Introduction
Hepatocellular carcinoma (HCC), a primary malignancy
of the liver, predominantly occurs in patients with chronic liver
disease and cirrhosis. HCC is the most common type of primary liver
cancer in adults, and the most common cause of mortality in
patients with cirrhosis (1). An
estimated 662,000 HCC-associated mortalities are reported annually
worldwide, and half of these cases have occurred in China (2).
The prognosis of HCC is poor, as only 10–20%
carcinomas can be completely removed by surgery. The patients
usually succumb to the disease within 3–6 months (3). With the approval of sorafenib for
advanced HCC treatment, the prognosis of metastatic or unresectable
HCC has improved (4). However,
more advanced and efficient therapies, and better prognostic
molecular markers are required.
Long-noncoding RNAs (lncRNAs), accounting for 80% of
noncoding RNAs, are defined as endogenous cellular RNAs longer than
200 nucleotides that lack an obvious open-reading frame (5). The role of lncRNAs in the
carcinogenesis, microvascular invasion and metastasis of HCCs has
been reported in a number of studies (6–8). For
instance, the overexpression of lncRNA-urothelial cancer associated
1 contributed to HCC progression through the inhibition of
miR-216b, and subsequent activation of the fibroblast growth factor
receptor 1/extracellular signal-regulated kinase signaling pathway
(9). lncRNA HOX transcript
antisense RNA (HOTAIR) overexpression was demonstrated to predict
tumor recurrence in patients with HCC following liver
transplantation (3). Furthermore,
upregulation of lncRNA ZEB1-antisense RNA 1 (AS1) and ANRIL
expression was reported to predict a poorer prognosis in HCC
(10,11). In comparison with HCC patients
without HOTAIR expression, those with high HOTAIR expression
exhibited significantly poorer prognosis (12). Downregulation of lncRNA growth
arrest specific 5 expression was also associated with HCC prognosis
(13). Despite these studies, the
functions of multiple lncRNAs in HCC remain unclear. Therefore, a
comprehensive analysis of HCC-associated lncRNAs is necessary in
order to reveal possible biomarkers and/or potential therapeutic
targets.
In the present study, RNA-sequencing (RNA-seq) data
of HCC samples were studied to select signature lncRNAs for the
prognosis of HCC. Differentially expressed lncRNAs (DELs) were
identified, and prognosis-associated lncRNAs were screened. Based
on a risk score calculated using prognosis-linked lncRNAs, a risk
assessment model was established. This model was tested by
performing a survival analysis of high-risk and low-risk samples
from the validation and entire datasets. Multivariate and
univariate analyses were performed to investigate
prognosis-associated clinical factors. Moreover, the genes
associated with signature lncRNAs were screened for co-expression
network and functional enrichment analyses.
Materials and methods
Data acquisition and processing
The RNA-seq data of HCC samples were downloaded from
The Cancer Genome Atlas database (https://gdc-portal.nci.nih.gov/; May 8, 2017). A total
of 424 samples were analyzed based on Illumina HiSeq 2000 RNA
Sequencing platform. Of these, 261 samples from early stage (stage
I and II) cancer were used in the present study. Among these
samples, 194 samples with available clinical information were
retained, and the samples with <6-month recurrence-free survival
(RFS) time and the non-recurrent samples were discarded. Hence, 167
samples remained for further analyses, and these were randomly
divided into training (83 samples) and validation datasets (84
samples).
Screening of DELs
The 83 training samples were divided into poor
prognosis samples (recurrence samples with RFS <24 months) and
good prognosis samples (non-recurrent samples with RFS >24
months). DEseq (14) and edgeR
(15) packages in R (version
3.1.0; R-project.org/) were used for the
screening of the DELs between poor and good prognosis samples. The
threshold was set as false discovery rate <0.05 and |log(fold
change)|>1.3. Overlapping lncRNAs screened using the two
packages were used for further analyses.
Screening of prognosis-associated
lncRNAs
Cox regression analysis was conducted for the
identification of prognosis-associated lncRNAs in the training set
using the survival package in R (16). The significance of the identified
lncRNAs was tested with log-rank test, and P<0.05 was considered
to indicate a statistically significant difference.
Construction of a risk assessment
model
The prognostic risk of each sample in the training
set was calculated using the regression coefficients of
prognosis-associated lncRNAs via multivariate Cox regression
analysis. The following formula was used for calculation, wherein
the regression coefficient of each lncRNA was weighted:
Riskscore=βlncRNA1xexprlncRNA1+βlncRNA2xexprlncRNA2+⋯+βlncRNAnxexprlncRNAn
where, β lncRNAn indicates the multivariate Cox
regression coefficient and exprlncRNAn indicates the
expression level of lncRNAn.
Association between risk assessment
model and clinical feature
The obtained formula was used to calculate the risk
score of patients in the validation set. The median risk score was
used to distinguish between high-risk and low-risk samples. The
survival information was compared using Kaplan-Meier survival
analysis and receiver operating characteristic (ROC) curve
(17). The area under the ROC
curve (AUC) indicated the accuracy of prognosis. An AUC >0.5
indicated a relatively high accuracy of prognosis, with higher AUC
values indicating a more accurate prognosis. The high-risk and
low-risk samples were also subjected to Cox regression analysis to
screen for prognosis risk-associated clinical features. The
independent prognosis factors were identified using both univariate
and multivariate Cox regression analyses. Moreover, the association
between high or low risk and prognosis under the same clinical
status was tested.
Co-expression network and functional
enrichment
The co-expression network between lncRNAs and the
corresponding mRNAs was constructed using the MEM package (18,19).
P<0.05 was used as the cut-off selection criterion. Gene pairs
associated with lncRNAs were obtained from the Search Tool for the
Retrieval of Interacting Genes/Proteins database (string-db.org/), using a connection score >0.4 as
threshold. A co-expression network of these genes was also
constructed. Functional enrichment of these lncRNA-associated genes
was conducted using the Database for Annotation, Visualization and
Integrated Discovery (david.ncifcrf.gov/), and P<0.05 was set as the
cut-off value (20).
