Introduction
PET with 18F-fluorodeoxyglucose (18FDG)
has been applied specifically to differentiate malignant and benign
nodules. Several reports have indicated that the number of patients
with pulmonary nodules who receive avoidable surgical biopsies is
reduced by PET (1). PET with
18FDG is a noninvasive and accurate method to diagnose
single nucleotide polymorphisms (SNPs), with an overall specificity
of 82% and sensitivity of 95% (2).
However, in a substantial number of patients, surgical resection is
required to differentiate malignant and benign nodules (3). The combining of PET and computed
tomography (CT) has demonstrated superior properties in identifying
a solitary pulmonary nodule (SPN) as malignant or benign, and the
specificity of PET and sensitivity of CT lead to markedly enhanced
accuracy overall (4).
The uptake of FDG on PET can be semi-quantitatively
and qualitatively evaluated. Visual assessment, which based on
comparisons of FDG uptake between the normal mediastinal blood pool
and lesion is the easiest method, however, it is difficult to
visually evaluate the nodules with comparable FDG uptake to the
mediastinum (5). Consequently, a
cut-off at the standard uptake value (SUVmax) is used to confirm
malignancy. However, the SUV is affected by a series of factors,
including lesion diameter, time following injection, blood glucose
concentration and body size (6).
Consequently, the SPN SUVmax may not represent true conditions.
As a group of non-coding small RNAs, microRNAs
(miRNAs) regulate the expression of genes at the
post-transcriptional level (7).
miRNAs induce translational repression or mRNA degradation by
targeting to the complementary sequences in the 3′untranslated
regions (3′UTRs) of their target RNAs (8). Numerous studies have demonstrated
that miRNAs are important in the maintenance, development and
progression of diseases, including cancer (9). Accumulating evidence reports that
miRNAs are involved in the progression of Ewing sarcoma, providing
novel perspectives for applications in Ewing sarcoma therapy and
diagnosis (10).
Hypoxia-inducible factor 1α (HIF1A)-activated
transcription pathways are involved in the regulation of vascular
endothelial growth factor (VEGF) genes and the expression of
downstream solute carrier family 2, member 1. Under HIF1A
induction-dependent hypoxic conditions, VEGF serves as the key
mediator of transcription, vascular permeability and angiogenesis
(11). VEGF plasma levels have
been confirmed to be associated with a C>T polymorphism located
at 936 in the 3′UTR (6). The T
variant, which is associated with lower levels of VEGF, has been
demonstrated to be associated with low FDG uptake (12) and colon cancer (13). These findings indicate that
variants, which can alter the expression of VEGF, may be involved
in the variability of FDG uptake in malignant tissues.
It has been shown that VEGF is a direct target of
miR-125a in colon cancer (14,15).
The rs3842530 SNP located in pri-miR-125a has been reported to
compromise the processing of the mature miRNA and reduce its
expression (16). Considering the
role of VEGF in the determination of 18FDG metabolism,
and the altered expression of miR-205 caused by the variant, the
present study hypothesized that the polymorphism may be associated
with 18FDG metabolism, and this was investigated in the
present study by examining associations.
Patients and methods
Patients
The present study involved a total of 270 patients
with breast cancer, all of which underwent a PET scan and donated 5
ml peripheral blood. All cases were diagnosed at Beijing Chaoyang
Hospital Affiliated to Capital Medical University (Beijing, China)
between September 2013 and December 2014. Patients suspected of
breast cancer were suggested to undergo PET-CT examinations. The
clinicopathological data of the participants are listed in Table I. Breast cancer tissue samples were
available in 39 patients, DNA was extracted from cancer tissue
samples and genotyped through direct sequencing. Written informed
consent was obtained from each participant prior to the
investigation. The study was performed according to the Helsinki
declaration and approved by the Ethical Committee of Capital
Medical University.
 | Table I.Demographic, clinicopathological and
genotypic parameters of the patients recruited in the present
study. |
Table I.
Demographic, clinicopathological and
genotypic parameters of the patients recruited in the present
study.
| miR205 rs3842530
genotype | INS/INS | INS/DEL +
DEL/DEL | P-value |
|---|
| Patients (n) | 165 | 97+8 |
|
| Sex |
|
|
|
|
Male | 98 | 62 |
|
|
Female | 67 | 43 | 0.857 |
| Age (years) | 61.35±13.4 | 59.71±12.8 | 0.413 |
| Grading |
|
|
|
|
G1/G2 | 111 | 65 |
|
|
G3/G4 | 54 | 40 | 0.213 |
| pT category |
|
|
|
| T0 | 93 | 59 |
|
|
T1/T2 | 38 | 28 |
|
|
T3/T4 | 34 | 18 | 0.811 |
| Metastases |
|
|
|
| M
(+) | 37 | 30 |
|
| M
(−) | 128 | 75 | 0.621 |
| FDG metabolism |
|
|
|
|
SUVmax | 9.33±3.86 | 13.41±3.94 | <0.001 |
|
SUVpvc | 8.25±2.89 | 12.97±3.22 | <0.001 |
18F-FDG PET/CT imaging
Each patient underwent FDG PET-CT examinations. In
brief, the patients were weighed and fasted for 12 h prior to the
PET-CT scan. The patients blood glucose levels were also measured,
and those with a blood glucose level of 150 mg/dl were excluded
from the study. Each patient was intravenously injected with
18F-FDG (37 MBq/10 kg; GE Healthcare Life Sciences,
Little Chalfont, UK). At 1 h post-injection, the patients were
instructed to raise their arms, and images were obtained from the
top of the skull to the middle of the thigh via PET-CT scans,
followed by a whole-body PET-CT scan (Discovery LS; GE Healthcare
Life Sciences). The PET-CT protocol, including a low dose CT scan
and a 3D PET whole body scan were performed at 2.5 min/bed
position.
