Introduction
Charcot-Marie-Tooth disease (CMT) is one of the most
common inherited neurological disorders presenting with
symmetrical, slowly progressive distal motor neuropathy (1). It results in varying degrees of
muscle weakness and atrophy in the feet and/or hands.
Electrophysiological examination may be used for classifying CMT
into CMT1 with demyelinating features or CMT2 with axonal features.
Certain patients exhibit central nervous system symptoms and
cerebral white matter lesions. The disease exhibits marked clinical
and genetic heterogeneity. At present, >80 unique genes are
known to be involved in CMT in autosomal-dominant,
recessive-dominant or X-linked manners (2).
X-linked CMT (CMTX), primarily caused by mutations
in the gap junction protein beta 1 (GJB1) gene, is the
second most common genetic variant of CMT (3). The GJB1 gene encodes the gap
junction beta 1 protein connexin 32 (Cx32) which is expressed in
myelinating Schwann cells and formed gap junctions. Gap junctions
provide a direct diffusion pathway for ions and small molecules and
are conductive to rapid and special communication between the
periaxonal and peripheral cytoplasm (4). Mutations in the GJB1 gene lead
to the dysfunction of Cx32 and cause damage to peripheral myelin.
In small families, the mode of inheritance of disease may be
incorrectly characterized using the pedigree analysis. In addition,
various severe clinical presentations of CMTX are not easily
differentiated from other types of CMTs. Therefore, next-generation
sequencing has the potential to quickly screen for mutations in the
diseases with extreme genetic heterogeneity, resulting in a precise
diagnosis.
The present study describes a two-generation Chinese
family with CMT. A novel mutation in the GJB1 gene and a
rare variation in the dehydrogenase E1 and transketolase domain
containing 1 (DHTKD1) gene were detected using
high-throughput sequencing followed by Sanger sequencing.
Materials and methods
Patients
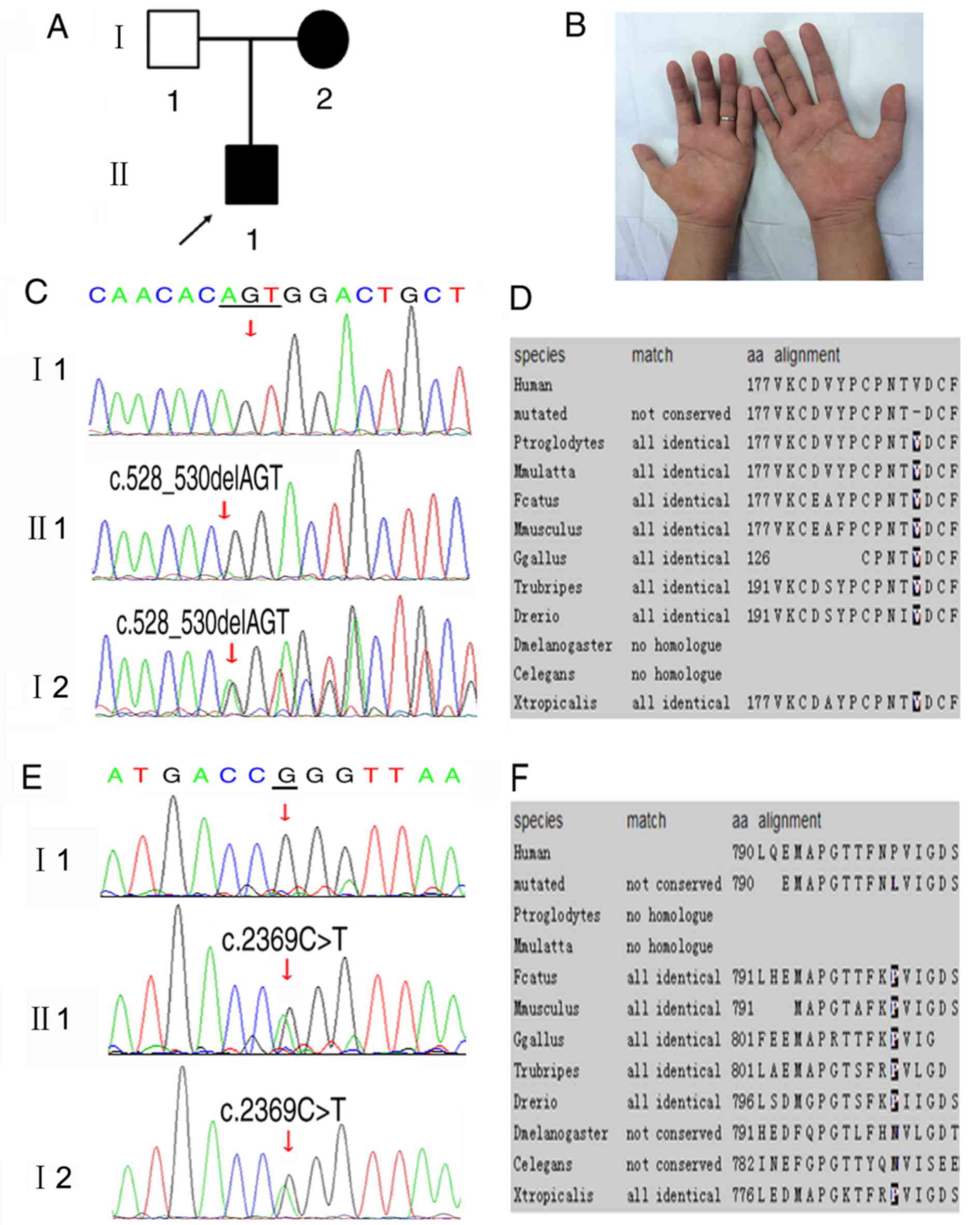
A two-generation Han Chinese CMT family with 3
family members, living in Southeast China, was studied (Fig. 1A). The proband was a 27-year-old
male, with a 48-year-old father and 53-year-old mother. Standard
clinical and electrophysiological evaluations were performed in all
the patients. Peripheral blood samples (10 ml) were collected for
mutational analysis from the elbow vein of each family member upon
admission to the Department of Neurology, Fujian Provincial
Hospital (Fuzhou, China) in December 2017. In addition, 500 healthy
individuals (>65 years, 221 males and 279 females) with no known
history of CMT were selected as a control group. The samples
provided from Fujian Key Laboratory of Molecular Neurology, Fujian
Medical University (Fuzhou, China) were collected from April 2014
to December 2016. The study was approved by the Ethics Committee of
Fujian Provincial Hospital, Fujian Medical University. Informed
consent was obtained from all individuals included in the
study.