Results
Identification and validation of five
lncRNA prognostic signatures in two datasets
The clinical information of the samples in the
training set, validation set and entire set is presented in
Table I. A total of 23 poor
prognosis samples and 18 good prognosis samples were reported in
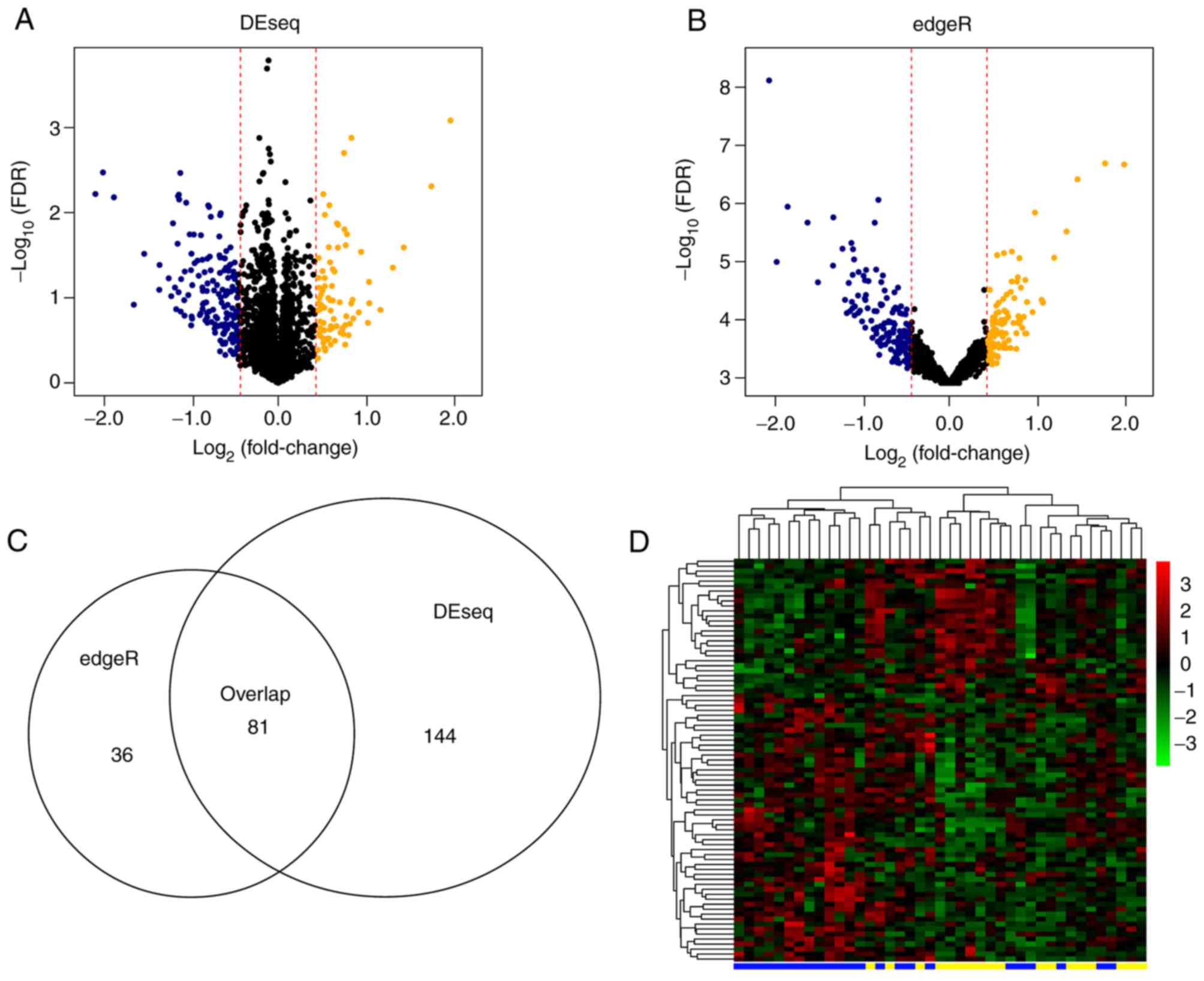
the training set. Using edgeR and DEseq packages for R, 117 and 225
DELs were screened comparing the poor and good prognosis samples,
respectively. The bidirectional hierarchical clustering results of
the overlapped 81 lncRNAs are displayed in Fig. 1.
 | Table I.Clinical information of samples in
training set, validation set and entire set. |
Table I.
Clinical information of samples in
training set, validation set and entire set.
|
| Stage I + II |
|---|
|
|
|
|---|
| Clinical
parameters | Training set
(n=83) | Testing set
(n=84) | Entire set
(n=167) |
|---|
| Age (years, mean ±
sd) | 58.99±11.82 | 58.42±13.35 | 59.14±12.59 |
| Gender
(male/female) | 61/22 | 56/28 | 117/50 |
| Pathologic M
(M0/-) | 64/19 | 70/14 | 134/33 |
| Pathologic N
(N0/-) | 61/22 | 63/21 | 124/43 |
| Pathologic T
(T1/T2) | 55/28 | 58/26 | 113/54 |
| Virus infection
(HBV/HCV/mixed/non) | 14/5/14/50 | 10/4/8/62 | 24/9/22/112 |
| Alcohol consumption
(yes/no/-) | 29/47/7 | 22/59/3 | 51/106/10 |
| Recurrence
(yes/no) | 31/52 | 32/52 | 63/104 |
| Recurrence free
survival time (months, mean ± sd) | 24.07±20.45 | 24.56±21.41 | 24.32±20.87 |
A total of 43 prognosis-associated lncRNAs were
obtained using univariate Cox regression analysis. Five of these
lncRNAs were selected for multivariate Cox regression analysis to
construct the risk assessment model. These five signature lncRNAs
were RP11-325L7.2, DKFZP434L187, RP11-100L22.4, DLX2-AS1 and
RP11-104L21.3 (Table II).
 | Table II.lncRNAs in the risk assessment
model. |
Table II.
lncRNAs in the risk assessment
model.
| lncRNA | Coefficient | HR | Lower .95 | Upper .95 | P-value |
|---|
| RP11-325L7.2 | −0.9283 | 0.3952 | 0.2193 | 0.7124 | 0.002 |
| DKFZP434L187 | −0.2867 | 0.7508 | 0.6238 | 0.9036 | 0.002 |
| RP11-100L22.4 | −0.6785 | 0.5074 | 0.2808 | 0.9167 | 0.025 |
| DLX2-AS1 | 0.3590 | 1.4319 | 1.0420 | 1.9678 | 0.027 |
| RP11-104L21.3 | 0.6786 | 1.9711 | 1.0793 | 3.5996 | 0.027 |
Riskscore=(-0.9283)xexprRP11-325L7.2+(-0.28667)xexprDKFZP434L187+(-0.67848)xexprRP11-100L22.4+(0.35902)xexprDLX2-AS1+(0.67857)xexprRP11-104L21.3
These five lncRNAs were used to evaluate the HCC
risk in each patient. The risk score distribution, RFS status and
expression of five signature lncRNAs in training set, validation
set and entire set are depicted in Fig. 2. Their distributions were similar,
supporting the robust prediction ability of the five lncRNA-based
risk score assessment model.