PET image quantitative analysis
Each PET image was quantitatively analyzed using
Xeleris software (version 1.1363; GE Healthcare Life Sciences). PET
images were initially processed and visually analyzed on sagittal,
transaxial and coronal displays by three experienced technicians,
who were all blind to the clinical data and the results of previous
imaging studies. The circular target region was drawn to the
abnormal 18F-FDG-uptake-increased areas in the tumor and
the standardized uptake values (including SUVmax and
SUVmean) were measured. For each slice, at least three
circular (1 cm diameter) target regions were drawn from the
corresponding normal breast tissues, and the highest
SUVmax was presented as the SUVmax of the
normal breast tissue. The radioactivities of each tumor and normal
breast tissue sample were assessed and the tumor/normal ratios were
calculated.
Genotyping by direct sequencing
DNAs were extracted from homogenized cancer tissue
samples and genotyped through direct sequencing. In brief, total
DNA was extracted from sample tissues using a
ChargeSwitch® DNA Extraction kit (Thermo Fisher
Scientific, Inc., Waltham, MA, USA). The DNA extracts were then
amplified through polymerase chain reaction (PCR) analysis. PCR
amplification was performed on an ABI 7300 Real-Time PCR system
(Applied Biosystems; Thermo Fisher Scientific Inc.) with a mixture
including reverse transcription product (3 µl), forward primer (1
µl), reverse primer (1 µl), 2XSYBR-Green I Master mix (12.5 µl;
Thermo Fisher Scientific Inc.) and water (7.5 µl). The reaction
settings were as follows: 20 sec at 95°C, 3 sec at 95°C, and 40
cycles of 3 sec at 95°C and 30 sec at 60°C. The primers, forward
5′-ACAGGCTGAGGTTGACATGC-3′, reverse 5′-GAGTTACTCTTGCTGCTGCTG-3′,
were used to amplify a 247 bp fragment. The amplified samples were
sent to Shanghai Shenggong Biology Engineering Technology Service,
Ltd. (Shanghai, China) for genotyping using a direct sequencing
method.
RNA isolation and reverse
transcription-quantitative PCR (RT-qPCR) analysis
Total RNA in the sample tissues and normal tissues
were isolated using an RNA extraction kit (Sigma-Aldrich; Merck
Millipore, Darmstadt, Germany) according to the manufacturer's
protocol. The cDNA strands of target RNAs were synthesized using
the high-capacity cDNA reverse transcription kit (Applied
Biosystems; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. The RNA expression was quantified and qPCR
analysis was performed. PCR amplification was performed on an ABI
7300 Real-Time PCR system (Applied Biosystems; Thermo Fisher
Scientific. Inc.). The relative expression of miR-205, VEGF mRNA
and β-actin were determined using the 2−∆∆Cq method
(17). The PCR conditions were as
follows: 2 min denaturation at 94°C, 28 cycles at 94°C for 30 sec
and 58°C for 30 sec and 72°C for 40 sec. The expression of β-actin
mRNA was determined as internal control.
MCF-7 cell culture and
transfection
The MCF-7 cells (Chinese Cell Bank of the Chinese
Academy of Sciences, Shanghai, China) were cultured
1×104 cell per well in Eagle's minimum essential medium
(Invitrogen; Thermo Fisher Scientific, Inc.) supplemented with 20%
FBS (Gibco; Thermo Fisher Scientific, Inc.) at 37°C in a 5%
CO2 environment. VEGF mimic, miR-205 mimics and scramble
control mimics (GenePharma, Inc., Suzhou, China) were transfected
into MCF-7 cells using Lipofectamine 2000 reagent (Invitrogen
Thermo Fisher Scientific, Inc.) for 4 h at 37°C.
Bioinformatics analysis
Bioinformatics analysis was performed using online
miRNA database (www.mirdb.org) (4).
Vector construction and
mutagenesis
The VEGF gene −3onstr, which contains a conserved
binding site for miR-205, was amplified from the human gene
extracted from cancer tissue samples using PCR. The PCR products
were subcloned into the pmiR-RB-REPORT™ vector (Promega
Corporation, Madison, WI, USA). In addition, mutant fragments were
generated from mutagenesis and introduced into the same sites of
the control vector.