Mutational analysis
DNA extraction
DNA extraction was performed using a Qiagen Blood
and Tissue kit according to the manufacturer's protocol (Qiagen
GmbH, Hilden, Germany). DNA concentrations were determined using a
NanoDrop 2000 UV/Vis Spectrophotometer (NanoDrop Technologies;
Thermo Fisher Scientific, Inc., Wilmington, DE, USA).
High-throughput sequencing, Sanger
sequencing and variant analysis
High-throughput sequencing was applied for the
mutation screening of the pedigree. Briefly, the coding exons and
flanking regions of the 110 genes (Table I) associated with inherited motor
never diseases in the panel were selected and captured with a
GenCap custom enrichment kit (MyGenostics, Inc., Beijing, China).
Then, the enriched libraries were sequenced by using an Illumina
NextSeq500 Sequencer with a sequencing depth of 100× (Illumina,
Inc., San Diego, CA, USA). The raw data were compared to the human
reference genome (hg19) using the Burrows Wheeler Aligner
Multi-Vision software package (version 0.7.10) (5). Duplicates were removed using Picard
software version 1.119 (http://broadinstitute.github.io/picard). Then, the
single nucleotide polymorphisms and insertions/deletions were
identified by the SOAPsnp version 1.03 (6) and Genome Analysis Toolkit programs
(version 3.3.0) (7). Synonymous
variants and the variants with allele frequencies 1>% in the
1000Genome (http://www.internationalgenome.org/), ESP6500
(https://esp.gs.washington.edu) and
inhouse databases were excluded. The Human Gene Mutation Database
was used to detect the known disease-causing mutations. To validate
the pathogenicity of the variant sites, Sanger sequencing was used
to amplify the regions of interest by polymerase chain reaction
performed using a GoldStar MasterMix PCR kit (Beijing CoWin Biotech
Co., Ltd., Beijing, China) under the following conditions: 95°C for
5 min; 35 cycles of 95°C for 30 sec, 58°C for 45 sec and 72°C for 1
min; and 72°C for 5 min. The primers are listed in Table II. To analyze the evolutionary
conservation of the two variation positions, sequence alignments
with the human genes were performed for 10 other species (Pan
troglodytes, Macaca mulatta, Mus musculus, Felis catus, Gallus
gallus, Xenopus tropicalis, Takifugu rubripes, Danio rerio,
Drosopila melanogaster and Caenorhabditis elegans) using
MutationTaster (http://www.mutationtaster.org/) (8). Matches were classed as ‘all
identical’, ‘conserved’ or ‘not conserved’, depending on whether
the aligned amino acid was the same, similar or different. A total
of 500 unrelated Chinese healthy controls were screened for the
de novo mutation position of the GJB1 gene by Sanger
sequencing. The effects of the mutations on protein function were
assessed using the Swiss-Model tool (http://swissmodel.expasy.org/) (9–13).
Mutation Taster (8) was used to
assess the biological relevance of the novel amino acid
changes.
 | Table I.Genes associated with inherited motor
never diseases in the panel for high-throughput sequencing. |
Table I.