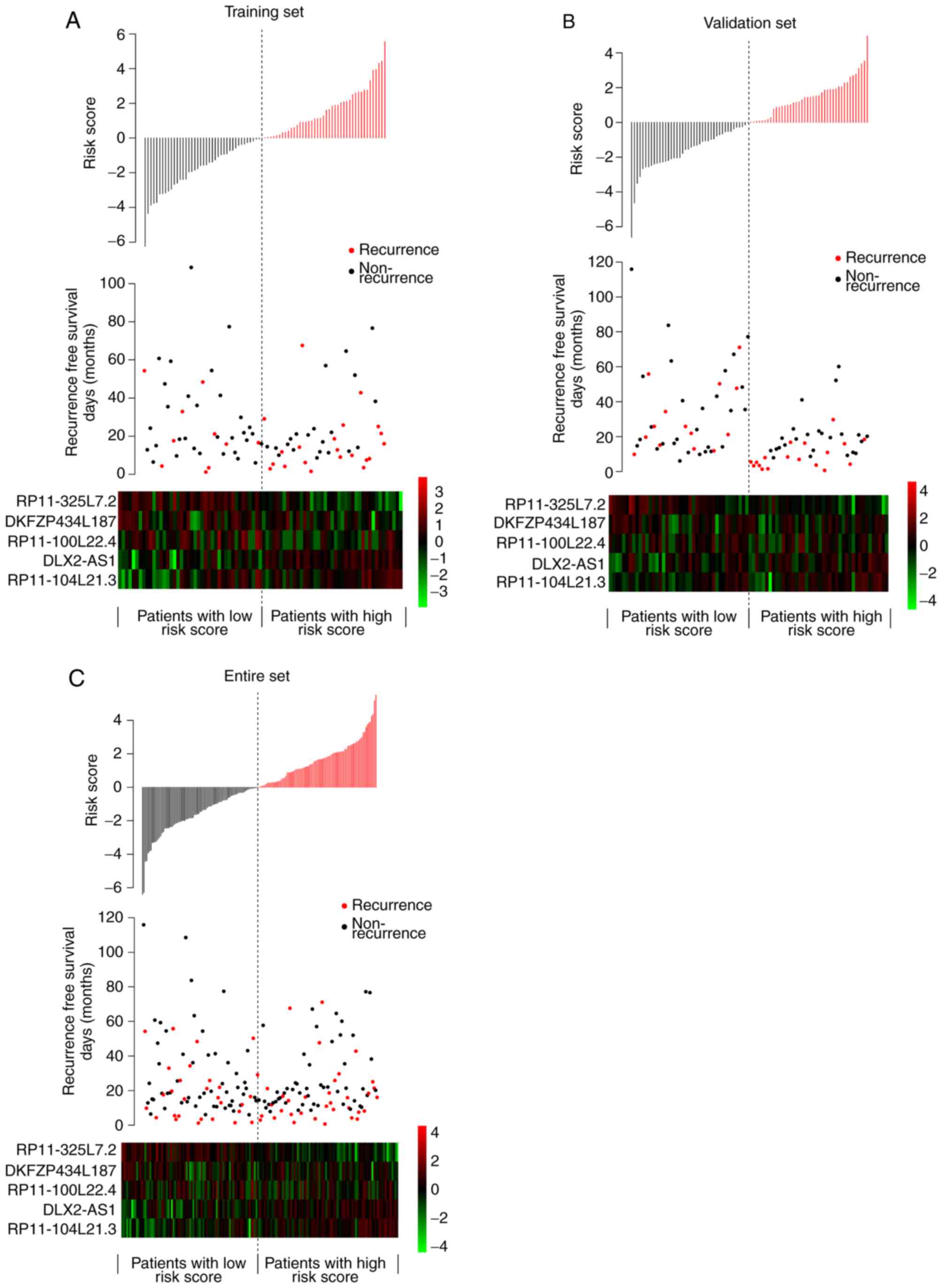 | Figure 2.Risk score distributions, RFS status,
and expressions of five signature lncRNAs in three datasets. Risk
score distributions, RFS status, and expressions of lncRNAs in the
(A) training set, (B) validation set and (C) entire set. In the
risk score distribution graphs, the horizontal axis represents
samples categorized between low and high-risk scores, while the
vertical axis represents risk scores. In the RFS status graphs, the
horizontal axis represents samples with low to high-risk scores,
while vertical axis represents RFS time. Recurrence samples and
non-recurrence samples are marked in red and black, respectively.
RFS, recurrence-free survival; lncRNAs, long noncoding RNAs. |
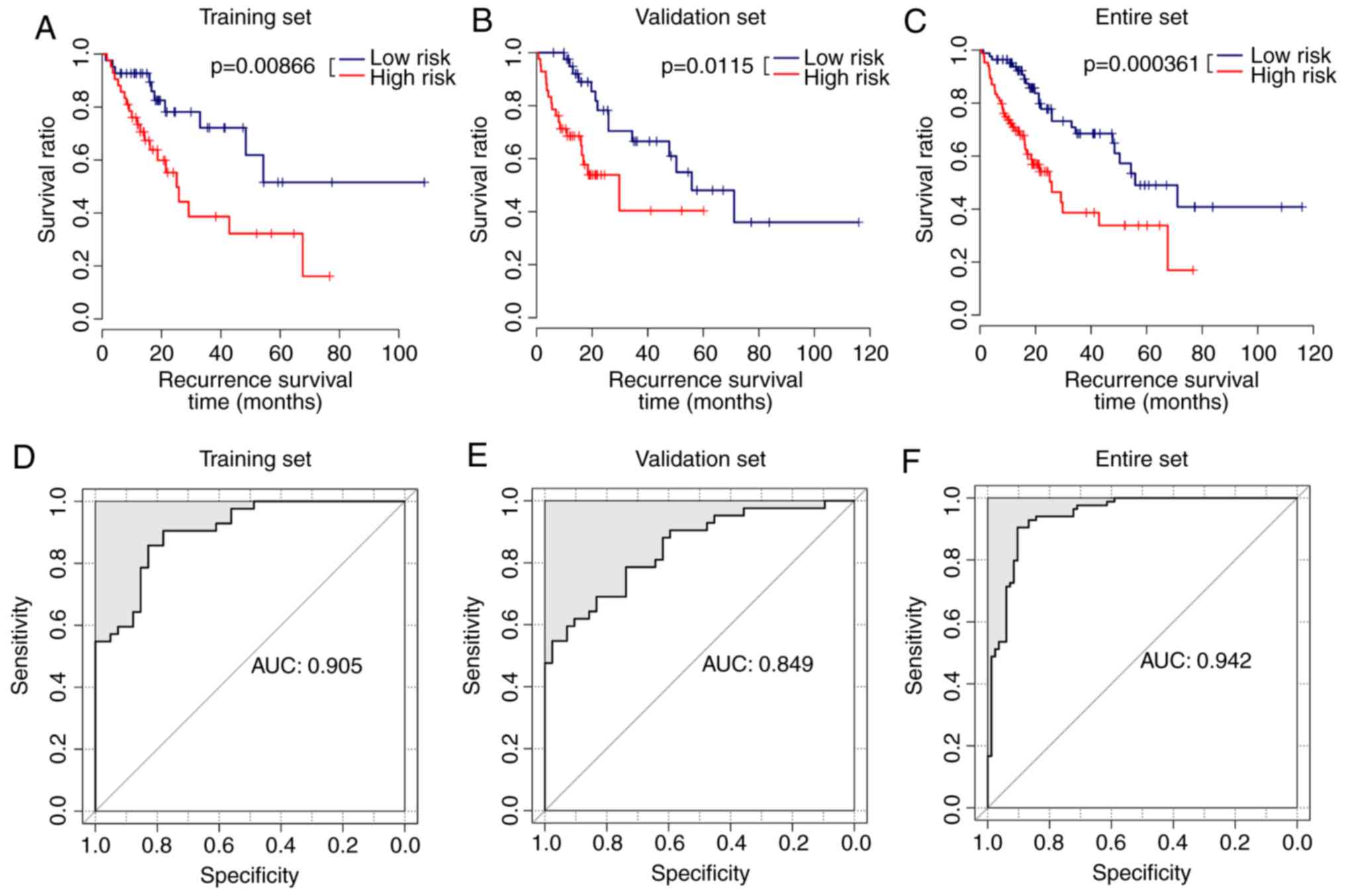
Survival analysis using the five
lncRNAs in the training, validation and entire sets
Using the risk assessment model, samples in the
training set were divided into high-risk group (42 samples) and
low-risk group (41 samples). The Kaplan-Meier survival curves
indicated that the samples from the low-risk group had longer
survival times compared with those from the high-risk group
(average RFS was 27.22±22.23 vs. 21.01±18.28 months) in the
training set (Fig. 3A). Similar
results were observed for the samples from the validation set.
