Introduction
Renal cell carcinoma (RCC) is a lethal urological
cancer (1,2) and accounts for ~3% of all adult
malignancies worldwide (3). RCC is
generally asymptomatic and >70% of RCCs are discovered
incidentally during an ultrasound or an abdominal scan requested
for other diseases. Due to the rareness of biomarkers in the early
diagnosis of RCC, >20% of patients with RCC exhibit distant
metastasis at their first diagnosis (4). Furthermore, RCC is resistant to
radiation therapy and chemotherapy, which are usually effective for
other types of adult malignancy. Surgery is the first choice of
treatment for the early or intermediate stage of RCC. However, ~30%
of the patients undergoing nephrectomy for clinically localized RCC
develop relapse at distant sites (5). The 5-year survival rate is 65–90% in
cases without cancer metastasis; however, this is reduced in cases
where the cancer has metastasized (6).
Numerous biomarkers have been recognized as
prognostic for RCC. Increased expression levels of colony
stimulating factor-1 (7) and
overexpressed Aurora A (8) are
predictors of poor prognosis in patients with clear-cell RCC
(7). In addition, RCCRT1, a long
non-coding (lnc) RNA, has been correlated with the RCC prognosis,
and demonstrated to promote migration and invasion of RCC cells
(9). Furthermore, previous studies
have investigated serum and urine biomarkers for RCC (10).
Constitutive photomorphogenic 1 (COP1) belongs to
the COP-de-etiolated (DET)-fusca (FUS) protein family and
has been identified as a tumor suppressor in mouse prostate
adenocarcinomas (11), and liver
(12) and gastric cancers
(13). Previous studies confirmed
that COP1 is deregulated in triple-negative breast cancers, and
thus provides prognostic value for these types of cancer (14,15).
Multiple proteins, including c-Jun and PEA3 family members, have
been identified as substrates for COP1 (16). In addition, COP1 has been confirmed
to promote the degradation of E26 transformation-specific (ETS)
family members, such as ets variant 1 (ETV1) (11). However, the underlying mechanism of
the tumor suppression effect of COP1 remains unknown. In the
present study, the expression levels of COP1 in RCC tissue samples
were investigated and the correlation of COP1 expression levels
with the clinicopathological characteristics of RCC were examined.
Furthermore, the role of COP1 in the proliferation and migration of
ACHN RCC cells was assessed. To the best of our knowledge, the
current study is the first to indicate the tumor suppressive effect
of COP1 in RCC.
Materials and methods
Cell lines, culture and treatment
In the present study, 49 RCC specimens and 49
peritumor specimens (control; ≥10 mm from the tumor edge) were
obtained by surgical resection from patients who were
pathologically confirmed for RCC, and admitted to the First
Affiliated Hospital of Inner Mongolia Medical University (Hohhot,
China) between 2011 and 2014. The ACHN human renal cell line was
purchased from the American Type Culture Collection (Manassas, VA,
USA). The cells were cultured in Gibco Eagle's minimum essential
medium (EMEM; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
supplemented with 10% fetal bovine serum (FBS; Sijiqing, Hangzhou,
China), 100 units/ml penicillin (CSPC Shijiazhuang Pharmaceutical
Group, Shijiazhuang, China) and 100 mg/ml streptomycin (CSPC) in a
humidified incubator under 5% CO2 at 37°C.
To overexpress COP1 in the ACHN cells, the COP1
coding sequence was amplified with a human COPI cDNA clone
(HG19467-CM; Sinobiological, Beijing, China) as template, and with
COP1-specific primer pairs (forward,
5′-TCTAAGCTTATGTCTGGTAGCCGCCAGGCC-3′; reverse,
5′-ATAGGATCCTCATACCAATTCTAGCACCT-3′) under the following
conditions: 95°C for 5 min, 35 cycles of 95°C for 1 min, 56°C for 1
min and 72°C for 2 min followed by 72°C for 10 min. Subsequently,
this was cloned into the eukaryotic pcDNA3.1(+) vector (Invitrogen;
Thermo Fisher Scientific, Inc.). The COP1-pcDNA3.1(+) and control
pcDNA3.1(+) plasmids were transfected into the ACHN cells using
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.).
The COP1-positive, ACHN(COP1+), and the control, ACHN(Con), cell
clones were selected in the presence of 1 mg/ml G418 (Thermo Fisher
Scientific, Inc.) and maintained in medium containing 0.7 mg/ml
G418. To knockdown the COP1 expression in the ACHN(COP1+) cells, 25
or 50 nM COP1-specific small interfering (si)RNA, siRNA-COP1 or
siRNA-Con (Santa Cruz Biotechnology, Inc., Dallas, TX, USA) was
transfected with a HiPerFect Transfection Reagent (Qiagen, GmbH,
Hilden, Germany) into the ACHN(COP1+) cells to abrogate COP1
expression.
RNA isolation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
RCC intra-tumor and peritumor tissue samples were
homogenized and centrifuged at 3,000 × g at 4°C for 15 min to
remove the non-homogenized tissue fraction. Each homogenate and
ACHN cell sample was subjected to the lysis buffer in the Oligotex
Direct mRNA Midi/Maxi kit (Qiagen, GmbH) for mRNA isolation and
total mRNA isolation was performed according to the manufacturer's
instructions. RT-qPCR was performed using the One-Step SYBR
PrimeScript™ RT-PCR kit II (Perfect Real Time; Takara, Tokyo,
Japan) to evaluate COP1, ETV1, matrix metalloproteinase 7 (MMP7)
and β-actin mRNA expression levels. Each reaction system (25
µl for total volume) contained 12.5 µl SYBR Green PCR
Master Mix, 2 µl template mRNA, 1 µl forward and
reverse primer and 8.5 µl ddH2O. The PCR reaction
was performed under the following conditions: An initial
denaturation for 5 min at 95°C, followed by 40 cycles of
denaturation for 20 sec at 94°C, annealing for 20 sec at 61°C and
extension for 20 sec at 72°C, then a final extension for 5 min at
72°C. The primer pairs (Sangon Biotech, Shanghai, China) were as
follows: COP1, forward 5′-ATGAAATGACCTGCAATTCG-3′ and reverse
5′-TCACTGCTAGCTAACAGGTTC-3′; ETV1, forward
5′-CATCCCAGCAGAACAGAAGG-3′ and reverse 5′-CAGGTGTCATCATAAAACTGC-3′;
MMP7, forward 5′-TCATAGAAATAATGCAGAAGC-3′ and reverse
5′-GTGAGTATTCTGCAACATCT-3′; β-actin, foward
5′-CATTAAGGAGAAGCTGTGCT-3′ and reverse 5′-GTTGAAGGTAGTTTCGTGGA-3′.