Genes associated with inherited motor
never diseases in the panel for high-throughput sequencing.
| No. | Gene | No. | Gene | No. | Gene | No. | Gene | No. | Gene | No. | Gene |
|---|
| 1 | AAAS | 20 | DHTKD1 | 39 | GJB3 | 58 | LAS1L | 77 | PLA2G6 | 96 | SLC25A46 |
| 2 | AARS | 21 | DNAJB2 | 40 | GNB4 | 59 | LITAF | 78 | PLEKHG5 | 97 | SLC5A7 |
| 3 | ABHD12 | 22 | DNM2 | 41 | HARS | 60 | LMNA | 79 | PMM2 | 98 | SPTLC1 |
| 4 | AIFM1 | 23 | DNMT1 | 42 | HEXA | 61 | LRSAM1 | 80 | PMP22 | 99 | SPTLC2 |
| 5 | ARHGEF10 | 24 | DPYSL5 | 43 | HINT1 | 62 | MARS | 81 | PRNP | 100 | SURF1 |
| 6 | ARSA | 25 | DST | 44 | HK1 | 63 | MED25 | 82 | PRPS1 | 101 | TDP1 |
| 7 | ASAH1 | 26 | DYNC1H1 | 45 | HNRNPA2B1 | 64 | MFN2 | 83 | PRX | 102 | TFG |
| 8 | ATL1 | 27 | EGR2 | 46 | HOXD10 | 65 | MPZ | 84 | RAB7A | 103 | TRIM2 |
| 9 | ATL3 | 28 | EXOSC3 | 47 | HSPB1 | 66 | MTMR2 | 85 | RBM28 | 104 | TRPV4 |
| 10 | ATP7A | 29 | FAM134B | 48 | HSPB3 | 67 | MYH14 | 86 | REEP1 | 105 | TTR |
| 11 | BICD2 | 30 | FBLN5 | 49 | HSPB8 | 68 | NAGLU | 87 | RNF170 | 106 | UBA1 |
| 12 | BSCL2 | 31 | FBXO38 | 50 | IFRD1 | 69 | NDRG1 | 88 | SBF1 | 107 | VPS13A |
| 13 | CCT5 | 32 | FGD4 | 51 | IGHMBP2 | 70 | NEFL | 89 | SBF2 | 108 | VRK1 |
| 14 | COX6A1 | 33 | FIG4 | 52 | IKBKAP | 71 | NGF | 90 | SCN1A | 109 | WNK1 |
| 15 | CTDP1 | 34 | FLVCR1 | 53 | INF2 | 72 | NTRK1 | 91 | SCN9A | 110 | YARS |
| 16 | CUL4B | 35 | GALC | 54 | KARS | 73 | PDK3 | 92 | SETX |
|
|
| 17 | DCAF8 | 36 | GARS | 55 | KIF1A | 74 | PEX1 | 93 | SH3TC2 |
|
|
| 18 | DCTN1 | 37 | GDAP1 | 56 | KIF1B | 75 | PEX7 | 94 | SIGMAR1 |
|
|
| 19 | DHH | 38 | GJB1 | 57 | KLHL9 | 76 | PHYH | 95 | SLC12A6 |
|
|
 | Table II.Primers for Sanger sequencing. |
Table II.
Primers for Sanger sequencing.
| Target gene
mutation | Forward
(5′-3′) | Reverse
(5′-3′) |
|---|
| DHTKD1
c.2369C>T |
TTGCGTCTATTAGGACATGGG |
GGGAGATCTGCATCACCTG |
| GJB1
c.258_530delAGT |
TGGTGGACCTATGTCATCAGC |
CGCAGTATGTCTTTCAGGGAG |
Results
Clinical manifestation, physical
examination and electromyography results
The proband (patient II1; Fig. 1A) was admitted for slowly
progressing limb atrophy (Fig.
1B). He had developed distal muscle weakness and limb atrophy
at 17 years of age. He had mild gait abnormalities, but was able to
walk without assistance. Neurological examination revealed no deep
tendon reflexes in the limbs. Electrophysiological examination
identified axonal and demyelinating features. His median and ulnar
motor nerve conduction velocities (MCVs) were 28.5 and 25.5 m/sec,
respectively. In addition, his bilateral tibial MCVs were not
detected. His median, ulnar and peroneal sensory nerve conduction
velocities (SCVs) were 31.1, 32.7 and 35.5 m/sec, respectively. The
compound motor action potentials (CAMPs) were markedly decreased in
all nerves, particularly in sensory nerves and in the lower legs.
His CMT disease neuropathy score was 8 points, which indicated mild
severity of the disease (14). The
proband's mother (Patient I2), a 48-year-old woman, was also
examined by a neurology chief physician in Fujian Provincial
Hospital in December 2017, and presented with no clinical symptoms.
Her median, ulnar and peroneal MCVs were 43.7, 48.3 and 40 m/sec,
respectively. Her median and ulnar SCVs were 43.1 and 41.1 m/sec,
respectively. However, her bilateral tibial SCVs were not detected.
The CAMPs were normal in her upper limbs, but were decreased in the
lower limbs. She scored 0 points on the CMT disease neuropathy
evaluation.