Samples from the low-risk group had longer survival times compared
with those from the high-risk group (average RFS was 33.50±24.67
vs. 15.62±12.45 months; Fig. 3B).
The samples from the low-risk group similarly exhibited longer
survival times compared with those from the high-risk group
(average RFS was 30.39±23.57 vs. 18.31±5.78 months; Fig. 3C) in the entire set. The average
AUC value of these five lncRNAs in the training set was 0.905
(Fig. 3D), suggestive of the
highly accurate prognosis using these five lncRNAs. The average AUC
value of these five lncRNAs in the validation set was 0.849
(Fig. 3E). The average AUC value
of the lncRNAs for the entire dataset was 0.942 (Fig. 3F).
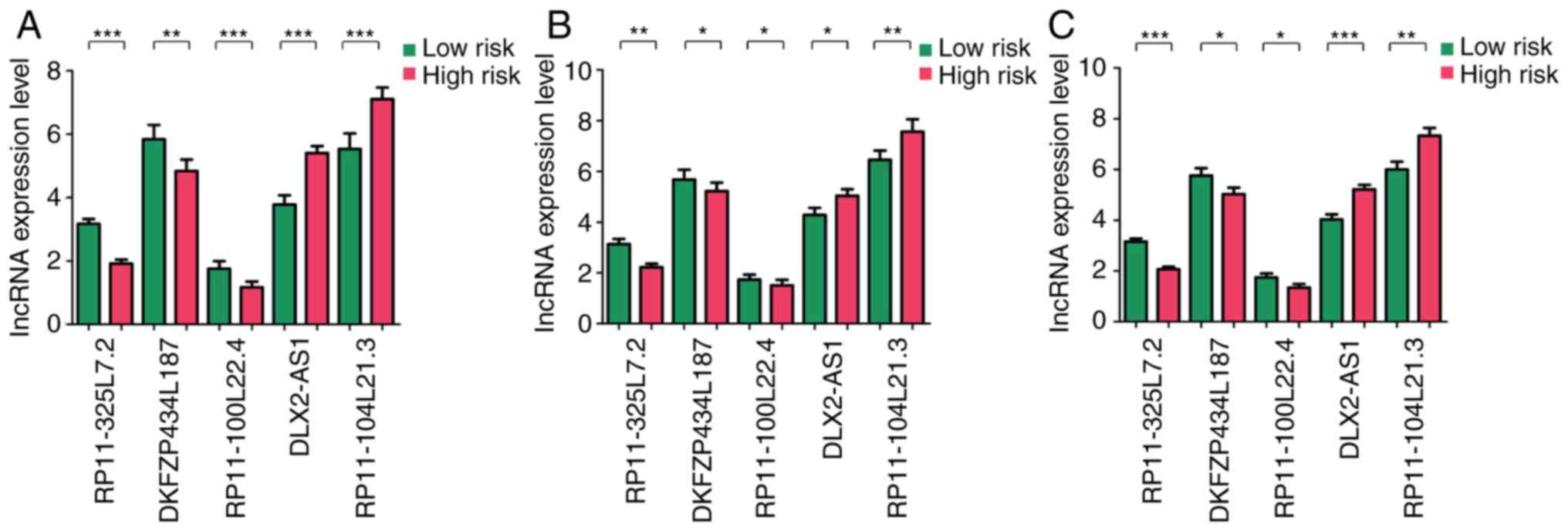
Significant differences were observed in the
expression levels of these five lncRNAs between samples from the
low- and high-risk groups in training set, validation set and
entire set (P<0.05; Fig. 4A-C,
respectively). Expression levels of DLX2-AS1 and RP11-104L21.3 were
significantly higher in the samples from the high-risk group
compared with those from the low-risk group, whereas the expression
levels of DKFZP434L187, RP11-100L22.4 and RP11-325L7.2 were
significantly lower in the samples from the high-risk group
compared with those from the low-risk group (P<0.05).
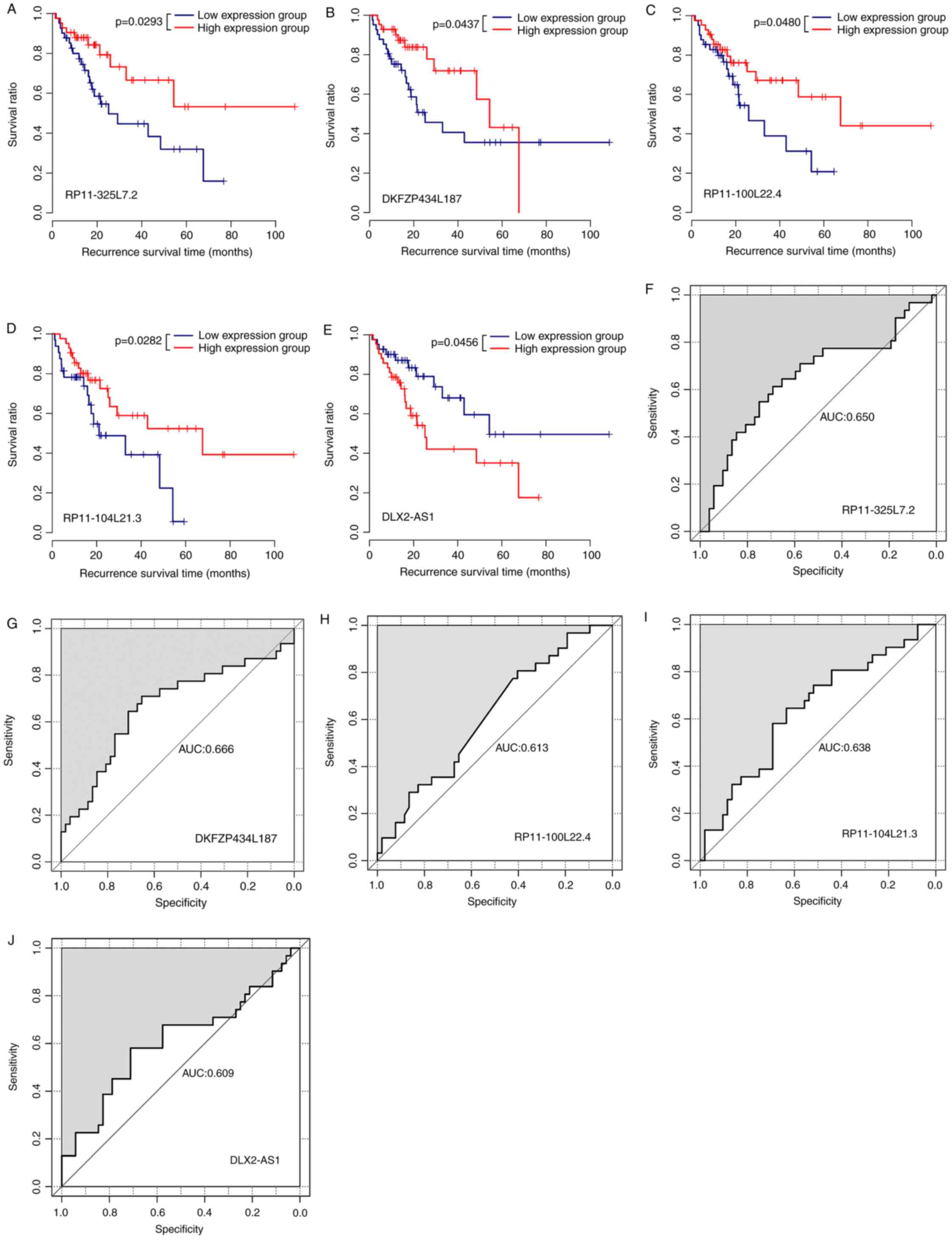
Association between survival information and
expression pattern was different for the five lncRNAs. In the
validation set, high expression samples had a longer survival time
compared with the low expression group for RP11-325L7.2,
DKFZP434L187, RP11-100L22.4 and RP11-104L21.3 (Fig. 5A-D, respectively), suggesting that
the high expression of these lncRNAs may be associated with good
prognosis. Low DLX2-AS1 expression was associated with longer
survival time compared with those from the high expression group
(Fig. 5E), indicating that high
expression of DLX2-AS1 may be associated with poor prognosis. The
AUC values for prognosis using RP11-325L7.2, DKFZP434L187,
RP11-100L22.4 and RP11-104L21.3 (Fig.