Data were normalized to β-actin and fold changes were calculated
using the 2−ΔΔCq normalization method (17).
Protein isolation and western blot
analysis
Western blot analysis was performed in RCC tissue
specimens or ACHN cells to detect the protein levels of COP1, ETV1,
MMP7 and β-actin. Briefly, each homogenized tissue sample or
post-treated ACHN cell sample was lysed with ice-cold Cell Lysis
Buffer (Cell Signaling Technology, Inc., Danvers, MA, USA) and was
centrifuged at 9,000 × g for 30 min at 4°C to remove the cellular
debris. Proteins (25 µg) were separated by 12% SDS-PAGE gel
(Sigma-Aldrich, St. Louis, MO, USA), and transferred to
polyvinylidene fluoridehydrophobic membranes (EMD Millipore,
Billerica, MA, USA), which were blocked with 2% bovine serum
albumin (Ameresco, Inc., Framingham, MA, USA) at 4°C overnight.
Subsequent to blocking, the membranes were incubated at 4°C for 2 h
with rabbit polyclonal antibodies against COP1 (ab56400; 1:400;
Abcam, Cambridge, UK), ETV1 (SAB2104467; 1:500; Sigma-Aldrich),
MMP7 (AV46075; 1:500; Sigma-Aldrich) and β-actin (ab8227; Abcam)
and washed three times. Following washes, the membranes were
incubated with horseradish peroxidase-conjugated anti-rabbit IgG
secondary antibody (7074; 1:1000; Cell Signaling Technology, Inc.,
Danvers, MA, USA) for 1 h at 4°C and washed three times. Finally,
the specific binding band was detected with a chemiluminescent
detection reagent (Amersham; GE Healthcare Life Sciences, Chalfont,
UK). The relative expression levels for each protein were
determined as a ratio to β-actin.
Cell counting and colony formation
assays
The growth curve of ACHN(COP1+) and ACHN(Con) cells
was determined using the cell counting assay (18). Briefly, ACHN(COP1+) or ACHN(Con)
cells were seeded in 12-well plates and were incubated at 37°C for
24, 48 or 72 h. The cells were trypsinized and counted using a
hemocytometer (Reicher, Buffalo, NY, USA) and trypan blue
(Sigma-Aldrich). A colony formation assay was conducted to evaluate
the growth of ACHN(COP1+) and ACHN(Con) cells. ACHN(COP1+) and
ACHN(Con) cells were seeded into a 12-well plate (200 cells/well)
and 4 days later, the cell colonies were stained with 0.5% Crystal
violet (Sigma-Aldrich) and counted directly by eye.
Migration and invasion assay
The migration and invasion capabilities of
ACHN(COP1+) and ACHN(Con) cells were assessed using the scratch and
transwell migration assays, respectively. For the scratch assay,
ACHN(COP1+) and ACHN(Con) cells were grown in 6-well plates and,
upon reaching 90% confluency, the plates were scratched using a
200-µl pipette tip. The healing process of the ACHN(COP1+)
and ACHN(Con) cells was observed daily, and the migratory cells
that crossed the wound area were counted at 48 h post inoculation.
For the invasion assay, the experimental procedure followed was the
same as the scratch assay. Briefly, cells were seeded
(1×105 cells) on the upper transwell chamber with the
non-coated membrane (8-µm pore size; Merck Millipore,
Darmstadt, Germany). Once the cells had grown to 2×105
cells, they were coated with Matrigel (Sigma-Aldrich) and the lower
chamber contained EMEM media with 20% FBS as a chemoattractant. The
cells in the upper chamber were discarded after 24 h and the
migratory cells in the lower chamber were counted using a
microscope (BX60; Olympus Corporation, Tokyo, Japan). All the
experiments were repeated in triplicate.
Statistical analysis
Statistical analysis was performed by unpaired
t-test, using the Graphpad Prism software, version 5 (GraphPad
Software, La Jolla, CA, USA) and P<0.05 was considered to
indicate a statistically significant difference.