Variant analysis results
Next-generation sequencing technology identified two
potentially pathogenic mutations in the proband. The
c.528_530delAGT mutation in the GJB1 gene on the X
chromosome was identified in the proband (II1) and his mother (I2;
Fig. 1C), but not in his father
(I1). The proband and his father presented homozygous genotype
sequencing results and his mother exhibited a heterozygous
sequencing result, as males carry 1 X chromosome and females carry
2 X chromosomes. The c.528_530delAGT mutation, passed from the
mother to her son, matches the X-linked recessive inheritance
disease pattern. This variation was not identified in 500 control
subjects and, to the best of our knowledge, it has not been
described previously. This mutation leads to a valine deletion at
amino acid position 177. The amino acid V177 in Cx32 protein is
highly conserved among species; the deletion mutation was
identified as ‘not conserved’ (Fig.
1D). MutationTaster programs predicted that the pathogenicity
of the c.528_530delAGT mutation in the GJB1 gene may be disease
causing. The mutation in the GJB1 gene was classified as ‘Like
Pathogenicity’, according to the criteria of the American College
of Medical Genetics and Genomics (ACMG) standards and guidelines
(12).
The second mutation, a heterozygous mutation, was
c.2369C>T in the DHTKD1 gene from the proband (II1) and
his mother (I2; Fig. 1E). This
mutation is listed in dbSNP (rs776497952) and was identified in the
Exome Aggregation Consortium with a minor allele frequency of
0.004118% among 60,706 individuals. This mutation leads to an amino
acid exchange of p.P790L. The amino acid P790 in the DHTKD1 protein
is highly conserved, with the exception of Drosophila
melanogaster and Caenorhabditis elegans; the p.P790L
mutation was identified as ‘not conserved’ (Fig. 1F). The pathogenicity of the
c.2369C>T variants in the DHTKD1 gene was also assessed
to be disease causing by Mutation Taster programs. The mutation in
the DHTKD1 gene was classified as ‘Uncertain Significance’,
according to the criteria of the ACMG standards and guidelines
(15).
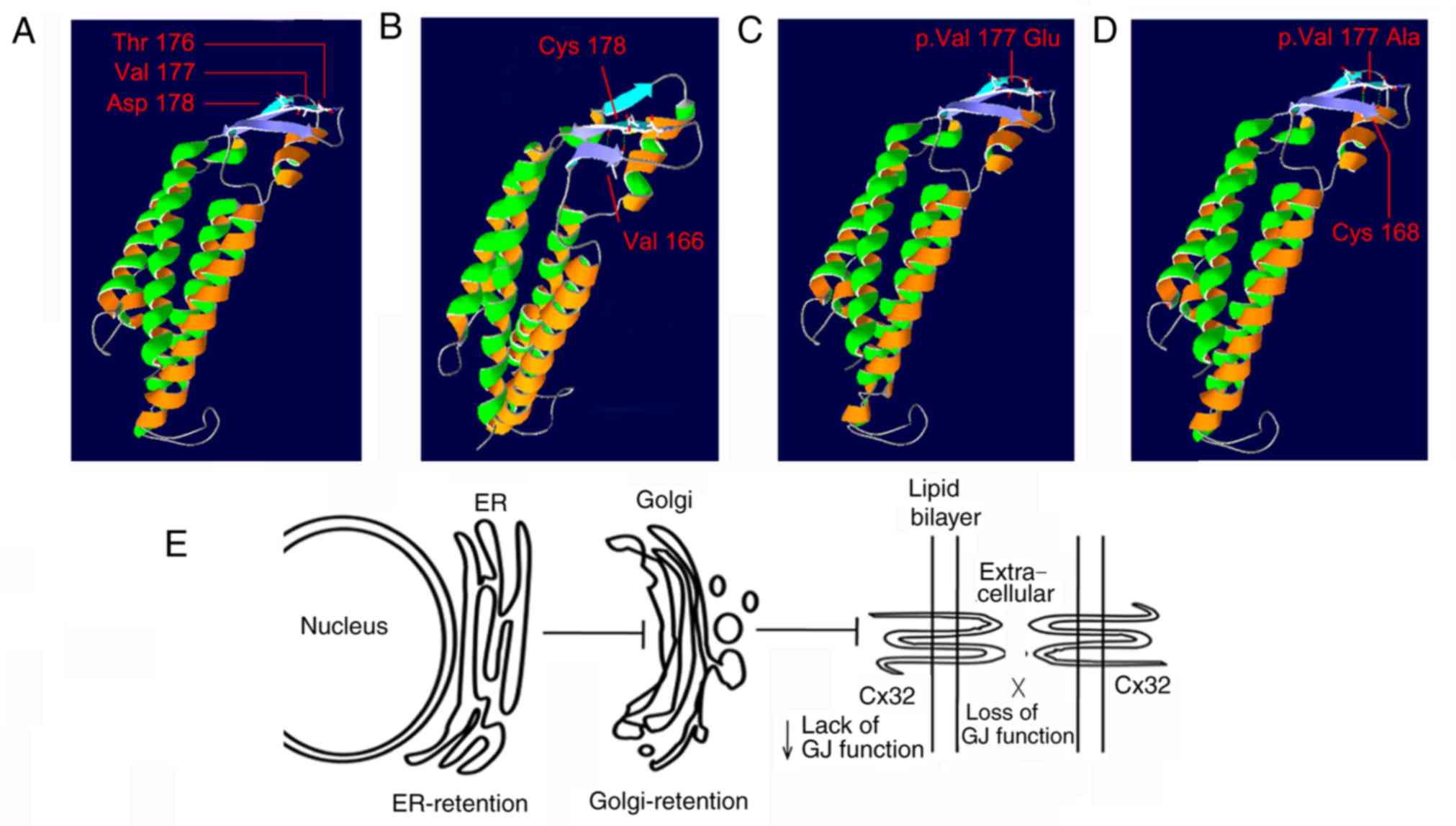
The Swiss-Model tool revealed that, compared with
the wild-type Cx32 (Fig. 2A), the
deletion of amino acid Val did not alter the 3-dimensional (3D)
structure of Cx32, and a new hydrogen bond was formed between amino
acid residues 178 and 166 (Fig.
