grant no.
14DJ1400100), the National Natural Science Foundation of China
(grant nos. 30971582, 81261120400, 81173127 and 31371274) and the
973 Program (no. 2011CB504501).
References
|
1
|
Westlind A, Malmebo S, Johansson I, Otter
C, Andersson TB, Ingelman-Sundberg M and Oscarson M: Cloning and
tissue distribution of a novel human cytochrome p450 of the CYP3A
subfamily, CYP3A43. Biochem Biophys Res Commun. 281:1349–1355.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sevrioukova IF and Poulos TL:
Understanding the mechanism of cytochrome P450 3A4: recent advances
and remaining problems. Dalton transactions. 42:3116–3126. 2013.
View Article : Google Scholar :
|
|
3
|
de Wildt SN, Kearns GL, Leeder JS and van
den Anker JN: Cytochrome P450 3A: Ontogeny and drug disposition.
Clin Pharmacokinet. 37:485–505. 1999. View Article : Google Scholar
|
|
4
|
Lamba JK, Lin YS, Schuetz EG and Thummel
KE: Genetic contribution to variable human CYP3A-mediated
metabolism. Adv Drug Deliv Rev. 54:1271–1294. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Urquhart BL, Tirona RG and Kim RB: Nuclear
receptors and the regulation of drug-metabolizing enzymes and drug
transporters: Implications for interindividual variability in
response to drugs. J Clin Pharmacol. 47:566–578. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Keshava C, McCanlies EC and Weston A:
CYP3A4 polymorphisms-potential risk factors for breast and prostate
cancer: A HuGE review. Am J Epidemiol. 160:825–841. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wojnowski L and Kamdem LK: Clinical
implications of CYP3A polymorphisms. Expert Opin Drug Metab
Toxicol. 2:171–182. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Perera MA: The missing linkage: What
pharmacogenetic associations are left to find in CYP3A? Expert Opin
Drug Metab Toxicol. 6:17–28. 2010. View Article : Google Scholar
|
|
9
|
Klein K and Zanger UM: Pharmacogenomics of
Cytochrome P450 3A4: Recent progress toward the ῾Missing
Heritability᾿ problem. Front Genet. 4:122013. View Article : Google Scholar
|
|
10
|
Kitazono M, Sumizawa T, Takebayashi Y,
Chen ZS, Furukawa T, Nagayama S, Tani A, Takao S, Aikou T and
Akiyama S: Multidrug resistance and the lung resistance-related
protein in human colon carcinoma SW-620 cells. J Natl Cancer Inst.
91:1647–1653. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Willson TM and Kliewer SA: PXR, CAR and
drug metabolism. Nat Rev Drug Discov. 1:259–266. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Timsit YE and Negishi M: CAR and PXR: The
xenobiotic-sensing receptors. Steroids. 72:231–246. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Klein K, Thomas M, Winter S, Nussler AK,
Niemi M, Schwab M and Zanger UM: PPARA: A novel genetic determinant
of CYP3A4 in vitro and in vivo. Clin Pharmacol Ther. 91:1044–1052.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Oda Y, Nakajima M, Tsuneyama K, Takamiya
M, Aoki Y, Fukami T and Yokoi T: Retinoid X receptor α in human
liver is regulated by miR-34a. Biochem Pharmacol. 90:179–187. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yang J, Hao C, Yang D, Shi D, Song X, Luan
X, Hu G and Yan B: Pregnane X receptor is required for
interleukin-6-mediated down-regulation of cytochrome P450 3A4 in
human hepatocytes. Toxicol Lett. 197:219–226. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Watanabe K, Sakurai K, Tsuchiya Y, Yamazoe
Y and Yoshinari K: Dual roles of nuclear receptor liver X receptor
α (LXRα) in the CYP3A4 expression in human hepatocytes as a
positive and negative regulator. Biochem Pharmacol. 86:428–436.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Goodwin B, Hodgson E, D'Costa DJ,
Robertson GR and Liddle C: Transcriptional regulation of the human
CYP3A4 gene by the constitutive androstane receptor. Mol Pharmacol.
62:359–365. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang K, Chen S, Xie W and Wan YJ:
Retinoids induce cytochrome P450 3A4 through RXR/VDR-mediated
pathway. Biochem Pharmacol. 75:2204–2213. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Thirumaran RK, Lamba JK, Kim RB, Urquhart
BL, Gregor JC, Chande N, Fan Y, Qi A, Cheng C, Thummel KE, et al:
Intestinal CYP3A4 and midazolam disposition in vivo associate with
VDR polymorphisms and show seasonal variation. Biochem Pharmacol.
84:104–112. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jones SA, Moore LB, Shenk JL, Wisely GB,
Hamilton GA, McKee DD, Tomkinson NC, LeCluyse EL, Lambert MH,
Willson TM, et al: The pregnane X receptor: A promiscuous
xenobiotic receptor that has diverged during evolution. Mol
Endocrinol. 14:27–39. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Aitken AE and Morgan ET: Gene-specific
effects of inflammatory cytokines on cytochrome p450 2c, 2b6 and
3a4 mrna levels in human hepatocytes. Drug Metab Dispos.
35:1687–1693. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Dickmann LJ, Patel SK, Wienkers LC and
Slatter JG: Effects of Interleukin 1β (IL-1β) and IL-1β/interleukin
6 (IL-6) combinations on drug metabolizing enzymes in human
hepatocyte culture. Curr Drug Metab. 13:930–937. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wolbold R, Klein K, Burk O, Nüssler AK,
Neuhaus P, Eichelbaum M, Schwab M and Zanger UM: Sex is a major
determinant of CYP3A4 expression in human liver. Hepatology.
38:978–988. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Vilalta A, Kickhoefer VA, Rome LH and
Johnson DL: The rat vault RNA gene contains a unique RNA polymerase
III promoter composed of both external and internal elements that
function synergistically. J Biol Chem. 269:29752–29759.
1994.PubMed/NCBI
|
|
25
|
Kickhoefer VA, Rajavel KS, Scheffer GL,
Dalton WS, Scheper RJ and Rome LH: Vaults are up-regulated in
multidrug-resistant cancer cell lines. J Biol Chem. 273:8971–8974.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
van Zon A, Mossink MH, Schoester M,
Scheffer GL, Scheper RJ, Sonneveld P and Wiemer EA: Multiple human
vault RNAs: expression and association with the vault complex. J
Biol Chem. 276:37715–37721. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kitazono M, Okumura H, Ikeda R, Sumizawa
T, Furukawa T, Nagayama S, Seto K, Aikou T and Akiyama S: Reversal
of LRP-associated drug resistance in colon carcinoma SW-620 cells.
Int J Cancer. 91:126–131. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Patel J and Mitra AK: Strategies to
overcome simultaneous P-glycoprotein mediated efflux and CYP3A4
mediated metabolism of drugs. Pharmacogenomics. 2:401–415. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Siva AC, Raval-Fernandes S, Stephen AG,
LaFemina MJ, Scheper RJ, Kickhoefer VA and Rome LH: Up-regulation
of vaults may be necessary but not sufficient for multidrug
resistance. Int J Cancer. 92:195–202. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Persson H, Kvist A, Vallon-Christersson J,
Medstrand P, Borg A and Rovira C: The non-coding RNA of the
multidrug resistance-linked vault particle encodes multiple
regulatory small RNAs. Nat Cell Biol. 11:1268–1271. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Michaelis L and Menten ML: Die kinetik der
invertinwirkung. Biochem Z. 49:333–369. 1913.
|
|
32
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|