Results
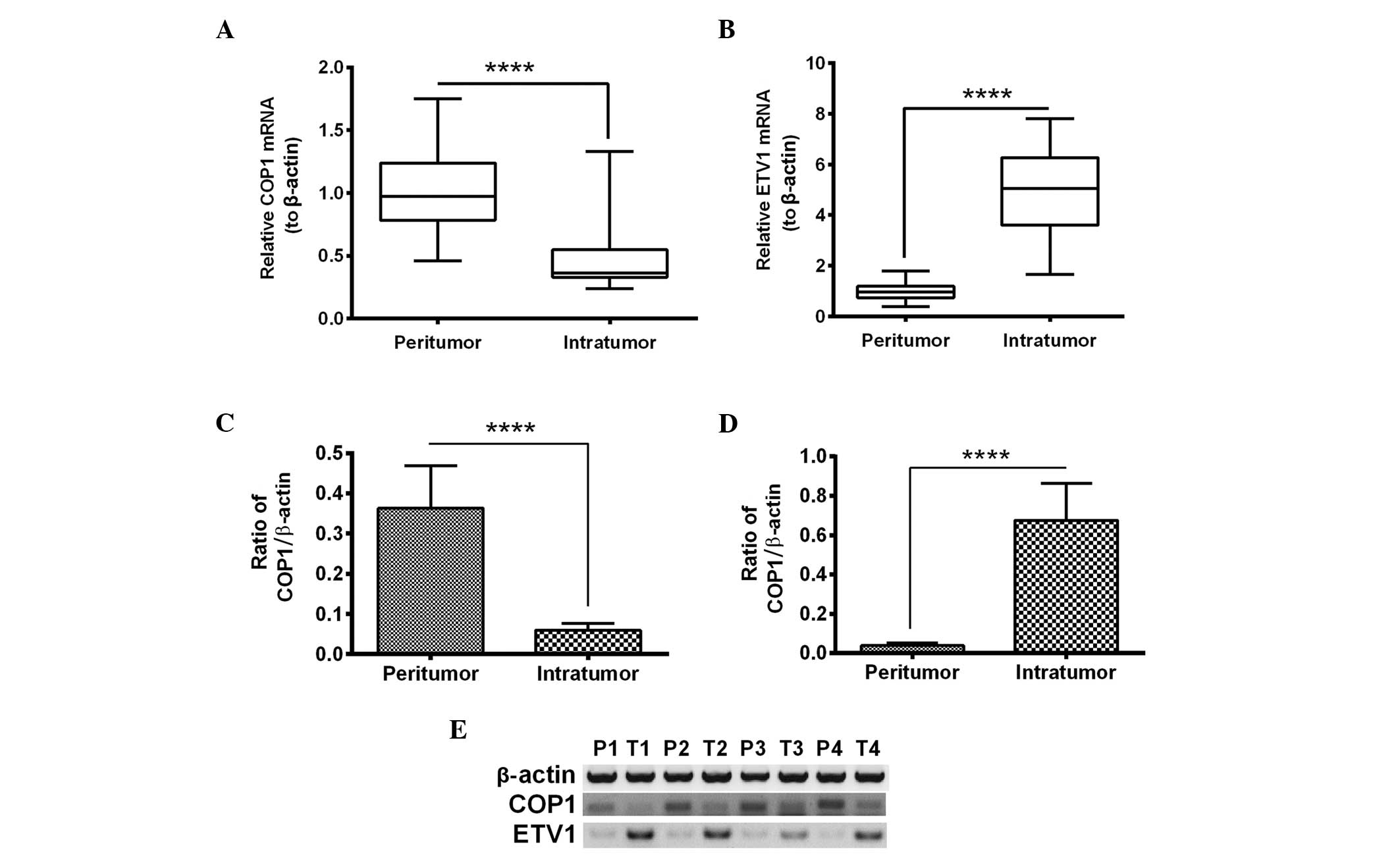
Reduced COP1 and upregulated ETV1 mRNA
and protein expression levels in RCC tissue samples are associated
with tumor malignancy
To investigate the role of COP1 and its downstream
factor, ETV1 in RCC, the expression levels of the two markers were
determined in 49 RCC specimens. The RT-qPCR analysis indicated that
the mRNA expression level of COP1 was significantly lower in the
intratumor tissue samples (0.4704±0.0530) compared with the
peritumor tissue samples (1.000±0.1208) of RCC (Fig. 1A; P<0.0001). The ETV1 mRNA
levels were significantly increased in the intratumor tissue
samples (4.931±0.637) when compared with the peritumor tissue
samples (1.000±0.120) of RCC (Fig.
1B; P<0.0001). The down- and upregulated expression levels
of COP1 and ETV1, respectively, were also confirmed at the protein
level by western blot analysis (P<0.0001; Fig. 1C–E).
Association of COP1 expression with the
clinicopathological characteristics of RCC patients
As demonstrated in Table I, no significant association was
observed between gender or age and COP1 expression level in
patients with RCC. However, COP1 expression was negatively
associated with the tumor size (P=0.006), tumor-node-metastasis
(TNM) stage (P=0.012), lymph node metastasis (P=0.010) and distant
metastasis (P=0.023; Table I). The
COP1 expression level in patients with larger tumors (>5 cm),
lymph node metastasis, increased TNM stage or distant metastasis
was significantly lower (Table
I).
 | Table IClinicopathological characteristics of
patients with renal cell carcinoma. |
Table I
Clinicopathological characteristics of
patients with renal cell carcinoma.