2B). 3-D structures of Cx32 were also simulated for two otjer
mutations, p.V177E (Fig. 2C) and
p.V177A (Fig. 2D). The two
mutations did not lead to any alterations in the 3-D structure, but
the latter formed a new hydrogen bond between the amino acid
residues 177 and 168. Potential mechanisms underlying the
hypothesized pathogenicity of mutations affecting the amino acid
residue 177 were presented in Fig.
2E.
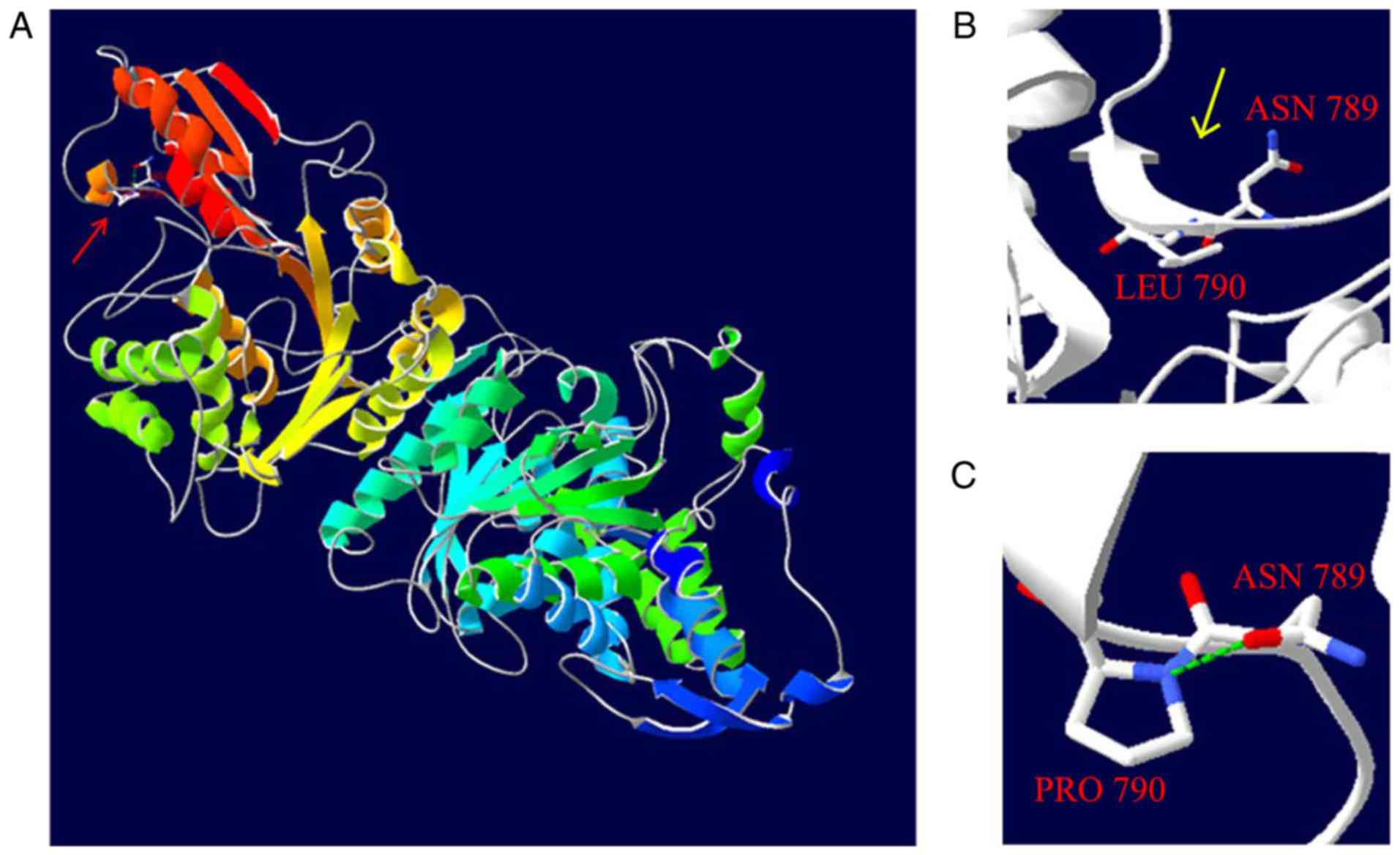
Computer simulation of 3-D structure using the
Swiss-Model tool revealed that the p.P790L mutation resulted in a
structure alternation from an α-helix (Fig. 3A) to a β-fold (Fig. 3B), and the removal of the hydrogen
bond between amino acid residues 790 and 789 (Fig. 3C).