5F-I, respectively) were 0.650, 0.666, 0.613, and 0.638,
respectively, indicating that these four lncRNAs demonstrated
relatively high accuracy for prognosis, and the best performance
was produced by DKFZP434L187. The AUC value for DLX2-AS1 prognosis
was 0.609 (Fig. 5J), indicative of
the relatively and equally high accuracy of prognosis using this
lncRNA.
Prognostic factors for HCC
The results of univariate and multivariate Cox
regression analyses are presented in Table III. In the training, validation
and entire sets, the risk score was significantly associated with
the prognosis of patients and served as an independent prognostic
factor (P<0.05). Alcohol consumption was also an independent
predictive index in training and entire sets (P<0.05). Virus
infection was not a significant factor (P>0.05).
 | Table III.Cox regression results between
clinical features and prognosis. |
Table III.
Cox regression results between
clinical features and prognosis.
| A, Training set
(n=83) |
|---|
|
|---|
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Variables | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Risk score
(high/low) | 2.557 | 1.200–5.447 | 0.009 | 2.452 | 0.544–3.048 | 0.024 |
| Age
(≤60/>60) | 0.943 | 0.464–1.914 | 0.869 | 2.172 | 0.526–3.964 | 0.283 |
| Gender
(male/female) | 0.938 | 0.429–2.050 | 0.872 | 0.528 | 0.495–1.291 | 0.265 |
| Virus infection
(HBV/HCV/mixed) | 1.317 | 0.723–2.398 | 0.369 | 1.283 | 0.653–2.519 | 0.469 |
| Alcohol consumption
(yes/no) | 1.402 | 0.752–1.963 | 0.046 | 1.103 | 0.713–1.933 | 0.043 |
|
| B, Validation
set (n=84) |
|
|
| Univariate
analysis | Multivariate
analysis |
|
|
|
|
|
Variables | HR | 95% CI | P-value | HR | 95% CI | P-value |
|
| Risk score
(high/low) | 2.723 | 1.289–5.751 | 0.012 | 2.992 | 1.196–3.458 | 0.034 |
| Age
(≤60/>60) | 1.107 | 0.547–2.241 | 0.777 | 0.265 | 0.040–1.767 | 0.170 |
| Gender
(male/female) | 0.645 | 0.319–1.300 | 0.216 | 0.561 | 0.090–3.522 | 0.538 |
| Virus infection
(HBV/HCV/mixed) | 1.098 | 0.467–2.059 | 0.958 | 2.243 | 0.678–7.430 | 0.186 |
| Alcohol consumption
(yes/no) | 1.132 | 0.298–1.802 | 0.495 | 1.358 | 0.520–2.466 | 0.296 |
|
| C, Entire set
(n=167) |
|
| Univariate
analysis | Multivariate
analysis |
|
|
|
|
|
Variables | HR | 95% CI | P-value | HR | 95% CI | P-value |
|
| Risk score
(high/low) | 2.569 | 1.530–4.315 | 0.001 | 1.906 | 0.697–5.205 | 0.021 |
| Age
(≤60/>60) | 1.018 | 0.619–1.672 | 0.944 | 0.215 | 0.437–3.518 | 0.686 |
| Gender
(male/female) | 0.773 | 0.462–1.294 | 0.328 | 0.424 | 0.566–4.132 | 0.402 |
| Virus infection
(HBV/HCV/mixed) | 1.178 | 0.746–1.860 | 0.481 | 1.180 | 0.715–1.948 | 0.517 |
| Alcohol consumption
(yes/no) | 1.542 | 0.803–1.648 | 0.047 | 1.209 | 0.554–1.792 | 0.021 |
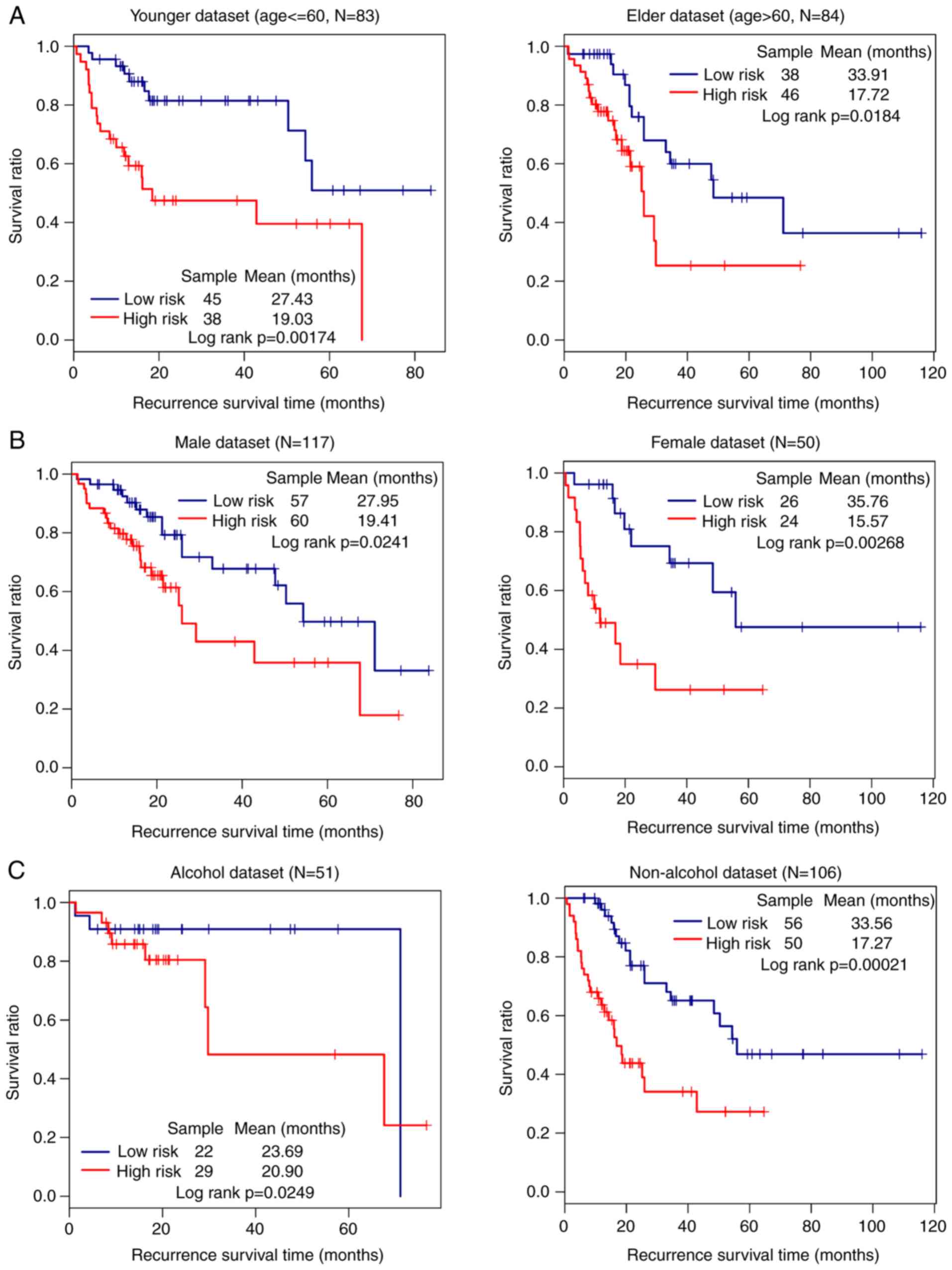
The impact of high and low risk on prognosis within
subgroups of the same clinical feature was evaluated. In the entire
set, the low-risk group had a significantly better prognosis
compared with the high-risk group in all age, sex and alcohol
subgroups (P<0.05; Fig. 6A-C,
respectively).