| Characteristic | Patients (n) | Fold-change of COP1
(mean ± standard deviation) | P-value |
|---|
| Gender | | 0.4612±0.0481 | 0.485 |
| Male | 27 | | |
| Female | 22 | 0.4816±0.0502 | |
| Age (years) | | | 0.228 |
| ≤55 | 21 | 0.4279±0.0432 | |
| >55 | 28 | 0.5023±0.0534 | |
| Size (cm) | | | 0.006 |
| ≤5.0 | 19 | 0.5791±0.0623 | |
| >5.0 | 30 | 0.4016±0.0452 | |
| TNM stage | | | 0.012 |
| I+II | 37 | 0.4992±0.0516 | |
| III+IV | 12 | 0.3815±0.0412 | |
| Lymph node
metastasis | | | 0.010 |
| Yes | 17 | 0.3903±0.0413 | |
| No | 32 | 0.5129±0.0530 | |
| Distant
metastasis | | | 0.023 |
| Yes | 15 | 0.3982±0.0413 | |
| No | 34 | 0.5023±0.0562 | |
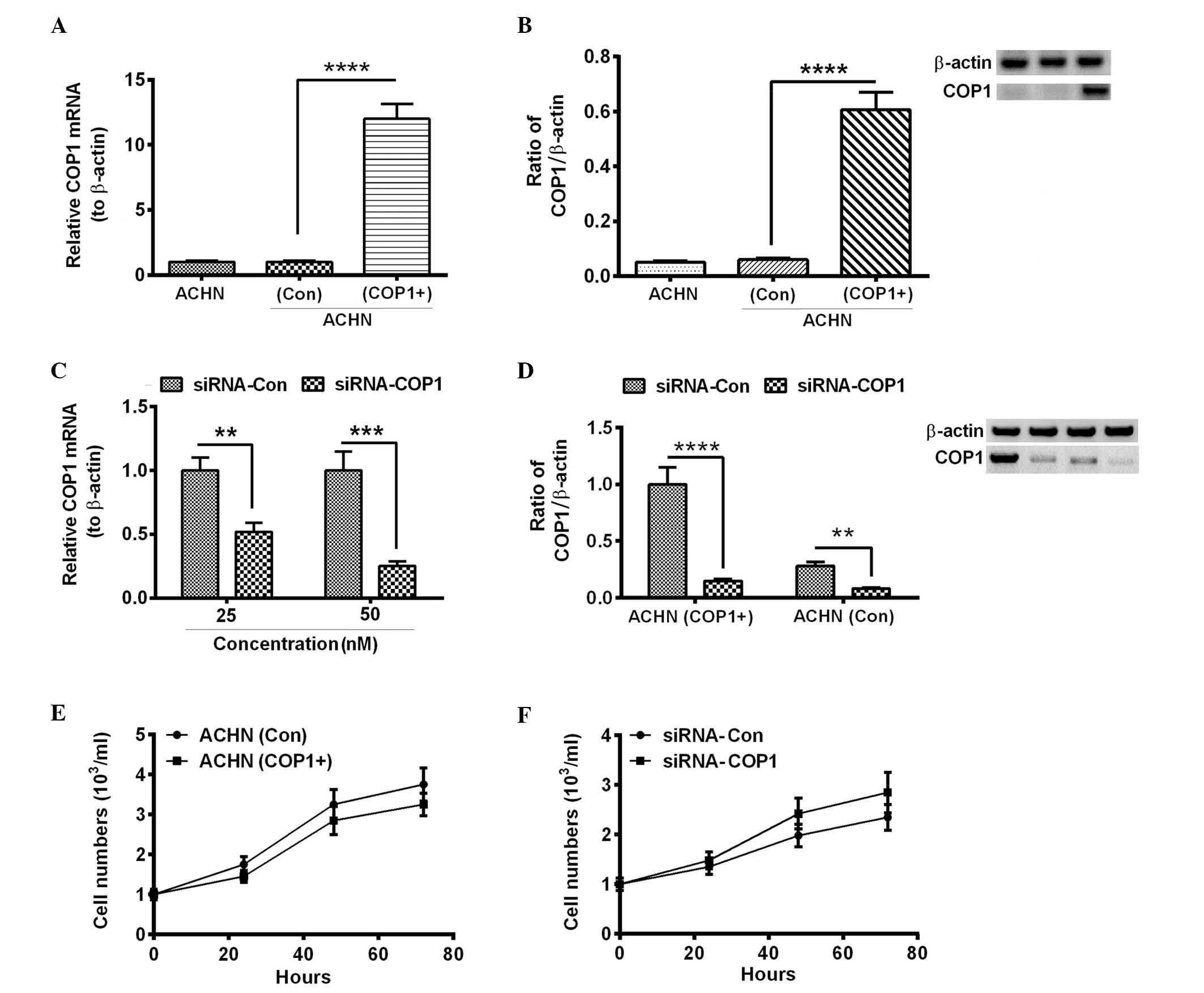
Influence of COP1 overexpression on the
growth of RCC AHCN cells
To further investigate the role of COP1 in the RCC
cells, COP1 was overexpressed via a eukaryotic vector, pcDNA3.1(+).
The coding sequence of COP1 was cloned into pcDNA3.1(+) and the
recombinant plasmid was transfected into the AHCN cells. As
demonstrated in Fig. 2A and B, the
relative COP1 mRNA and protein levels were significantly increased
in the AHCN cells transfected with the recombinant COP1-pcDAN3.1(+)
plasmid compared with the AHCN(Con) cells (P<0.0001). The effect
of COP1 on the growth of RCC AHCN cells was investigated by
silencing COP1 expression. To knockdown COP1, AHCN(COP1+) cells
were transfected with COP1-specific siRNA (siRNA-COP1). As
demonstrated in Fig. 2C, the
relative mRNA levels of COP1 were significantly reduced by
transfection with 25 and 50 nM siRNA-COP1 compared with the
siRNA-Con (P<0.01 and P<0.001, respectively). As demonstrated
in Fig. 2D, the relative protein
levels of COP1 were significantly reduced by the siRNA-COP1
transfection compared with the siRNA-Con in the AHCN(COP1+) and
AHCN(Con) groups (P<0.0001 and P<0.01, respectively).