Discussion
The GJB1 gene is the primary cause of X
chromosome linkage dominant inherited type 1 Charcot-Marie-Tooth
disease (16). GJB1 gene
mutation usually results in classical CMT, characterized by slowly
progressive distal muscle weakness, atrophy, sensory abnormalities,
pes cavus and hammer toes, usually starting during the teenage
years (17). In pedigrees with
CMTX, female patients always present with milder symptoms compared
with males, due to the random X chromosome inactivation in each
myelinating cell (18). In the
present study, the proband presented with the classical clinical
and electrophysiological symptoms of CMT caused by GJB1
mutation (19). It was identified
that the c.528_530delAGT mutation, leading to a valine deletion at
amino acid position 177, was a novel pathogenicity mutation. Valine
at amino acid position 177 is an important site for the function of
the GJB1 gene, which is supported not only by ortholog
analysis of GJB1 among species, but also by the other 2 mutations
that have been identified at this position, p.V177E (20) and p.V177A (21). Although these mutations do not
result in any change in the 3-D structure of the GJB1 gene,
the c.528_530delAGT and the p.V177A mutations lead to the formation
of new hydrogen bonds in the neighbor amino acid residues. A number
of potential mechanisms have been suggested for the pathological
effects of changes in the extracellular loop where the amino acid
residue 177 is located, as demonstrated in Fig. 2E. In vitro studies have
suggested that certain Cx32 mutants affecting the extracellular
loop, including p.E186K, p.T55I, p.A40V and p.R183S, interfered
with their trafficking between the endoplasmic reticulum (ER) and
Golgi apparatus (22–24). As a result, these abnormal proteins
were retained in the ER and/or Golgi and degraded by proteasomal or
lysosomal pathways (25). Although
a fraction of certain mutants, including R183S, may reach the cell
membrane, the amount of protein is too low to allow sufficient gap
junction formation (3). In
addition, they are not able to induce functional gap junction
formation for normal communication between cells (26). Therefore, the present study
confirmed that the c.528_530delAGT mutation is the pathogenic
mutation in this family.
Allelic heterogeneity and phenotypic variability
complicate CMT genetic analysis. In the study by Williams et
al (20), there was a mutation
of p.V177E detected in a 51-year-old European male with the
peripheral hallmarks of CMT, optic atrophy and cerebellar ataxia.
In the study by Ikegami et al (21), a mutation of p.V177A was identified
in a 15-year-old Japanese boy with CMTX, and in his mother and
maternal uncle. The patient presented with foot pain and exhibited
distal muscle weakness and atrophy of the lower limbs, with pes
cavus and hammer toe deformities, without detection of his motor
NCV. His maternal uncle had a similar presentation. His mother only
had a mild pes cavus deformity, but with decreased motor NCVs. The
clinical presentations of the patients in the present study were
also different from these previous cases. The proband in the
present study presented with more severe symptoms compared with his
mother. Therefore, there may be a particular genotype-phenotype
association with respect to the GJB1 gene.
The GJB1 gene encodes the gap junction beta 1
protein Cx32, a member of the large connexin family that encodes
homologous proteins and forms gap junctions (27). Cx32 is highly expressed in Schwann
cells, and cannot be functionally compensated by other connexin
family proteins as they are not expressed in peripheral nerves. A
total of 6 connexins compose a hemichannel, 2 of which form a gap
junction channel allowing the diffusion of ions and small molecules
(28). CMTX with GJB1
mutations has complex and diverse mechanisms of action, including
failure to synthesize Cx32 protein, Cx32 protein transport failure
to the plasma membrane and formation of gap junctions with abnormal
biophysical properties (29).
Certain amino acid changes, including p.E208K (30,31)
and p.R75Q (24,32), are associated with >1 of these
mechanisms. Loss of function is hypothesized to be a common factor
across GJB1 mutations. This may be the reason why the type
of mutation and the domain wherein the mutation was located were
not identified to be associated with clinical severity in previous
statistical analyses (33,34). However, patients carrying the same
mutation, including p.S26L and p.M34T, may present with severe or
mild clinical symptoms (33).
Therefore, the precise phenotype-genotype association in this
disease requires additional investigation.