Luciferase assay
The VEGF gene −3se as, which contains a miR-205
binding site, was amplified from the human gene using PCR. PCR
amplification was performed on an ABI 7300 Real-Time PCR system
(Applied Biosystems; Thermo Fisher Scientific. Inc.) with a mixture
including reverse transcription product (3 µl), forward primer (1
µl), reverse primer (1 µl), 2XSYBR-Green I Master mix (12.5 µl;
Thermo Fisher Scientific. Inc.) and water (7.5 µl). The reaction
setting were as follows: 20 sec at 95°C, 3 sec at 95°C, 40 cycles
of 3 sec at 95°C and 30 sec at 60°C. The primers used to amplify
VEGF were: Forward: 5′-CCTTTGGGTTTTGCCAGA−3′ and Reverse:
5′-CCAAGTTTGTGGAGCTGA−3′. The PCR products were subcloned into the
pmiR-RB-REPORT™ vector (Promega Corporation). Mutant fragments were
also generated from mutagenesis and introduced into the same sites
of the control vector. For the analysis of luciferase activity,
MCF-7 cells were seeded 1×104 cell per well in 96-well
plates overnight prior to transfection. The cells were
cotransfected with miR-205 mimic/mimic control and wild-type/mutant
vector (Promega Corporation) using the Lipofectamine 2000
Transfection system (Invitrogen; Thermo Fisher Scientific, Inc.).
At 48 h post-transfection, luciferase activity was measured using
the Dual-Luciferase Reporter Assay system (Promega Corporation)
according to the manufacturer's protocol. A Renilla luciferase
plasmid was used as an internal control. Each experiment was
repeated three times.
Western blot analysis
Western blot analysis was performed to determine the
protein expression levels of miR-205 and VEGF in the sample tissues
and cultured cells. The sample tissues were rinsed with 4°C PBS
(Beijing Zhongshan Golden Bridge Biotechnology Co., Ltd., Beijing,
China) and total proteins were extracted using cell lysis buffer,
which contained 50 mmol/l Tris HCl, 150 mmol/l NaCl, 0.1% SDS, 100
µg/ml phenylmethylsulfonyl fluoride, 1% NP-40 and 1 µg/ml aprotinin
(pH 8.0). The concentrations of protein extracts were determined
using the Bradford method. The protein extracts (30 µg) were loaded
on 10% SDS-PAGE gels. Following electrophoresis, the proteins were
transblotted onto nitrocellulose membranes (Bio-Rad Laboratories,
Inc., Hercules, CA, USA). The blots were then blocked and washed to
avoid unspecific binding. The membranes were then incubated with
primary antibodies (anti-VEGF antibody; sc-4570; 1:1,000;
anti-β-actin antibody; sc-418965; 1:10,000) and secondary antibody
