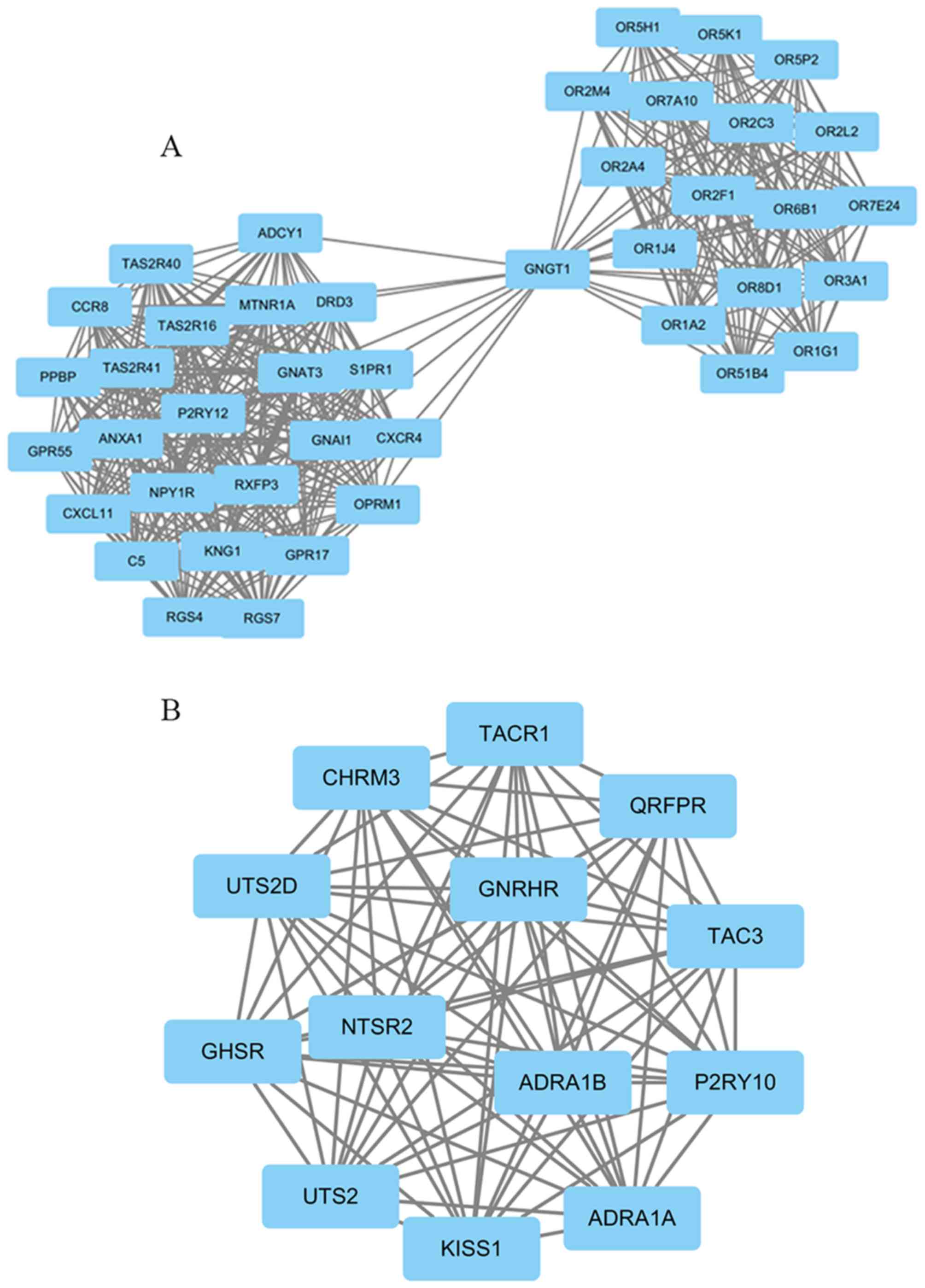
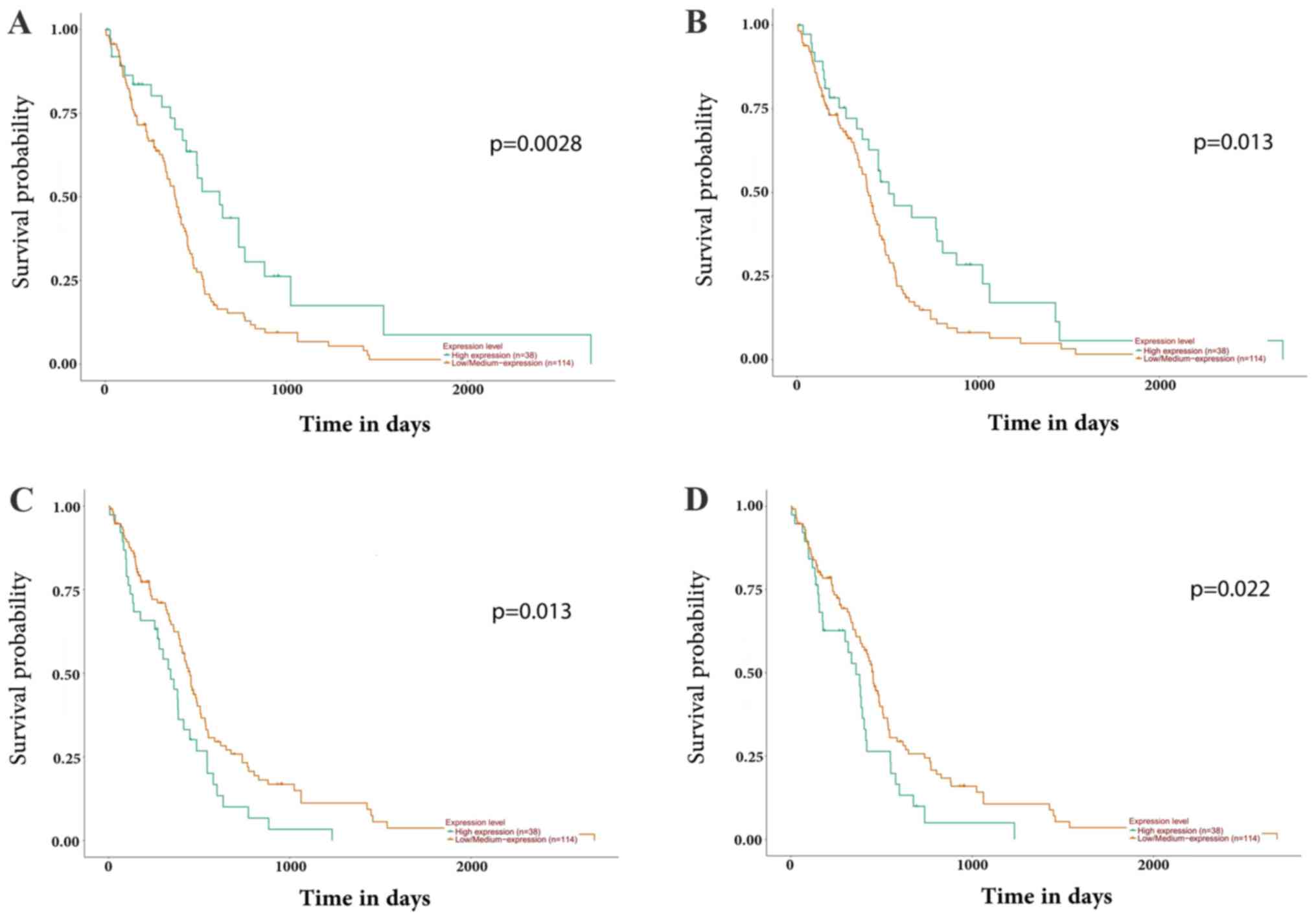
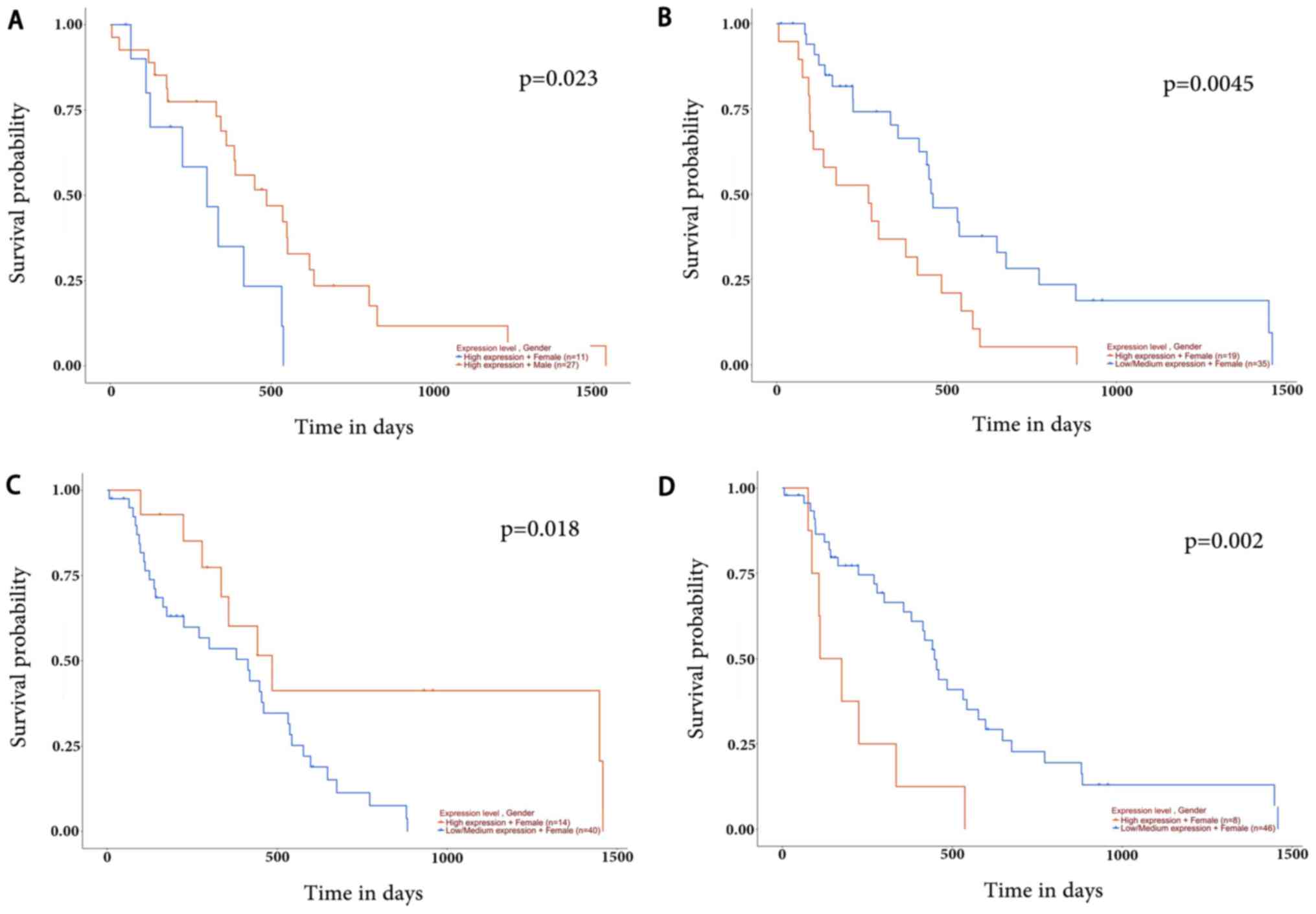
|
1
|
Steiner HH, Herold-Mende C, Bonsanto M,
Geletneky K and Kunze S: Prognosis of brain tumors: Epidemiology,
survival time and clinical course. Versicherungsmedizin.
50:173–179. 1998.(In French). PubMed/NCBI
|
|
2
|
Fisher JL, Schwartzbaum JA, Wrensch M and
Wiemels JL: Epidemiology of brain tumors. Neurol Clin. 25:867–890.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Louis DN, Ohgaki H, Wiestler OD, Cavenee
WK, Burger PC, Jouvet A, Scheithauer BW and Kleihues P: The 2007
WHO classification of tumours of the central nervous system. Acta
Neuropathol. 114:97–109. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Asthagiri AR, Pouratian N, Sherman J,
Ahmed G and Shaffrey ME: Advances in brain tumor surgery. Neurol
Clin. 25:975–1003. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Grzmil M, Morin P Jr, Lino MM, Merlo A,
Frank S, Wang Y, Moncayo G and Hemmings BA: MAP kinase-interacting
kinase 1 regulates SMAD2-dependent TGF-β signaling pathway in human
glioblastoma. Cancer Res. 71:2392–2402. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Walsh AM, Kapoor GS, Buonato JM, Mathew
LK, Bi Y, Davuluri RV, Martinez-Lage M, Simon MC, O'Rourke DM and
Lazzara MJ: Sprouty2 drives drug resistance and proliferation in
glioblastoma. Mol Cancer Res. 13:1227–1237. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gerber NK, Goenka A, Turcan S, Reyngold M,
Makarov V, Kannan K, Beal K, Omuro A, Yamada Y, Gutin P, et al:
Transcriptional diversity of long-term glioblastoma survivors.