There was a rare variation, c.2369C>T, in the
DHTKD1 gene identified in this family. DHTKD1 was
demonstrated to be the pathogenic gene in a Chinese CMT disease
type 2Q (CMT2Q) pedigree with the c.1455T>G mutation (35). These patients presented with
progressive and severe weakness and atrophy of the distal muscles.
The electrophysiological and morphological results were
characteristic of axonal features without demyelinating damage.
There has been no other CMT2Q patient or pedigree described, to the
best of our knowledge. Indeed, in the small pedigree in the present
study, an autosomal-dominant linked inheritance mode may be a
possibility. However, the electrophysiological and clinical
presentations of the patients were not consistent with CMT2Q. In
addition, the DHTKD1 mutation has been more commonly
identified in alpha-aminoadipic and alpha-ketoadipic aciduria, an
autosomal-recessive linked inherited disease whose symptoms exhibit
apparent heterogeneity, ranging from psychomotor retardation,
hypotonia, ataxia, epilepsy and failure to thrive, to no clinical
phenotype at all. Therefore, we hypothesized that the c.2369C>T
variation in the DHTKD1 gene was not the pathogenic mutation
in this pedigree for CMT2Q. The fact that the same protein may be
affected in 2 inherited diseases indicates that there may be a
particular genotype-phenotype association with respect to the
DHTKD1 gene (36).
In summary, the present study described a novel
mutation in the GJB1 gene in a Chinese CMTX pedigree with a
rare variation in the DHTKD1 gene. The results provided
additional information regarding the range of mutations of the
GJB1 gene, which may facilitate the understanding of the
genotype-phenotype association of CMTX.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated and analyzed during the present
study are included in this published article.
Authors' contributions
ZHZ drafted the manuscript. ZHZ and ZTC performed
the genetic experiments. RLZ and YZW made significant contributions
towards the design of the study, collected clinical samples and
obtained the clinical data of patients.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
Fujian Provincial Hospital, Fujian Medical University (Fuzhou,
China). Informed consent was obtained from all individuals included
in the study.
Patient consent for publication
Informed consent was obtained from all individuals
included in the study.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Kazamel M and Boes CJ: Charcot marie tooth
disease (CMT): Historical perspectives and evolution. J Neurol.
262:801–805. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Timmerman V, Strickland AV and Zuchner S:
Genetics of Charcot-Marie-Tooth (CMT) disease within the frame of
the human genome project success. Genes (Basel). 5:13–32. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ionasescu VV, Ionasescu R and Searby C:
Screening of dominantly inherited Charcot-Marie-Tooth neuropathies.
Muscle Nerve. 16:1232–1238. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bennett MV, Barrio LC, Bargiello TA, Spray
DC, Hertzberg E and Sáez JC: Gap junctions: New tools, new answers,
new questions. Neuron. 6:305–320. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li H and Durbin R: Fast and accurate short
read alignment with burrows-wheeler transform. Bioinformatics.
25:1754–1760. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li R, Li Y, Fang X, Yang H and Wang J,
Kristiansen K and Wang J: SNP detection for massively parallel
whole-genome resequencing. Genome Res. 19:1124–1132. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
McKenna A, Hanna M, Banks E, Sivachenko A,
Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly
M and DePristo MA: The genome analysis toolkit: A MapReduce
framework for analyzing next-generation DNA sequencing data. Genome
Res. 20:1297–1303. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Schwarz JM, Cooper DN, Schuelke M and
Seelow D: MutationTaster2: Mutation prediction for the
deep-sequencing age. Nat Methods. 11:361–362. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Waterhouse A, Bertoni M, Bienert S, Studer
G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C,
Bordoli L, et al: SWISS-MODEL: Homology modelling of protein
structures and complexes. Nucleic Acids Res. 46:W1. W296–W303.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bienert S, Waterhouse A, de Beer TA,
Tauriello G, Studer G, Bordoli L and Schwede T: The SWISS-MODEL
Repository-new features and functionality. Nucleic Acids Res.
45(D1): D313–D319. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Guex N, Peitsch MC and Schwede T:
Automated comparative protein structure modeling with SWISS-MODEL
and Swiss-PdbViewer: A historical perspective. Electrophoresis. 30
(Suppl 1):S162–S173. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Benkert P, Biasini M and Schwede T: Toward
the estimation of the absolute quality of individual protein
structure models. Bioinformatics. 27:343–350. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bertoni M, Kiefer F, Biasini M, Bordoli L
and Schwede T: Modeling protein quaternary structure of homo- and
hetero-oligomers beyond binary interactions by homology. Sci Rep.
7:104802017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shy ME, Blake J, Krajewski K, Fuerst DR,
Laura M, Hahn AF, Li J, Lewis RA and Reilly M: Reliability and
validity of the CMT neuropathy score as a measure of disability.