(sc-51948; 1:10,000; all from Santa Cruz Biotechnology, Inc.). The
bands were visualized by chemiluminescence (ECL-Plus; Santa Cruz
Biotechnology, Inc.).
Statistical analysis
Statistical analysis was performed using the
Statistical Package for the Social Sciences software (SPSS) for
Windows 13.0 (SPSS, Inc., Chicago, IL, USA). The data for SUVmean,
SUVmax and tumor/normal are expressed as the mean ± standard
deviation, and a two-sample t-test was used for comparison between
tumors and normal tissues. A χ2 test was performed to
compare categorical data in different groups. The odds ratios and
95% confidence intervals were calculated using univariate and
multivariate logistic regression analyses to determine potential
associations. Differences between the groups were determined using
a Mann-Whitney test for nonparametric data. Group differences for
dichotomous data were determined using a χ2 test or
Fishers exact test. P<0.05 was considered to indicate a
statistically significant difference.
Results
miR-205 represses the expression of
VEGF
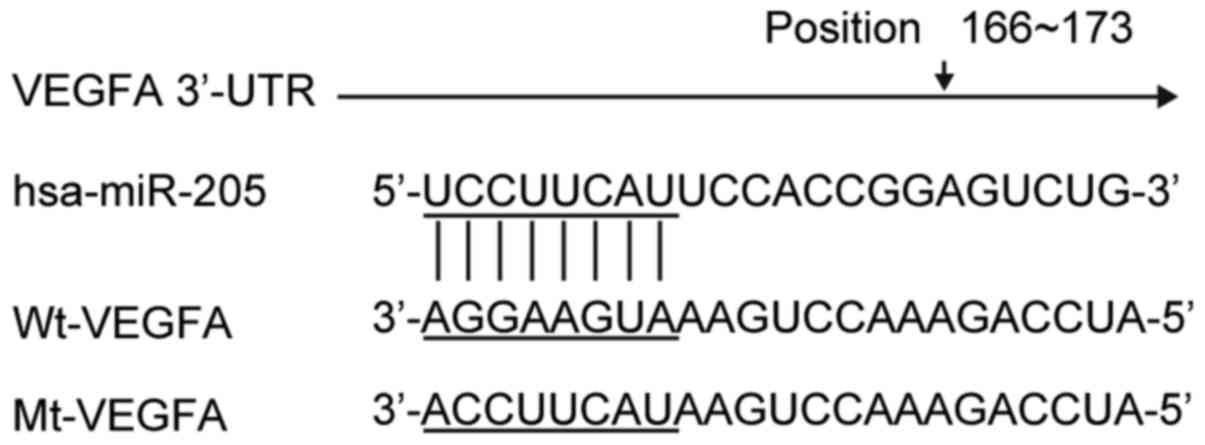
Using bioinformatics analysis, miR-205 was predicted
to target the human VEGF 3UTR at position 166–173 (Fig. 1). To investigate whether the
expression level of miR-205 was correlated with the expression
level of VEGF, luciferase reporter vectors were constructed, which
contained a wild-type or mutated VEGF 3UTR downstream of Firefly
luciferase (Fig. 1). The miR-205
mimic and reporter vectors were co-transfected into MCF-7 cells. A
dual-luciferase assay was performed 48 h post-transfection. The
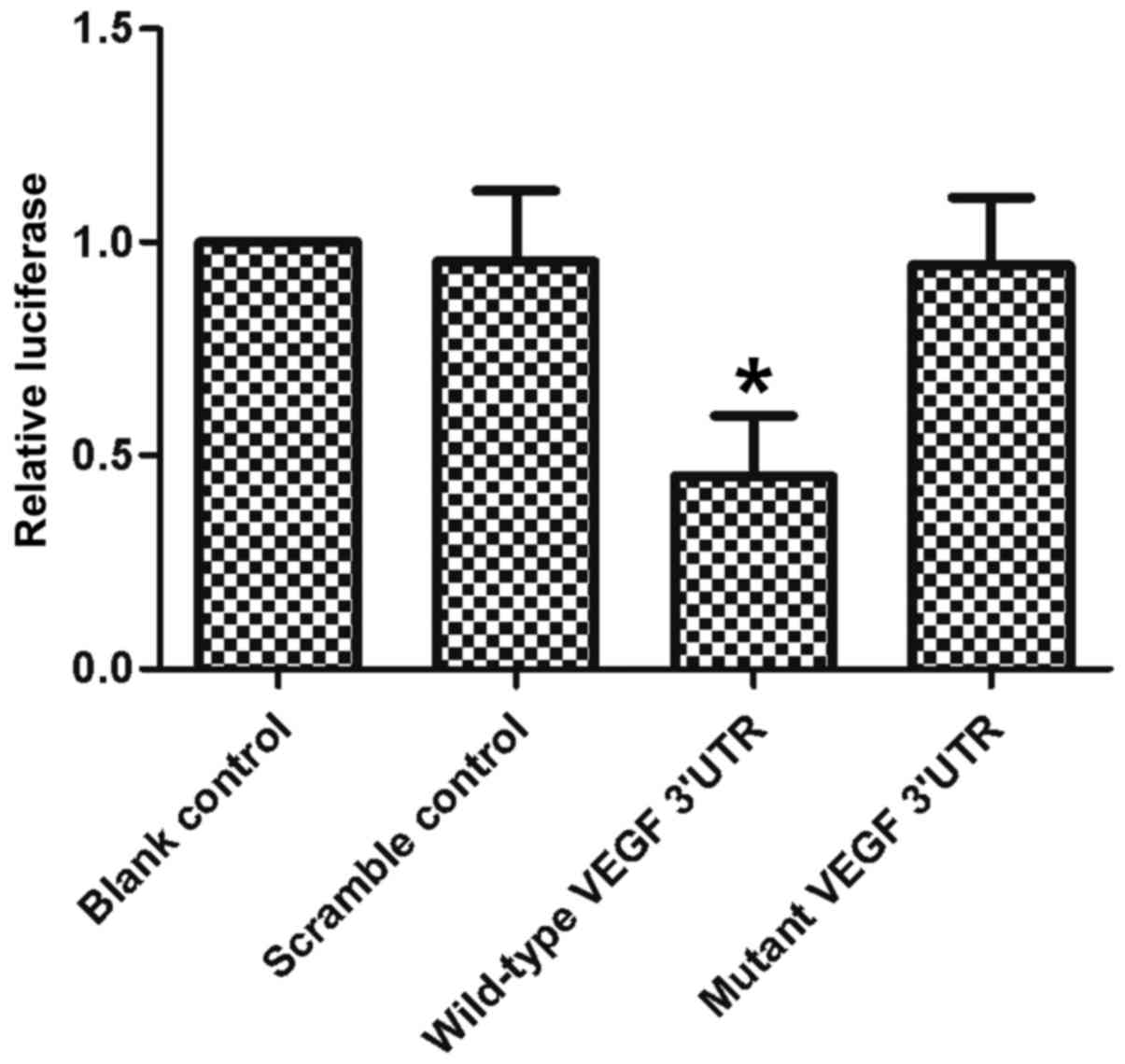
results of the dual-luciferase assay showed that when the reporter
vector contained a wild-type VEGF 3UTR, miR-205 suppressed the
expression of luciferase. By contrast, mutation of the seed regions
completely eliminated this inhibition (Fig. 2). These results showed that miR-205
targeted the VEGF 3′UTR directly and suppressed the expression of
VEGF.
To determine whether miR-205 disrupted the
endogenous expression of VEGF in breast cancer cells, miR-205
mimics were transfected into MCF-7 cells, with blank and scramble
as negative controls, and VEGF small interfering (si)RNA as a
positive control. The data showed that mRNA and protein expression
levels of VEGF were suppressed by the miR-205 mimic (Fig. 3A and B). Similar experiments were
performed involving miR-205 inhibitor treatment, and the decrease
of miR-205 increased the mRNA and protein levels of VEGF (Fig. 3A and B).