Following establishment of a successful knockdown
model, the influence of COP1 overexpression on the growth of RCC
AHCN cells was assessed using cell counting and colony formation
assays. AHCN(COP1+) and AHCN(Con) cells were seeded with an initial
titer of 103 cells/ml in 12-well plates, and the cell
number was counted every 24 h post-seeding. In a separate
experiment, the seeded cells were transfected with siRNA-COP1 or
siRNA-Con and the cell numbers were counted. As demonstrated in
Fig. 2E, the titer of the
AHCN(COP1+) cells was lower when compared with the AHCN(Con) cells;
however, no significant difference was identified between the two
groups. Furthermore, the transfection of siRNA-COP1 did not
significantly influence the proliferation of AHCN(COP1+) cells
(Fig. 2F; P>0.05). In addition,
the overexpression of COP1 did not demonstrate a significant change
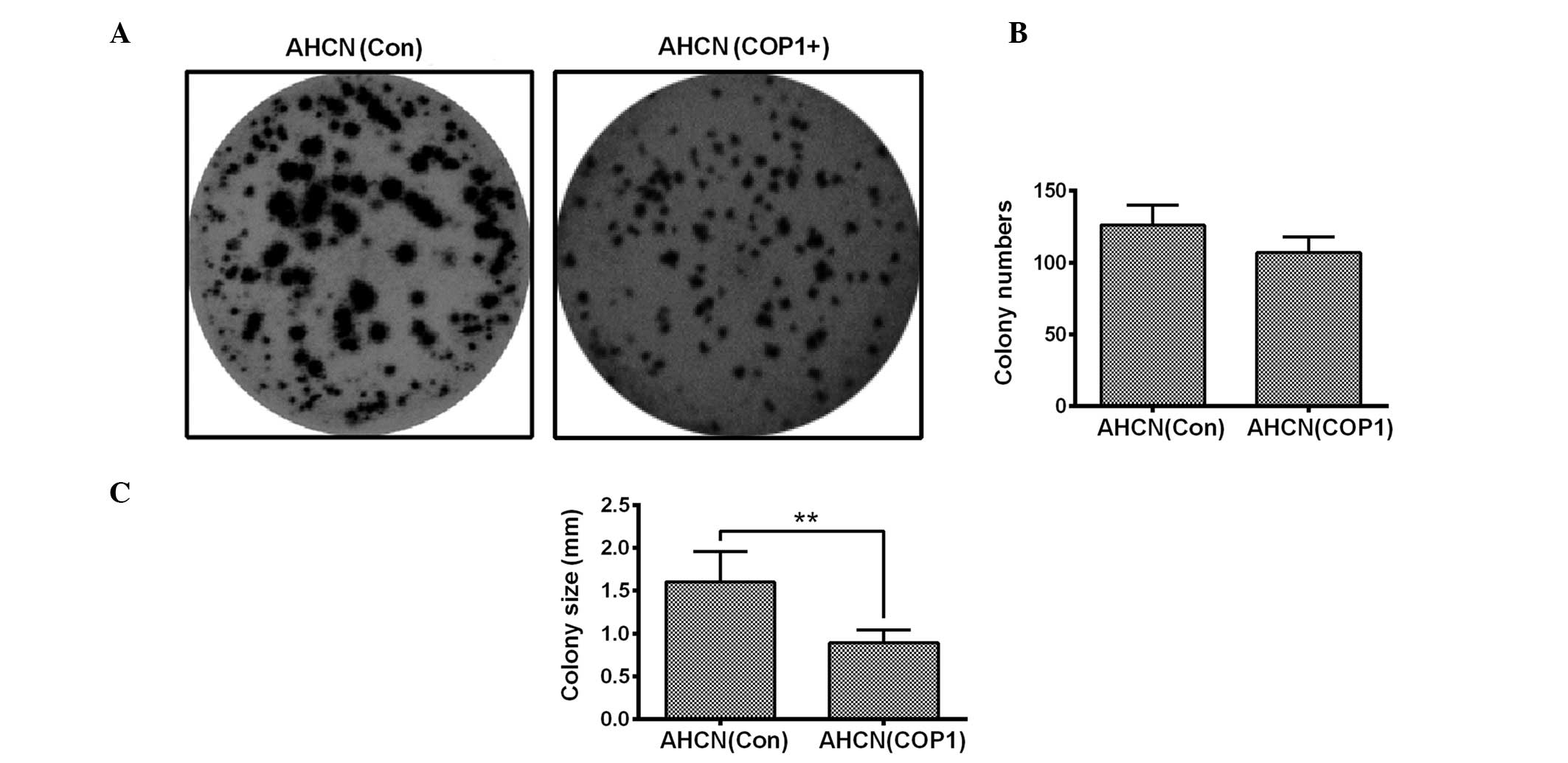
in the colony numbers of AHCN cells (Fig. 3A and B). However, COP1
overexpression resulted in a significantly reduced colony size when
compared with those of the AHCN(Con) group (Fig. 3C; P<0.05). These data indicate
that COP1 overexpression exerts a limited influence on the growth
of RCC cells.