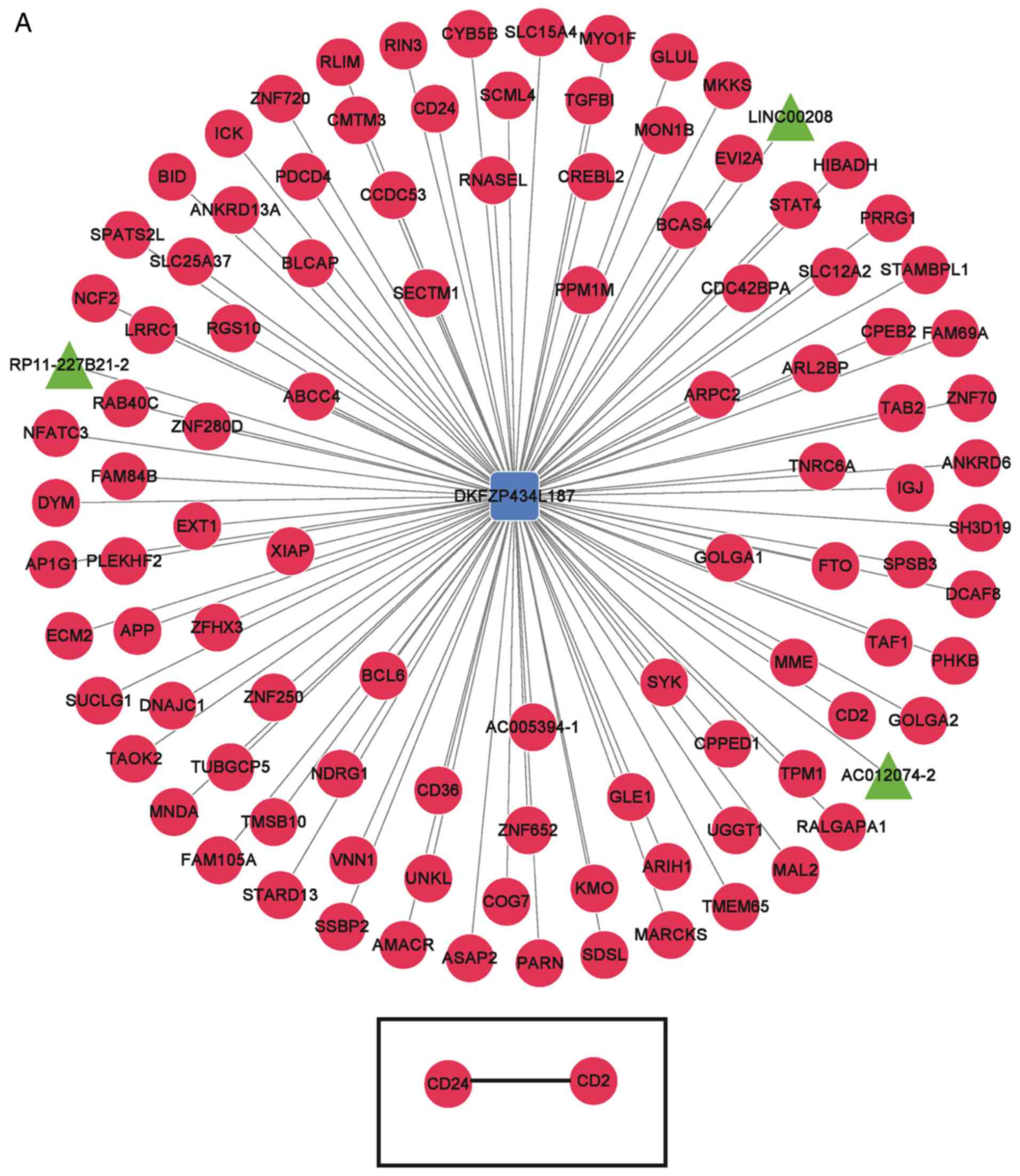
Functional enrichment for mRNAs
co-expressed with signature lncRNAs
The top 100 mRNAs associated with the five signature
lncRNAs were determined for the construction of signature
lncRNA-mRNA co-expression networks, and gene-gene co-expression
networks of the genes connected with signature lncRNAs (Fig. 7). WD repeat domain 5B (WDR5B) was
determined to be a target gene of RP11-100L22.4 and RP11-104L21.3
lncRNAs. Functional enrichment analysis demonstrated that
‘nucleoside-triphosphatase regulator activity’, ‘GTPase regulator
activity’, ‘enzyme binding’, ‘oxidation-reduction’ and ‘protein
amino acid phosphorylation’ were the most significantly enriched
functions, while ‘peroxisome proliferator-activated receptor (PPAR)
signaling pathway’, ‘fatty acid metabolism’, ‘Fc-γ-R-mediated
phagocytosis’, ‘leukocyte transendothelial migration’ and
‘adipocytokine signaling pathway’ were the most significantly
enriched pathways (Table IV).
 | Table IV.Significantly enriched functions and
pathways of genes correlated with signature long noncoding
RNAs. |
Table IV.