Neuro Oncol. 16:1186–1195. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Barrett T, Troup DB, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, et al: NCBI GEO: Archive for functional genomics data
sets-10 years on. Nucleic Acids Res. 39:D1005–D1010. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
NIH: The Cancer Genome Atlas, . TCGA Data
Portal. https://tcga-data.nci.nih.gov/docs/publications/tcga/?June
30–2016
|
|
10
|
Louis DN, Perry A, Reifenberger G, von
Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD,
Kleihues P and Ellison DW: The 2016 world health organization
classification of tumors of the central nervous system: A summary.
Acta Neuropathol. 131:803–820. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Diboun I, Wernisch L, Orengo CA and
Koltzenburg M: Microarray analysis after RNA amplification can
detect pronounced differences in gene expression using limma. BMC
Genomics. 7:2522006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gautier L, Cope L, Bolstad BM and Irizarry
RA: affy-analysis of Affymetrix GeneChip data at the probe level.
Bioinformatics. 20:307–315. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Irizarry RA, Hobbs B, Collin F,
Beazer-Barclay YD, Antonellis KJ, Scherf U and Speed TP:
Exploration, normalization and summaries of high density
oligonucleotide array probe level data. Biostatistics. 4:249–264.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gene Ontology Consortium, . The gene
ontology (GO) project in 2006. Nucleic Acids Res. 34:D322–D336.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The gene
ontology consortium. Nature Genetics. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dennis G Jr, Sherman BT, Hosack DA, Yang
J, Gao W, Lane HC and Lempicki RA: DAVID: Database for annotation,
visualization and integrated discovery. Genome Biol. 4:P32003.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Franceschini A, Szklarezyk D, Frankild S,
Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C
and Jensen LJ: STRING v9.1: Protein-protein interaction networks,
with increased coverage and integration. Nucleic Acids Res.
41:D808–D815. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Smoot ME, Ono K, Ruscheinski J, Wang PL
and Ideker T: Cytoscape 2.8: New features for data integration and
network visualization. Bioinformatics. 27:431–432. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Rodriguez I, Chakravarthi BVSK and Varambally S:
UALCAN: A portal for facilitating tumor subgroup gene expression
and survival analyses. Neoplasia. 19:649–658. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu TT, Achrol AS, Mitchell LA, Rodriguez
SA, Feroze A, Iv M, Kim C, Chaudhary N, Gevaert O, Stuart JM, et
al: Magnetic resonance perfusion image features uncover an
angiogenic subgroup of glioblastoma patients with poor survival and
better response to antiangiogenic treatment. Neuro Oncol.
19:997–1007. 2017.PubMed/NCBI
|
|
22
|
Brandes A, Franceschi E, Tosoni A,
Fioravanti A, Agati R, Andreoli A, Mazzocchi V, Morandi L,
Bartolini S and Ermani M: Change in MGMT methylation status between
first and second surgery for recurrence: Clinical implications. Eur
J Cancer Suppl. 7:4952009. View Article : Google Scholar
|
|
23
|
Akbari A, Farahnejad Z, Akhtari J,
Abastabar M, Mobini GR and Mehbod AS: Staphylococcus aureus
enterotoxin B down-regulates the expression of transforming growth
factor-beta (TGF-β). Signaling Transducers in Human Glioblastoma.
Jundishapur J Microbiol. 9:e272972016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jiang C, Shen F, Du J, Hu Z, Li X, Su J,
Wang X and Huang X: MicroRNA-564 is downregulated in glioblastoma
and inhibited proliferation and invasion of glioblastoma cells by
targeting TGF-β1. Oncotarget. 7:56200–56208. 2016.PubMed/NCBI
|
|
25
|
Li J, Ren S, Liu Y, Lian Z, Dong B, Yao Y
and Xu Y: Knockdown of NUPR1 inhibits the proliferation of
glioblastoma cells via ERK1/2, p38 MAPK and caspase-3. J Neuro
Oncol. 132:15–26. 2017. View Article : Google Scholar
|
|
26
|
Kim EH, Song HS, Yoo SH and Yoon M: Tumor
treating fields inhibit glioblastoma cell migration, invasion and
angiogenesis. Oncotarget. 7:65125–65136. 2016.PubMed/NCBI
|
|
27
|
Firat E and Niedermann G: FoxO proteins or
loss of functional p53 maintain stemness of glioblastoma stem cells
and survival after ionizing radiation plus PI3K/mTOR inhibition.