Rs3842530 decreases the expression of
VEGF via miR-205
An SNP exists in the miR-205 gene, termed rs3842530.
To investigate whether the expression of miR-205 can be eliminated
by this SNP in breast cancer, the present study compared the
expression levels of miR-205 in a number of breast tumor tissue
samples with different miR-205 genotypes. The results showed that
the expression level of miR-205 was reduced in samples containing
rs3842530 (Fig. 4A). It was
hypothesized that a reduction of miR-205 mediated by rs3842530
leads to a further increase in the expression of VEGF. To evaluate
this, RT-qPCR analysis was used to determine the mRNA expression
level of VEGF. In addition, western blot analysis was used to
determine the protein expression level of VEGF (Fig. 4B and C). The results showed that
the mRNA and protein levels of VEGF were significantly increased in
samples with rs3842530 (Fig. 4B and
C). Of note, the changes in miR-205 and the level of VEGF were
inversely correlated.
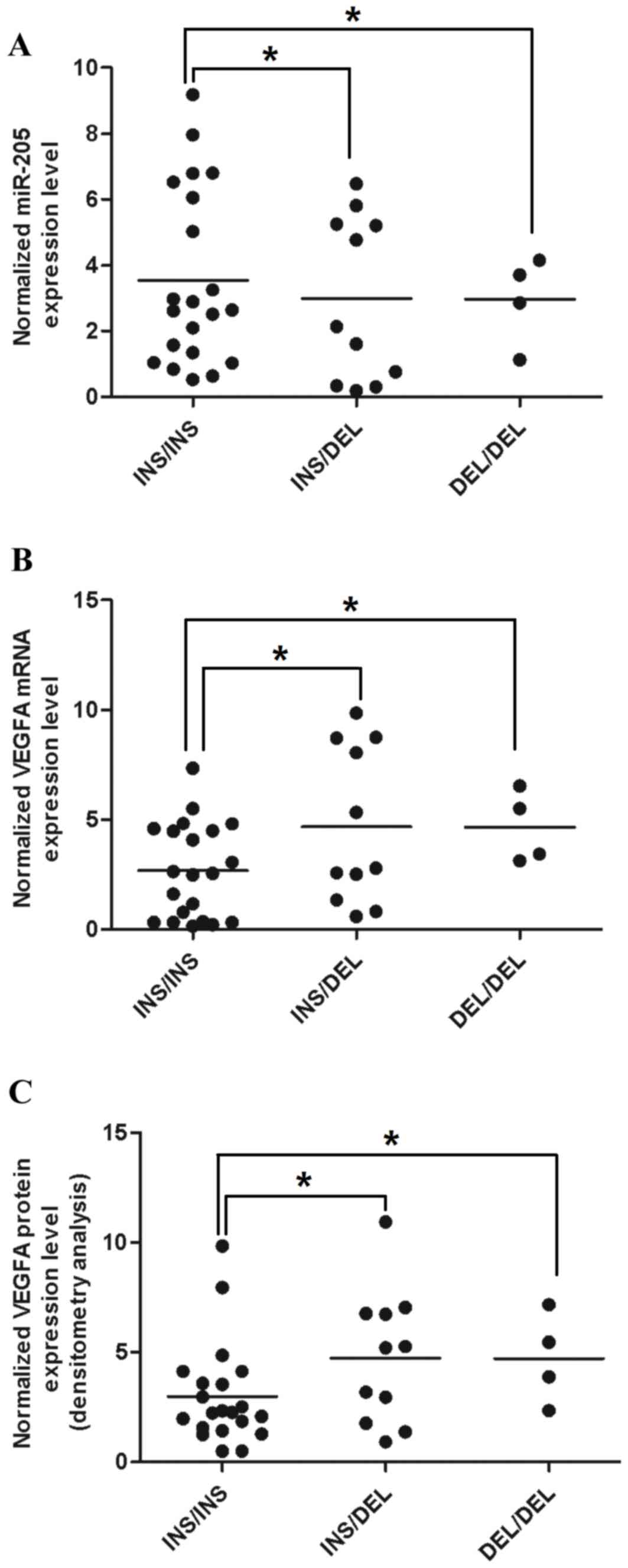 | Figure 4.DEL can reduce the expression level of
miR-205 and increase the expression level of VEGF. The expression
levels of (A) mature miR-205 (*P<0.05 vs. INS/INS group), (B)
VEGF mRNA (*P<0.05 vs. INS/INS group) and (C) VEGF protein
(*P<0.05 vs. INS/INS group) (INS/INS, 21; INS/DEL, 11; DEL/DEL,
4). Single DEL resulted in the reduced expression level of mature
miR-205, thereby releasing the expression of its target gene, VEGF.
miR, microRNA; VEGF, vascular endothelial growth factor; INS,
insertion; DEL, deletion. |
Rs3842530 affects 18FDG metabolism
mediated by miR-205 and VEGF
Correlation analyses were performed to determine
whether rs3842530 was associated with certain breast tumor
characteristics. It was found that patients with the minor allele
of rs3842530 had significantly higher SUVmax and SUVpvc values,
compared with patients with a wild-type genotype (Table I). As is already known, SUVmax and
SUVpvc are important parameters in 18FDG-PET analysis,
and they can be used to reflect glucose metabolism. A previous
study showed that a high level of glucose metabolism is an
important characteristic in malignant tumors and that dysregulation
of the expression of VEGF has been reported to be associated with
altered glucose metabolism (4).