Significantly enriched functions and
pathways of genes correlated with signature long noncoding
RNAs.
| A, Enriched
functions (GO terms) |
|---|
|
|---|
| Category | Term | Gene count | P-value |
|---|
| GOTERM_MF_FAT | GO:
0060589~nucleoside-triphosphatase regulator activity | 24 | 3.52×10-4 |
| GOTERM_MF_FAT | GO: 0030695~GTPase
regulator activity | 23 | 6.35×10-4 |
| GOTERM_BP_FAT | GO:
0055114~oxidation reduction | 28 | 2.81×10-3 |
| GOTERM_BP_FAT | GO: 0006468~protein
amino acid phosphorylation | 28 | 4.93×10-3 |
| GOTERM_MF_FAT | GO: 0019899~enzyme
binding | 26 | 1.79×10-3 |
|
| B, Enriched
pathways (KEGG analysis) |
|
|
Category | Term | Gene
count | P-value |
|
| KEGG_PATHWAY | hsa03320: PPAR
signaling pathway | 9 | 7.24×10-4 |
| KEGG_PATHWAY | hsa00071: Fatty
acid metabolism | 7 | 8.66×10-4 |
| KEGG_PATHWAY | hsa04666:
Fc-γ-R-mediated phagocytosis | 9 | 5.62×10-3 |
| KEGG_PATHWAY | hsa04670: Leukocyte
transendothelial migration | 10 | 6.48×10-3 |
| KEGG_PATHWAY | hsa04920:
Adipocytokine signaling pathway | 7 | 1.20×10-2 |
Discussion
In the present study, a series of complex
bioinformatics analyses identified five lncRNAs associated with HCC
prognosis, namely RP11-325L7.2, DKFZP434L187, RP11-100L22.4,
DLX2-AS1 and RP11-104L21.3. A risk-model comprising these five
lncRNAs was able to distinguish low-risk and high-risk samples with
a relatively high prognosis accuracy. The evaluation of the target
genes of these lncRNAs further revealed that
‘nucleoside-triphosphatase regulator activity’, ‘GTPase regulator
activity’, ‘enzyme binding’, ‘PPAR signaling pathway’ and ‘fatty
acid metabolism’ were potentially the most affected pathways.
To the best of our knowledge, no studies have yet
reported a direct correlation between RP11-325L7.2, RP11-100L22.4
or RP11-104L21.3 expression and HCC prognosis. Other
RP11-associated lncRNAs have been reported to be implicated in HCC.
For instance, high expression levels of RP11-589N15.2,
RP11-343N15.5 and RP11-479G22.8 were reported to be associated with
the malignant phenotypes of HCC (21). In addition, RP11.404P21.3 and
RP11.488L18.10 were identified as two prognostic lncRNA biomarkers
for HCC (22). The lncRNA
RP11-513I15.6, a member of the three exosomal RNA-based panel, has
high sensitivity and specificity in the identification of HCC cases
among patients with chronic hepatitis C virus infection and healthy
individuals (23). In the present
study, the samples with higher expression of the three
RP11-associated lncRNAs exhibited longer survival times compared
with those which had lower expression, indicating that the high
expression of these lncRNAs may be associated with a good HCC
prognosis.
DLX2-AS1 is also termed DLX2 divergent transcript.
To the best of our knowledge, no studies have confirmed a link
between this lncRNA and HCC prognosis. However, the expression of
distal-less homeobox 2 (DLX2) protein has been linked with HCC. In
a previous study evaluating the response of HCC cells to
Actinidia chinensis root (a traditional Chinese medicine)
treatment, treatment with acRoots inhibited proliferation, invasion
and migration, clonality and epithelial-to-mesenchymal transition,
and promoted HCC cells apoptosis by downregulating DLX2 expression
(24). Moreover, high expression
level of DLX2 was linked to poor prognosis in patients with HCC
(24). Another study revealed the
association between DLX2 overexpression and poor prognosis in HCC
(25). In the present study,
samples with high DLX2-AS1 expression had shorter survival times
compared with those with low expression. The present results are
inconsistent with previous studies, and further experiments are
required to understand the discrepancy between the present and
previous results.
The function of DKFZP434L187 (also termed LINC02249)
is yet unknown. Based on the present results, this lncRNA may serve
as a novel prognostic lncRNA biomarker for HCC. However, further
studies are required in order to understand the role of this lncRNA
in HCC.
It is known that lncRNAs participate in various
biological processes such as transcription, translation, cellular
differentiation, chromatin modification, regulation of gene
expression, cell cycle and nuclear-cytoplasmic trafficking
(26–28). lncRNAs guide chromatin-modifying
complexes, thus allowing epigenetic modifications in cancer
(29). According to the results
presented in this study, WDR5B is a target gene of both
RP11-100L22.4 and RP11-104L21.3. WDR5 is associated with the lncRNA
HOXA distal transcript antisense RNA (HOTTIP), and its expression
is associated with disease progression and predictive outcomes in
HCC (30). HOTTIP binds directly
to WDR5, and targets WDR5/mixed linear leukemia complexes across
the HOXA gene cluster, thereby driving histone H3 lysine 4
trimethylation and gene transcription (31). Downregulated expression of HOX is
also reported to be involved in the development of HCC (31). Therefore, RP11-100L22.4 and
RP11-104L21.3 may participate in the prognosis of HCC through
interactions with WDR5.
Lipid metabolism is an essential function of the
liver, and any anomalies in this function may cause liver diseases,
including fibrosis (32). Defects
and/or deregulation of fatty acid metabolism have also been
reported to be associated with HCC (33,34).
Fatty acid metabolism also serves an important role in determining
the function of extra-mitochondrial pathways (35). In the present study, fatty acid
metabolism was revealed as a potentially dysfunctional pathway
associated with the signature lncRNA differentially expressed in
the HCC samples studied. Thus, fatty acid metabolism may be a
potential pathway influencing the prognosis of patients with
HCC.
Although these predictive results are valuable, the
present study has certain limitations. The expression of these
lncRNAs should be experimentally validated. In addition,
substantial experiments should be performed to confirm the
prognostic accuracy of these five lncRNAs in HCC.
In conclusion, the signature lncRNAs screened in
this study (RP11-325L7.2, DKFZP434L187, RP11-100L22.4, DLX2-AS1 and
RP11-104L21.3) may serve as novel lncRNA biomarkers to predict the
prognosis of HCC. Further studies of these lncRNAs and associated
genes may contribute to a deeper understanding of the underlying
mechanism of HCC development.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Presidential
Foundation of the 302 Military Hospital, China (grant no.
YNKT2014027).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YQS analyzed the data and wrote the manuscript. LW,
FFZ and XS collected and analyzed the data. JJ, FZ and SY drafted
the manuscript and contributed to the experimental design. HHL
conceived, designed the study and revised the manuscript. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
HCC
|
hepatocellular carcinoma
|
|
lncRNA
|
long noncoding RNA
|
|
DEL
|
differentially expressed lncRNA
|
|
PPAR
|
peroxisome proliferator-activated
receptor
|
|
RFS
|
recurrence-free survival
|
|
ROC
|
receiver operating characteristic
|
References
|
1
|
Forner A, Llovet JM and Bruix J:
Hepatocellular carcinoma. Lancet. 379:1245–1255. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
McGlynn KA and London WT: The global
epidemiology of hepatocellular carcinoma: Present and future. Clin
Liver Dis. 15:223–243, vii-x. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yang Z, Zhou L, Wu LM, Lai MC, Xie HY,
Zhang F and Zheng SS: Overexpression of long non-coding RNA HOTAIR
predicts tumor recurrence in hepatocellular carcinoma patients
following liver transplantation. Ann Surg Oncol. 18:1243–1250.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Llovet JM, Ricci S, Mazzaferro V, Hilgard
P, Gane E, Blanc JF, de Oliveira AC, Santoro A, Raoul JL, Forner A,
et al: Sorafenib in advanced hepatocellular carcinoma. N Engl J
Med. 359:378–390. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhou J, Li X, Wu M, Lin C, Guo Y and Tian
B: Knockdown of long noncoding RNA GHET1 inhibits cell
proliferation and invasion of colorectal cancer. Oncol Res.