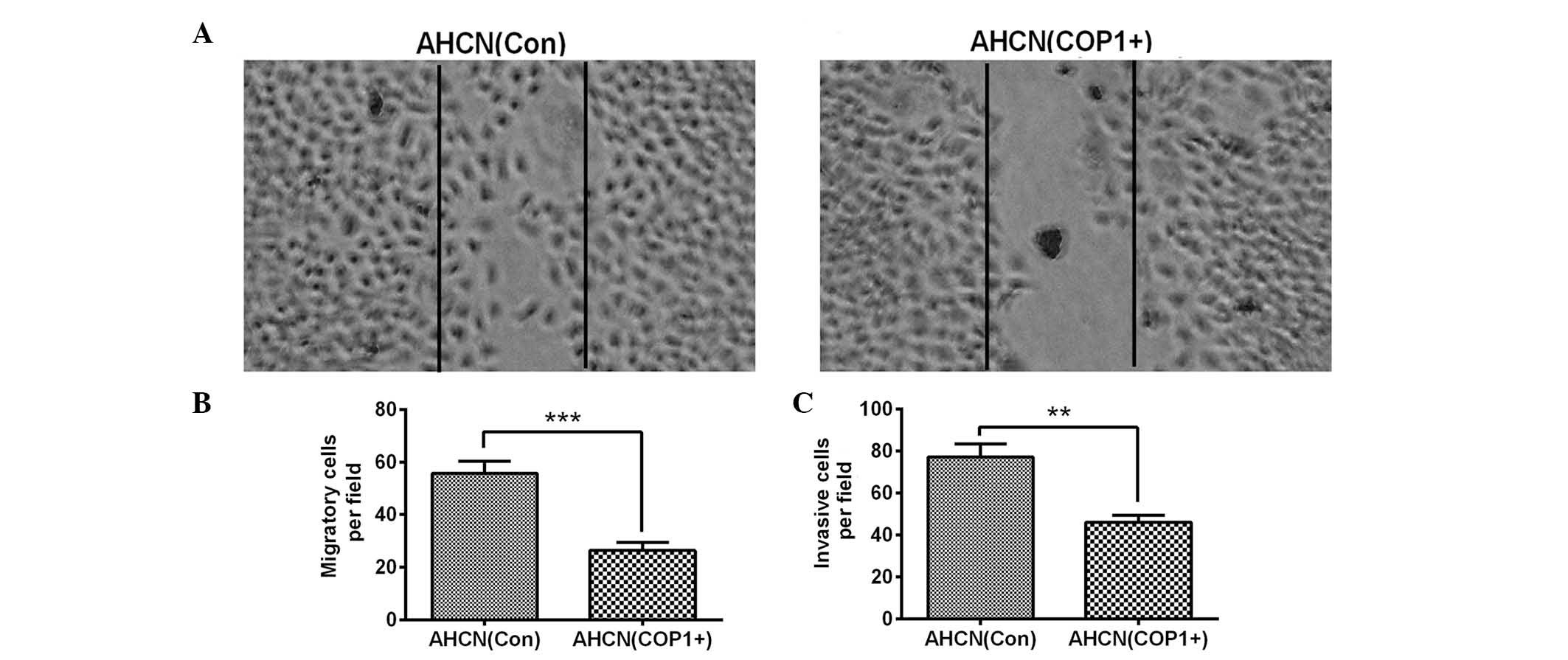
COP1 overexpression inhibits the
migration of RCC AHCN cells in vitro
It has been proposed that cell migration contributes
to tumor metastasis (19). To
further investigate the regulatory role of COP1 in the progression
of RCC, the migration of AHCN(COP1+) and AHCN(Con) cells was
assessed via scratch assay. The results demonstrated fewer
migratory cells in the AHCN(COP1+) group when compared with the
AHCN(Con) group (Fig. 4A and B;
P<0.001). To further evaluate the invasion of AHCN(COP1+) and
AHCN(Con) cells, the Matrigel-coated transwell assay was utilized.
The results demonstrated a reduced number of invasive cells per
field in the AHCN(COP1+) group when compared with the AHCN(Con)
group (Fig. 4C; P<0.01). The
results from the assays indicated that COP1 overexpression reduced
the migration and invasion of RCC cells in vitro.
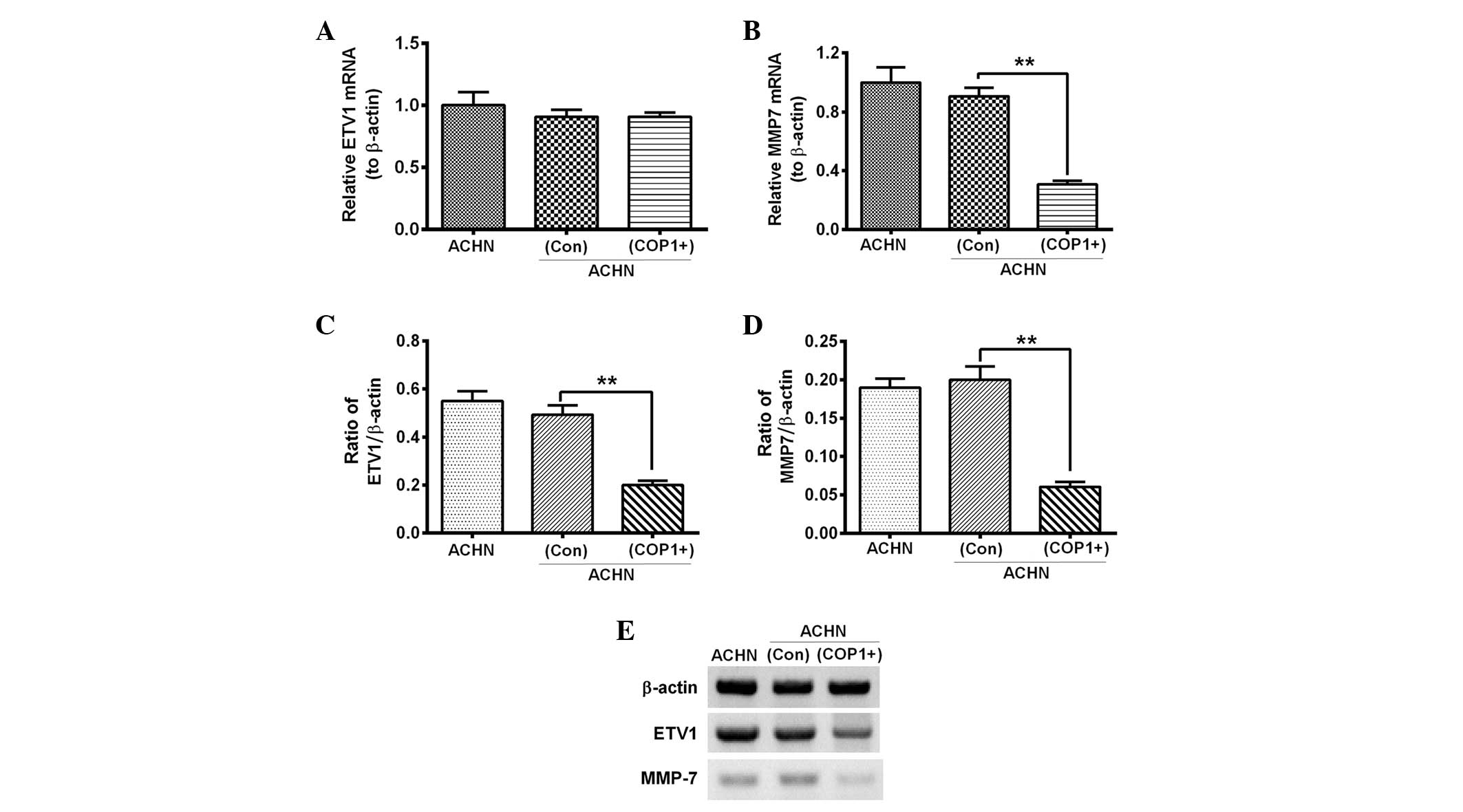
Overexpressed COP1 reduces MMP7 via the
degradation of ETV1 in RCC AHCN cells
COP1 has been indicated to promote the degradation
of ETV1 (14). To deduce the
inhibition to the migration of AHCN cells by overexpressed COP1,
the ETV1 and MMP7 expression levels in the AHCN(COP1+) and
AHCN(Con) cells were assessed. No significant difference was
observed in the ETV1 mRNA expression levels between the AHCN(COP1+)
and AHCN(Con) groups (Fig. 5A;
P<0.05). However, the MMP7 mRNA expression levels were
significantly downregulated in the AHCN(COP1+) group compared with
the AHCN(Con) group (Fig. 5B;
P<0.01). Furthermore, the protein levels of the two markers were
significantly downregulated in the AHCN(COP1+) group compared with
the AHCN(Con) group (Fig. 5C–E;
P<0.01). These data indicate that MMP7 is significantly
inhibited by the overexpression of COP1.