Consequently, the present study hypothesized that patients with the
minor allele of rs3842530 have associated higher glucose
metabolism. These data suggested that the minor allele of rs3842530
accelerated glucose metabolism by compromising the expression of
miR-205, which indicates it as a promising biomarker for the
metabolism of 18FDG.
Discussion
As non-coding small RNAs, miRNAs negatively regulate
the expression of genes via the degradation of mRNA or via
translational repression. In humans, >700 miRNAs have been
identified and registered, and each of these can bind to several
genes on the bases of the seed sequence matches in their 3′UTRs
(18). miRNAs are involved in
pathologic and biological processes, including cell apoptosis,
proliferation, differentiation and metabolism (19), and they have shown potential as
tissue-specific biomarkers, which may be used to identify cancer
origin and type (20). Increasing
evidence demonstrates that the deregulation of miRNAs is involved
in human cancer, and suggests a causative role of miRs in cancer
progression and initiation as they can serve as tumor suppressors
or oncogenes (21). Marked
differences in the expression patterns of miRNAs have been reported
among adenocarcinoma, in esophageal and squamous cell carcinoma and
in other types of cancer (22).
High expression levels of miR-205 in malignant and benign squamous
epithelia, including esophageal squamous cell carcinoma (ESCC),
were reported by Kimura et al (23), whereas the expression level was
lower in tissues and cell lines other than squamous epithelia. By
contrast, the expression level of miR-21, which acts as an
oncogenic miRNA in several types of cancer, was induced in ESCC,
compared with paired normal squamous epithelia. However,
elucidation of the roles of functional VEGF specific-miRs remained
limited (23). In the present
study, it was predicted that miR-205 targeted the human VEGF 3UTR
at position 166–173 using bioinformatics analysis (Fig. 1). Additionally, a dual-luciferase
assay was performed, which found that in reporter vectors
containing a wild-type VEGF 3UTR, miR-205 suppressed the expression
of luciferase. By contrast, seed region mutations completely
eliminated this inhibition (Fig.
2). These results confirmed that miR-205 targeted the VEGF 3UTR
directly.
VEGF serves as an essential factor in the
angiogenesis of tumors, and VEGF pathway activation requires the
binding of ligands, including VEGFA, to one of its receptors,
including kinase insert domain receptor (KDR) or fms-like tyrosine
kinase 1 (FLT1). Consequently, downstream signals are generated to
stimulate the structural reorganization and proliferation of
endothelial cells (24). There are
four members in the VEGF family, including VEGFD, VEGFC, VEGFB and
VEGFA, and VEGF receptors are comprised of three subtypes,
including VEGFR3, VEGFR2 (KDR) and VEGFR1 (FLT1) (25). VEGFA acts as major regulator in the
development of angiogenesis and vasculogenesis, predominantly by
interacting with KDR and FLT1 to produce downstream signaling
(25). As a tyrosine kinase
receptor, FLT1 demonstrates ~10-fold higher affinity, compared with
KDR in binding VEGFA. Although VEGF ligands poorly activate FLT1,
it has been found to accelerate tumor metastasis and growth
(25). By contrast, potent
downstream signals are induced when the VEGF ligand binds KDR,
leading to endothelial cell migration and growth (25). There has been limited success in
targeted therapy for malignant glioma due to the converging complex
interactions and parallels among certain critical pathways,
including pathways involved in the regulation of angiogenesis of
tumors (26). In the present
study, miR-205 mimics were transfected into MCF-7 cells, with blank
and scramble as negative controls, and VEGF siRNA as a positive
control. The data showed that the mRNA and protein expression
levels of VEGF were suppressed by the miR-205 mimic (Fig. 3A and B). Similar experiments were
performed using miR-205 inhibitor, and the decrease in miR-205
increased the mRNA and protein levels of VEGF (Fig. 3A and B).
SNPs are risk factors of breast cancer as nine large
genome-wide association studies reported (27). However, the commercial exploitation
of SNPs in clinical use remains controversial despite considerable
progress (28). Further
investigations are required to investigate the potential functional
effect of specific SNPs on PET uptake in human cancer. A previous
study was performed involving 37 patients with breast cancer
without metastases, which demonstrated the association between FDG
uptake in breast cancer and a human SNP (rs3025039 of VEGFA)
(12). miRs have been identified
as gene regulators, which are expressed at aberrant levels and are
involved in virtually all subtypes of cancer (29). miRs target the 3UTR of target
genes, re gions which show a high level of evolutionarily
conservation (30), indicating the
importance of these regions in natural selection. As each miRNA
manipulates numerous mRNAs at the same time (31), the potential for cellular
transformation originating from a single miRNA dysfunction is high.