23:303–309. 2016. View Article : Google Scholar
|
|
6
|
Yang F, Huo XS, Yuan SX, Zhang L, Zhou WP,
Wang F and Sun SH: Repression of the long noncoding RNA-LET by
histone deacetylase 3 contributes to hypoxia-mediated metastasis.
Mol Cell. 49:1083–1096. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhu Y, Yu M and Li Z, Kong C, Bi J, Li J,
Gao Z and Li Z: NcRAN, a newly identified long noncoding RNA,
enhances human bladder tumor growth, invasion, and survival.
Urology. 77:510.e1–5. 2011. View Article : Google Scholar
|
|
8
|
Prensner JR and Chinnaiyan AM: The
emergence of lncRNAs in cancer biology. Cancer Discov. 1:391–407.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang F, Ying HQ, He BS, Pan YQ, Deng QW,
Sun HL, Chen J, Liu X and Wang SK: Upregulated lncRNA-UCA1
contributes to progression of hepatocellular carcinoma through
inhibition of miR-216b and activation of FGFR1/ERK signaling
pathway. Oncotarget. 6:7899–7917. 2015.PubMed/NCBI
|
|
10
|
Li T, Xie J, Shen C, Cheng D, Shi Y, Wu Z,
Deng X, Chen H, Shen B, Peng C, et al: Upregulation of long
noncoding RNA ZEB1-AS1 promotes tumor metastasis and predicts poor
prognosis in hepatocellular carcinoma. Oncogene. 35:1575–1584.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hua L, Wang CY, Yao KH, Chen JT, Zhang JJ
and Ma WL: High expression of long non-coding RNA ANRIL is
associated with poor prognosis in hepatocellular carcinoma. Int J
Clin Exp Pathol. 8:3076–3082. 2015.PubMed/NCBI
|
|
12
|
Ishibashi M, Kogo R, Shibata K, Sawada G,
Takahashi Y, Kurashige J, Akiyoshi S, Sasaki S, Iwaya T, Sudo T, et
al: Clinical significance of the expression of long non-coding RNA
HOTAIR in primary hepatocellular carcinoma. Oncol Rep. 29:946–950.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tu ZQ, Li RJ, Mei JZ and Li XH:
Down-regulation of long non-coding RNA GAS5 is associated with the
prognosis of hepatocellular carcinoma. Int J Clin Exp Pathol.
7:4303–4309. 2014.PubMed/NCBI
|
|
14
|
Anders S and Huber W: Differential
expression analysis for sequence count data. Genome Biol.
11:R1062010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Robinson MD, McCarthy DJ and Smyth GK:
edgeR: A Bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang P, Wang Y, Hang B, Zou X and Mao JH:
A novel gene expression-based prognostic scoring system to predict
survival in gastric cancer. Oncotarget. 7:55343–55351.
2016.PubMed/NCBI
|
|
17
|
Hanley JA: The robustness of the
‘binormal’ assumptions used in fitting ROC curves. Med Decis
Making. 8:197–203. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Adler P, Kolde R, Kull M, Tkachenko A,
Peterson H, Reimand J and Vilo J: Mining for coexpression across
hundreds of datasets using novel rank aggregation and visualization
methods. Genome Biol. 10:R1392009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Harrow J, Frankish A, Gonzalez JM,
Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa
A, Searle S, et al: GENCODE: The reference human genome annotation
for The ENCODE Project. Genome Res. 22:1760–1774. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wu Y, Wang PS, Wang BG, Xu L, Fang WX, Che
XF, Qu XJ, Liu YP and Li Z: Genomewide identification of a novel
six-LncRNA signature to improve prognosis prediction in resectable
hepatocellular carcinoma. Cancer Med. 7:6219–6233. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang J, Pu J, Yao T, Lu X and Deng Y: Four
long noncoding RNAs as potential prognostic biomarkers for
hepatocellular carcinoma. J Cell Physiol. 234:8709–8716. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Abd El Gwad A, Matboli M, El-Tawdi A,
Habib EK, Shehata H, Ibrahim D and Tash F: Role of exosomal
competing endogenous RNA in patients with hepatocellular carcinoma.
J Cell Biochem. 119:8600–8610. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fang T, Fang Y, Xu X, He M, Zhao Z, Huang
P, Yuan F, Guo M, Yang B and Xia J: Actinidia chinensis
Planch root extract attenuates proliferation and metastasis of
hepatocellular carcinoma by inhibiting epithelial-mesenchymal
transition. J Ethnopharmacol. 231:474–485. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu J, Cui X, Qu L, Hua L, Wu M, Shen Z,
Lu C and Ni R: Overexpression of DLX2 is associated with poor
prognosis and sorafenib resistance in hepatocellular carcinoma. Exp
Mol Pathol. 101:58–65. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kim ED and Sung S: Long noncoding RNA:
Unveiling hidden layer of gene regulatory networks. Trends Plant
Sci. 17:16–21. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yoon JH, Abdelmohsen K and Gorospe M:
Posttranscriptional gene regulation by long noncoding RNA. J Mol
Biol. 425:3723–3730. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wilusz JE, Sunwoo H and Spector DL: Long
noncoding RNAs: Functional surprises from the RNA world. Genes Dev.
23:1494–1504. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Harries LW: Long non-coding RNAs and human
disease. Biochem Soc Trans. 40:902–906. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Quagliata L, Matter MS, Piscuoglio S,
Arabi L, Ruiz C, Procino A, Kovac M, Moretti F, Makowska Z,
Boldanova T, et al: Long noncoding RNA HOTTIP/HOXA13 expression is
associated with disease progression and predicts outcome in
hepatocellular carcinoma patients. Hepatology. 59:911–923. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang KC, Yang YW, Liu B, Sanyal A,
Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta
RA, et al: A long noncoding RNA maintains active chromatin to
coordinate homeotic gene expression. Nature. 472:120–124. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Moustafa T, Fickert P, Magnes C, Guelly C,
Thueringer A, Frank S, Kratky D, Sattler W, Reicher H, Sinner F, et
al: Alterations in lipid metabolism mediate inflammation, fibrosis,
and proliferation in a mouse model of chronic cholestatic liver
injury. Gastroenterology. 142:140–151, e12. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li Z, Guan M, Lin Y, Cui X, Zhang Y, Zhao
Z and Zhu J: Aberrant lipid metabolism in hepatocellular carcinoma
revealed by liver lipidomics. Int J Mol Sci. 18:25502017.
View Article : Google Scholar :
|
|
34
|
Zhou L, Wang Q, Yin P, Xing W, Wu Z, Chen
S, Lu X, Zhang Y, Lin X and Xu G: Serum metabolomics reveals the
deregulation of fatty acids metabolism in hepatocellular carcinoma
and chronic liver diseases. Anal Bioanal Chem. 403:203–213. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ockner RK, Kaikaus RM and Bass NM:
Fatty-acid metabolism and the pathogenesis of hepatocellular
carcinoma: Review and hypothesis. Hepatology. 18:669–676. 1993.
View Article : Google Scholar : PubMed/NCBI
|