Discussion
The present study investigated the expression of
COP1 in RCC cells and demonstrated that the mRNA and protein
expression levels of COP1 were significantly downregulated in 49
RCC tissue samples. ETV1, the targeting marker of COP1, was
significantly upregulated in these 49 RCC specimens. Despite being
deregulated in other types of tumor, such as prostate
adenocarcinomas (11), and liver
(12) and gastric (13) cancers, as well as being associated
with the prognosis of these types of tumor, the exact role of COP1
in tumorigenesis is not fully understood. The present study
indicated that the overexpression of COP1 in RCC ACHN cells did not
have a regulatory effect on the growth of ACHN cells, implying only
a marginal influence on the proliferation of RCC cells. However,
the colony formation assay demonstrated that COP1 significantly
inhibited the size of the colonies formed by ACHN cells. In
addition, the scratch and transwell assays confirmed the
significant inhibitory effect of COP1 on ACHN cell migration and
invasion.
COP1 belongs to the COP-DET-FUS protein family,
possesses E3 ubiquitin ligase activity and is involved in the
ubiquitylation of various protein substrates to trigger their
proteasomal degradation (20). The
ubiquitin ligase, COP1 negatively regulates ETV1, 4 and 5 via
degrading ubiquitinated ETVs in prostate epithelial cells (11). COP1 has been recognized to regulate
the stability and the transcriptional activity of PEA3 factor,
which is dependent on the RING finger domain of COP1 (21). In the present study, a upregulated
ETV1 in the mRNA and protein levels was demonstrated in the 49 RCC
specimens, which was associated with downregulated COP1 expression.
Furthermore, the in vitro experiments confirmed the
downregulation of ETV1 by COP1 in the ACHN cells. These findings
indicate that the downregulated ETV1 expression may have
contributed to the reduced migration and invasion of the ACHN
cells. This further implies that reduced COP1 expression and the
upregulated ETV1 may contribute to the aggressive nature of RCC
cells in vivo.
MMP7 is a zinc- and calcium-dependent endopeptidase
that cleaves a broad range of extracellular matrix macromolecules,
such as fibronectin, laminin, proteoglycan, elastin, gelatin, type
IV collagen and 1-antitrypsin (22,23).
The over-expression of MMP7 has been reported in numerous types of
cancerous tissue, such as colorectal (24), ovarian (25) and pancreatic (26) cancer. Furthermore, an increased
expression level of MMP7 was demonstrated in high-grade RCC tumors
(27). A previous study
demonstrated that ETV1 bound to the MMP7 gene promoter and
stimulated MMP7 expression (28).
The present study demonstrated that overexpressed COP1 reduced the
MMP7 expression levels, along with the downregulation of ETV1.
Thus, the reduced expression levels of MMP7 may contribute to the
reduced migration of ACHN cells that overexpressed COP1.
In conclusion, the current study confirmed the
reduced expression of COP1, along with the upregulated ETV1 in the
RCC tissue specimens, which was associated with a high TNM stage of
RCC. In addition, the overexpression of COP1 in the RCC ACHN cells
inhibited the migration and invasion of the ACHN cells, and
downregulated the expression levels of ETV1 and MMP7. Thus, the
present study confirmed the tumor suppressive role of COP1 in RCC
via inhibiting cell migration, and suggests that COP1 may be a
valuable target for the anti-cancer treatment of RCC.
References
|
1
|
Rini BI, Campbell SC and Escudier B: Renal
cell carcinoma. Lancet. 373:1119–1132. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rini BI, Rathmell WK and Godley P: Renal
cell carcinoma. Curr Opin Oncol. 20:300–306. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Motzer RJ, Bander NH and Nanus DM:
Renal-cell carcinoma. N Engl J Med. 335:865–875. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Itsumi M and Tatsugami K: Immunotherapy
for renal cell carcinoma. Clin Dev Immunol. 2010:2845812010.
View Article : Google Scholar
|
|
5
|
Pantuck AJ, Zisman A and Belldegrun AS:
The changing natural history of renal cell carcinoma. J Urol.
166:1611–1623. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Singer EA, Gupta GN, Marchalik D and
Srinivasan R: Evolving therapeutic targets in renal cell carcinoma.