The roles of the miRNA SNPs in disorders have been identified and
their importance has been confirmed. mir-125a, which shows aberrant
expression levels in breast cancer (32) has an SNP variant allele in the
mature miRNA sequence, which reduces the expression level (33). It has been demonstrated that SNPs
in binding sites of RNAs may be associated with disease, for
example, miR-189 binding is disrupted by a point mutation located
in the 3UTR of SLIT and NTRK like family member 1 in certain
patients with Tourettes syndrome (34). In addition, SNPs in miRNA target
spots in human cancer genes were reported in a study in which
differences in allele frequencies between normal and cancerous
tissues were found (35). Finally,
it has been reported that an SNP existing in the binding site of
miRNA in the kit oncogene was associated with induced expression of
genes in papillary thyroid cancer (36). In the present study, breast cancer
tissue samples were collected and genotyped for rs3842530 (37). It was found that the expression
level of miR-205 was reduced in samples containing rs3842530
(Fig. 4A), and the mRNA and
protein levels of VEGF were significantly increased in samples with
rs3842530 (Fig. 4B and C).
Furthermore, an association experiment was performed in the breast
cancer patient population (n=270), and it was identified that
patients with the minor allele of rs3842530 had significantly
higher SUVmax and SUVpvc, compared with those patients with a
wild-type genotype (Table I).
Taken together, the findings of the present study
showed that VEGF was a direct target of miR-205 and the presence of
rs3842530 reduced the expression of VEGF, rs3842530 was associated
with 18FDG metabolism in patients with breast cancer and
may be a promising biomarker for PET scan parameter
predictions.
References
|
1
|
Nomori H, Watanabe K, Ohtsuka T, Naruke T,
Suemasu K and Uno K: Visual and semiquantitative analyses for F-18
fluorodeoxyglucose PET scanning in pulmonary nodules 1 to 3 cm in
size. Ann Thorac Surg. 79:984–989. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cronin P, Dwamena BA, Kelly AM and Carlos
RC: Solitary pulmonary nodules: Meta-analytic comparison of
cross-sectional imaging modalities for diagnosis of malignancy.
Radiology. 246:772–782. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wahidi MM, Govert JA, Goudar RK, Gould MK
and McCrory DC: American College of Chest Physicians: Evidence for
the treatment of patients with pulmonary nodules: When is it breast
cancer? ACCP evidence-based clinical practice guidelines. Chest.
132 Suppl 3:S94–S107. 2007. View Article : Google Scholar
|
|
4
|
Wong N and Wang X: miRDB: An online
resource for microRNA target prediction and functional annotations.
Nucleic Acids Res. 43:(Database Issue). D146–D152. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Herder GJ, van Tinteren H, Golding RP,
Kostense PJ, Comans EF, Smit EF and Hoekstra OS: Clinical
prediction model to characterize pulmonary nodules: Validation and
added value of 18F-fluorodeoxyglucose positron emission tomography.
Chest. 128:2490–2496. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kim SH, Cho YR, Kim HJ, Oh JS, Ahn EK, Ko
HJ, Hwang BJ, Lee SJ, Cho Y, Kim YK, et al: Antagonism of
VEGF-A-induced increase in vascular permeability by an integrin
α3β1-Shp-1-cAMP/PKA pathway. Blood. 120:4892–4902. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yanokura M, Banno K, Kobayashi Y, Kisu I,
Ueki A, Ono A, Masuda K, Nomura H, Hirasawa A, Susumu N and Aoki D:
MicroRNA and endometrial cancer: Roles of small RNAs in human
tumors and clinical applications (Review). Oncol Lett. 1:935–940.
2010.PubMed/NCBI
|
|
8
|
Engels BM and Hutvagner G: Principles and
effects of microRNA-mediated post-transcriptional gene regulation.
Oncogene. 25:6163–6169. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Philippe L, Alsaleh G, Bahram S, Pfeffer S
and Georgel P: The miR-17~92 cluster: A key player in the control
of inflammation during rheumatoid arthritis. Front Immunol.
4:702013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dylla L, Moore C and Jedlicka P: MicroRNAs
in Ewing sarcoma. Front Oncol. 3:652013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Renner W, Kotschan S, Hoffmann C,
Obermayer-Pietsch B and Pilger E: A common 936 C/T mutation in the
gene for vascular endothelial growth factor is associated with
vascular endothelial growth factor plasma levels. J Vasc Res.
37:443–448. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wolf G, Aigner RM, Schaffler G,
Langsenlehner U, Renner W, Samonigg H, Yazdani-Biuki B and Krippl
P: The 936C> T polymorphism of the gene for vascular endothelial
growth factor is associated with 18F-fluorodeoxyglucose uptake.
Breast Cancer Res Treat. 88:205–208. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bae SJ, Kim JW, Kang H, Hwang SG, Oh D and
Kim NK: Gender-specific association between polymorphism of
vascular endothelial growth factor (VEGF 936 C>T) gene and colon
cancer in Korea. Anticancer Res. 28:1271–1276. 2008.PubMed/NCBI
|
|
14
|
Li J, Li L, Li Z, Gong G, Chen P, Liu H,
Wang J, Liu Y and Wu X: The role of miR-205 in the VEGF-mediated
promotion of human ovarian cancer cell invasion. Gynecol Oncol.