Oncotarget. 7:54883–54896. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Masui K, Tanaka K, Akhavan D, Babic I,
Gini B, Matsutani T, Iwanami A, Liu F, Villa GR, Gu Y, et al: mTOR
complex 2 controls glycolytic metabolism in glioblastoma through
FoxO acetylation and upregulation of c-Myc. Cell Metab. 18:726–739.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Al-Khafaji AS, Davies MP, Risk JM, Marcus
MW, Koffa M, Gosney JR, Shaw RJ, Field JK and Liloglou T: Aurora B
expression modulates paclitaxel response in non-small cell lung
cancer. Br J Cancer. 116:592–599. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Borges KS, Castro-Gamero AM, Moreno DA, da
Silva Silveira V, Brassesco MS, de Paula Queiroz RG, de Oliveira
HF, Carlotti CG Jr, Scrideli CA and Tone LG: Inhibition of Aurora
kinases enhances chemosensitivity to temozolomide and causes
radiosensitization in glioblastoma cells. J Cancer Res Clin Oncol.
138:405–414. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bornschein J, Nielitz J, Drozdov I,
Selgrad M, Wex T, Jechorek D, Link A, Vieth M and Malfertheiner P:
Expression of aurora kinase A correlates with the Wnt-modulator
RACGAP1 in gastric cancer. Cancer Med. 5:516–526. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yousef EM, Furrer D, Laperriere DL, Tahir
MR, Mader S, Diorio C and Gaboury LA: MCM2: An alternative to Ki-67
for measuring breast cancer cell proliferation. Mod Pathol.
30:682–697. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Su Z, Zheng X, Zhang X, Wang Y, Zhu S, Lu
F, Qu J and Hou L: Sox10 regulates skin melanocyte proliferation by
activating the DNA replication licensing factor MCM5. J Dermatol
Sci. 85:216–225. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
He X, Liu Z, Peng Y and Yu C:
MicroRNA-181c inhibits glioblastoma cell invasion, migration and
mesenchymal transition by targeting TGF-β pathway. Biochem Biophys
Res Commun. 469:1041–1085. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wu ZB, Cai L, Lin SJ, Lu JL, Yao Y and
Zhou LF: The miR-92b functions as a potential oncogene by targeting
on Smad3 in glioblastomas. Brain Res. 1529:16–25. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Carpenter RL, Paw I, Zhu H, Sirkisoon S,
Xing F, Watabe K, Debinski W and Lo HW: The gain-of-function GLI1
transcription factor TGLI1 enhances expression of VEGF-C and TEM7
to promote glioblastoma angiogenesis. Oncotarget. 6:22653–22655.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Sheik Y, Qureshi SF, Mohhammed B and
Nallari P: FOXC2 and FLT4 gene variants in lymphatic filariasis.
Lymphat Res Biol. 13:112–119. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Cornejo KM, Deng A, Wu H, Cosar EF, Khan
A, St Cyr M, Tomaszewicz K, Dresser K, O'Donnell P and Hutchinson
L: The utility of MYC and FLT4 in the diagnosis and treatment of
postradiation atypical vascular lesion and angiosarcoma of the
breast. Hum Pathol. 46:868–875. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Rani N, Aichem A, Schmidtke G, Kreft SG
and Groettrup M: FAT10 and NUB1L bind to the VWA domain of Rpn10
and Rpn1 to enable proteasome-mediated proteolysis. Nat Commun.
3:7492012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhang J, Yan B, Späth SS, Qun H, Cornelius
S, Guan D, Shao J, Hagiwara K, Van Waes C, Chen Z, et al:
Integrated transcriptional profiling and genomic analyses reveal
RPN2 and HMGB1 as promising biomarkers in colorectal cancer. Cell
Biosci. 5:532015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Liau CT, Chou WC, Wei KC, Chang CN, Toh CH
and Jung SM: Female sex, good performance status and
bevacizumab-induced hypertension associated with survival benefit
in Asian patients with recurrent glioblastoma treated with
bevacizumab. Asia Pac J Clin Oncol. 14:e8–e14. 2018. View Article : Google Scholar : PubMed/NCBI
|