Curr Opin Oncol. 25:273–280. 2013.PubMed/NCBI
|
|
7
|
Yang L, Wu Q, Xu L, Zhang W, Zhu Y, Liu H,
Xu J and Gu J: Increased expression of colony stimulating factor-1
is a predictor of poor prognosis in patients with clear-cell renal
cell carcinoma. BMC Cancer. 15:672015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ferchichi I, Kourda N, Sassi S, Romdhane
KB, Balatgi S, Cremet JY, Prigent C and Elgaaied AB: Aurora A
overexpression and pVHL reduced expression are correlated with a
bad kidney cancer prognosis. Dis Markers. 33:333–340. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Song S, Wu Z, Wang C, Liu B, Ye X, Chen J,
Yang Q, Ye H, Xu B and Wang L: RCCRT1 is correlated with prognosis
and promotes cell migration and invasion in renal cell carcinoma.
Urology. 84(3): 7302014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Pastore AL, Palleschi G, Silvestri L,
Moschese D, Ricci S, Petrozza V, Carbone A and Di Carlo A: Serum
and urine biomarkers for human renal cell carcinoma. Dis Markers.
2015:2514032015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Vitari AC, Leong KG, Newton K, Yee C,
O'Rourke K, Liu J, Phu L, Vij R, Ferrando R, Couto SS, et al: COP1
is a tumour suppressor that causes degradation of ETS transcription
factors. Nature. 474:403–406. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xu S, Tong M, Huang J, Zhang Y, Qiao Y,
Weng W, Liu W, Wang J and Sun F: TRIB2 inhibits Wnt/β-Catenin/TCF4
signaling through its associated ubiquitin E3 ligases, β-TrCP, COP1
and Smurf1, in liver cancer cells. FEBS Lett. 588:4334–4341. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sawada G, Ueo H, Matsumura T, Uchi R,
Ishibashi M, Mima K, Kurashige J, Takahashi Y, Akiyoshi S, Sudo T,
et al: Loss of COP1 expression determines poor prognosisin patients
with gastric cancer. Oncol Rep. 30:1971–1975. 2013.PubMed/NCBI
|
|
14
|
Ouyang M, Wang H, Ma J, Lü W, Li J, Yao C,
Chang G, Bi J, Wang S and Wang W: COP1, the negative regulator of
ETV1, influences prognosis in triple-negative breast cancer. BMC
Cancer. 15:1322015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shao J, Teng Y, Padia R, Hong S, Noh H,
Xie X, Mumm JS, Dong Z, Ding HF, Cowell J, et al: COP1 and GSK3β
cooperate to promote c-Jun degradation and inhibit breast cancer
cell tumorigenesis. Neoplasia. 15:1075–1085. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wei W and Kaelin WG Jr: Good COP1 or bad
COP1? In vivo veritas. J Clin Invest. 121:1263–1265. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
18
|
Li Z, Tian T, Lv F, Chang Y, Wang X, Zhang
L, Li X, Li L, Ma W, Wu J and Zhang M: Six1 promotes proliferation
of pancreatic cancer cells via upregulation of cyclin D1
expression. PLoS One. 8:e592032013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Marine JC: Spotlight on the role of COP1
in tumorigenesis. Nat Rev Cancer. 12:455–464. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Papoutsopoulou S and Janknecht R:
Phosphorylation of ETS transcription factor ER81 in a complex with
its coactivators CREB-binding protein and p300. Mol Cell Biol.
20:7300–7310. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Murphy G, Cockett MI, Ward RV and Docherty
AJ: Matrix metalloproteinase degradation of elastin, type IV
collagen and proteoglycan. A quantitative comparison of the
activities of 95 kDa and 72 kDa gelatinases, stromelysins-1 and -2
and punctuated metalloproteinase (PUMP). Biochem J. 277:277–279.
1991. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sires UI, Murphy G, Baragi VM, Fliszar CJ,
Welgus HG and Senior RM: Matrilysin is much more efficient than
other matrix metalloproteinases in the proteolytic inactivation of
alpha 1-anti-trypsin. Biochem Biophys Res Commun. 204:613–620.
1994. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Adachi Y, Yamamoto H, Itoh F, Arimura Y,
Nishi M, Endo T and Imai K: Clinicopathologic and prognostic
significance of matrilysin expression at the invasive front in
human colorectal cancers. Int J Cancer. 95:290–294. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tanimoto H, Underwood LJ, Shigemasa K,
Parmley TH, Wang Y, Yan Y, Clarke J and O'Brien TJ: The matrix
metalloprotease pump-1 (MMP-7, Matrilysin): A candidate
marker/target for ovarian cancer detection and treatment. Tumour
Biol. 20:88–98. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yamamoto H, Itoh F, Iku S, Adachi Y,
Fukushima H, Sasaki S, Mukaiya M, Hirata K and Imai K: Expression
of matrix metalloproteinases and tissue inhibitors of
metalloproteinases in human pancreatic adenocarcinomas:
Clinicopathologic and prognostic significance of matrilysin
expression. J Clin Oncol. 19:1118–1127. 2001.PubMed/NCBI
|
|
27
|
Sumi T, Nakatani T, Yoshida H, Hyun Y,
Yasui T, Matsumoto Y, Nakagawa E, Sugimura K, Kawashima H and
Ishiko O: Expression of matrix metalloproteinases 7 and 2 in human
renal cell carcinoma. Oncol Rep. 10:567–570. 2003.PubMed/NCBI
|
|
28
|
Rahim S, Minas T, Hong SH, Justvig S,
Çelik H, Kont YS, Han J, Kallarakal AT, Kong Y, Rudek MA, et al: A
small molecule inhibitor of ETV1, YK-4-279, prevents prostate
cancer growth and metastasis in a mouse xenograft model. PLoS ONE.
9:e1142602014. View Article : Google Scholar : PubMed/NCBI
|