Neurology. 64:1209–1214. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Richards S, Aziz N, Bale S, Bick D, Das S,
Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, et al:
Standards and guidelines for the interpretation of sequence
variants: A joint consensus recommendation of the American college
of medical genetics and genomics and the association for molecular
pathology. Genet Med. 17:405–424. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yuan JH, Sakiyama Y, Hashiguchi A, Ando M,
Okamoto Y, Yoshimura A, Higuchi Y and Takashima H: Genetic and
phenotypic profile of 112 patients with X-linked
Charcot-Marie-Tooth disease type 1. Eur J Neurol. 25:1454–1461.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang Y and Yin F: A review of X-linked
Charcot-Marie-Tooth disease. J Child Neurol. 31:761–772. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Scherer SS, Xu YT, Nelles E, Fischbeck K,
Willecke K and Bone LJ: Connexin32-null mice develop demyelinating
peripheral neuropathy. Glia. 24:8–20. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hahn AF, Brown WF, Koopman WJ and Feasby
TE: X-linked dominant hereditary motor and sensory neuropathy.
Brain. 113:1511–1525. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Williams MM, Tyfield LA, Jardine P, Lunt
PW, Stevens DL and Turnpenny PD: HMSN and HNPP. Laboratory service
provision in the south west of England-two years' experience. Ann N
Y Acad Sci. 883:500–503. 1999. View Article : Google Scholar
|
|
21
|
Ikegami T, Lin C, Kato M, Itoh A, Nonaka
I, Kurimura M, Hirayabashi H, Shinohara Y, Mochizuki A and Hayasaka
K: Four novel mutations of the connexin 32 gene in four Japanese
families with Charcot-Marie-Tooth disease type 1. Am J Med Genet.
80:352–355. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Deschênes SM, Walcott JL, Wexler TL,
Scherer SS and Fischbeck KH: Altered trafficking of mutant
connexin32. J Neurosci. 17:9077–9084. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kleopa KA, Yum SW and Scherer SS: Cellular
mechanisms of connexin32 mutations associated with CNS
manifestations. J Neurosci Res. 68:522–534. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yum SW, Kleopa KA, Shumas S and Scherer
SS: Diverse trafficking abnormalities of connexin32 mutants causing
CMTX. Neurobiol Dis. 11:43–52. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
VanSlyke JK, Deschenes SM and Musil LS:
Intracellular transport, assembly, and degradation of wild-type and
disease-linked mutant gap junction proteins. Mol Biol Cell.
11:1933–1946. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Oh S, Ri Y, Bennett MV, Trexler EB,
Verselis VK and Bargiello TA: Changes in permeability caused by
connexin 32 mutations underlie X-linked Charcot-Marie-Tooth
disease. Neuron. 19:927–938. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Nakagawa S, Maeda S and Tsukihara T:
Structural and functional studies of gap junction channels. Curr
Opin Struct Biol. 20:423–430. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Nualart-Marti A, Solsona C and Fields RD:
Gap junction communication in myelinating glia. Biochim Biophys
Acta. 1828:69–78. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bortolozzi M: What's the function of
connexin 32 in the peripheral nervous system? Front Mol Neurosci.
11:2272018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang HL, Chang WT, Yeh TH, Wu T, Chen MS
and Wu CY: Functional analysis of connexin-32 mutants associated
with X-linked dominant Charcot-Marie-Tooth disease. Neurobiol Dis.
15:361–370. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Castro C, Gómez-Hernandez JM, Silander K
and Barrio LC: Altered formation of hemichannels and gap junction
channels caused by C-terminal connexin-32 mutations. J Neurosci.
19:3752–3760. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Abrams CK, Islam M, Mahmoud R, Kwon T,
Bargiello TA and Freidin MM: Functional requirement for a highly
conserved charged residue at position 75 in the gap junction
protein connexin 32. J Biol Chem. 288:3609–3619. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Shy ME, Siskind C, Swan ER, Krajewski KM,
Doherty T, Fuerst DR, Ainsworth PJ, Lewis RA, Scherer SS and Hahn
AF: CMT1X phenotypes represent loss of GJB1 gene function.
Neurology. 68:849–855. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Panosyan FB, Laura M, Rossor AM, Pisciotta
C, Piscosquito G, Burns J, Li J, Yum SW, Lewis RA, Day J, et al:
Cross-sectional analysis of a large cohort with X-linked
Charcot-Marie-Tooth disease (CMTX1). Neurology. 89:927–935. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xu WY, Gu MM, Sun LH, Guo WT, Zhu HB, Ma
JF, Yuan WT, Kuang Y, Ji BJ, Wu XL, et al: A nonsense mutation in
DHTKD1 causes Charcot-Marie-Tooth disease type 2 in a large Chinese
pedigree. Am J Hum Genet. 91:1088–1094. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hagen J, te Brinke H, Wanders RJ, Knegt
AC, Oussoren E, Hoogeboom AJ, Ruijter GJ, Becker D, Schwab KO,
Franke I, et al: Genetic basis of alpha-aminoadipic and
alpha-ketoadipic aciduria. J Inherit Metab Dis. 38:873–879. 2015.
View Article : Google Scholar : PubMed/NCBI
|