137:125–133. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang L, Shan M, Liu Y, Yang F, Qi H, Zhou
L, Qiu L and Li Y: miR-205 suppresses the proliferative and
migratory capacity of human osteosarcoma Mg-63 cells by targeting
VEGFA. Onco Targets Ther. 16:2635–2642. 2015.
|
|
16
|
Leng S, Bernauer AM, Zhai R, Tellez CS, Su
L, Burki EA, Picchi MA, Stidley CA, Crowell RE, Christiani DC and
Belinsky SA: Discovery of common SNPs in the miR-205/200
family-regulated epithelial to mesenchymal transition pathway and
their association with risk for non-small cell lung cancer. Int J
Mol Epidemiol Genet. 2:145–155. 2011.PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Carthew RW and Sontheimer EJ: Origins and
mechanisms of miRNAs and siRNAs. Cell. 136:642–655. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Schmittgen TD: Regulation of microRNA
processing in development, differentiation and cancer. J Cell Mol
Medicine. 12:1811–1819. 2008. View Article : Google Scholar
|
|
20
|
Rosenfeld N, Aharonov R, Meiri E,
Rosenwald S, Spector Y, Zepeniuk M, Benjamin H, Shabes N, Tabak S,
Levy A, et al: MicroRNAs accurately identify cancer tissue origin.
Nat Biotechnol. 26:462–469. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Croce CM: Causes and consequences of
microRNA dysregulation in cancer. Nat Rev Genet. 10:704–714. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mathé EA, Nguyen GH, Bowman ED, Zhao Y,
Budhu A, Schetter AJ, Braun R, Reimers M, Kumamoto K, Hughes D, et
al: MicroRNA expression in squamous cell carcinoma and
adenocarcinoma of the esophagus: Associations with survival. Clin
Cancer Res. 15:6192–6200. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kimura S, Naganuma S, Susuki D, Hirono Y,
Yamaguchi A, Fujieda S, Sano K and Itoh H: Expression of microRNAs
in squamous cell carcinoma of human head and neck and the
esophagus: miR-205 and miR-21 are specific markers for HNSCC and
ESCC. Oncol Rep. 23:1625–1633. 2010.PubMed/NCBI
|
|
24
|
Ferrara N: VEGF and the quest for tumour
angiogenesis factors. Nat Rev Cancer. 2:795–803. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Takahashi S: Vascular endothelial growth
factor (VEGF), VEGF receptors and their inhibitors for
antiangiogenic tumor therapy. Biol Pharm Bull. 34:1785–1788. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Omuro AM, Faivre S and Raymond E: Lessons
learned in the development of targeted therapy for malignant
gliomas. Mol Cancer Ther. 6:1909–1919. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Thomas G, Jacobs KB, Kraft P, Yeager M,
Wacholder S, Cox DG, Hankinson SE, Hutchinson A, Wang Z, Yu K, et
al: A multistage genome-wide association study in breast cancer
identifies two new risk alleles at 1p11. 2 and 14q24. 1 (RAD51L1).
Nat Genet. 41:579–584. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Goldstein DB: Common genetic variation and
human traits. N Engl J Med. 360:1696–1698. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Esquela-Kerscher A and Slack FJ:
Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer.
6:259–269. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen K and Rajewsky N: Natural selection
on human microRNA binding sites inferred from SNP data. Nat Genet.
38:1452–1456. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Iorio MV, Ferracin M, Liu CG, Veronese A,
Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M,
et al: MicroRNA gene expression deregulation in human breast
cancer. Cancer Res. 65:7065–7070. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Duan R, Pak C and Jin P: Single nucleotide
polymorphism associated with mature miR-125a alters the processing
of pri-miRNA. Hum Mol Genet. 16:1124–1131. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Abelson JF, Kwan KY, O'Roak BJ, Baek DY,
Stillman AA, Morgan TM, Mathews CA, Pauls DL, Rasin MR, Gunel M, et
al: Sequence variants in SLITRK1 are associated with Tourette's
syndrome. Science. 310:317–320. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Landi D, Gemignani F, Barale R and Landi
S: A catalog of polymorphisms falling in microRNA-binding regions
of cancer genes. DNA Cell Biol. 27:35–43. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
He H, Jazdzewski K, Li W, Liyanarachchi S,
Nagy R, Volinia S, Calin GA, Liu CG, Franssila K, Suster S, et al:
The role of microRNA genes in papillary thyroid carcinoma. Proc
Natl Acad Sci USAmerica. 102:19075–19080. 2005. View Article : Google Scholar
|
|
37
|
Domigan CK, Warren CM, Antanesian V,
Happel K, Ziyad S, Lee S, Krall A, Duan L, Torres-Collado AX,
Castellani LW, et al: Autocrine VEGF maintains endothelial survival
through regulation of metabolism and autophagy. J Cell Sci.
128:2236–2248. 2015. View Article : Google Scholar : PubMed/NCBI
|