Introduction
Inflammatory airway diseases, including asthma,
chronic obstructive pulmonary disease (COPD), and other pulmonary
inflammatory diseases, are characterized by limited airflow and
airway hyperresponsiveness, which can aggravate the process of lung
disorders further (1,2). As these lung diseases are associated
with abnormal immune responses of the airway to inhaled allergens
or toxic substances, an improved understanding of their
physiopathology is required, particularly that of the first-line
epithelial tissues (3). As innate
immune sensors and modulators, airway epithelial cells play a
pivotal role in mediating local innate and adaptive immune
responses in airway microenvironments (4). Damaged airway epithelium-instigated
abnormal airway barrier function responses trigger inflammatory
airway diseases (5,6). When injured, airway epithelial cells
release cytokines, including IL-25, IL-33 and thymic stromal
lymphopoietin, that mediate the inflammatory response and airway
remodeling of asthma (7–9). Therefore, airway epithelial
cell-produced functional molecules mediate intercellular
interactions and communications (10).
Intercellular informational communication plays a
critical role in modulating physiopathological functions in
organisms (11). Exosomes have
recently drawn widespread attention for facilitating cell-to-cell
communication and participating in various pathophysiological
processes, including immune responses (12), antigen presentation (13), inflammatory responses (14), cell migration and cell proliferation
(15). Various cell types, such as
alveolar macrophages, stem cells, airway epithelial cells and
eosinophils, secrete 30 to 120 nm-sized exosomes (3,16).
Exosomes are also widely present in body fluids, such as plasma,
serum, urine, breast milk and bronchoalveolar lavage fluids (BALF)
(17,18). These nano-vesicles are formed by the
inward budding of late endosomes that fuse with the cytoplasmic
membrane and release intracellular vesicles into the extracellular
space (19). Receptor cells can
uptake exosomes in various ways to alter their phenotypic
appearances and functions (20).
Therefore, exosomes, comprising proteins, lipids and nucleic acids
can release their contents to participate in the intercellular
transfer of information (19). In
particular, exosomes can alter the biological functions of
recipient cells via the transfer of mitochondria (21).
Recent studies have shown close associations between
exosomes and inflammatory airway diseases (22–24).
Exosomes are crucial in informational communication between asthma
microenvironments and various cells (25). Eosinophils secrete exosomes in
healthy subjects and patients with asthma, the latter of which is
particularly enhanced (16), but
stable patients with asthma and healthy subjects express exosomal
microRNAs (miRNAs/miRs) differently (26). Additionally, miR-34a, miR-92b and
miR-210 are enriched in exosomes, altering airway microenvironments
upon asthma development (27). One
study found exosome secretion in mice BALF to be significantly
enhanced upon contact with ovalbumin compared with controls
(28). Paredes et al
(29) noted that exosomes from
asthmatics induced leukotriene C4 and interleukin-8 (IL-8)
secretions in airway epithelial cells (29). These extracellular vesicles can also
mediate the pathogenesis of COPD (30). Plasma exosome concentrations are
higher in patients with acute exacerbations of COPD and stable COPD
than in healthy individuals (31).
Reportedly, exosomal miR-21 from bronchial epithelial cells in
patients with COPD promotes myofibroblast differentiation through
hypoxia-inducible factor 1α (32).
Clearly, exosomes from various sources mediate the pathogenesis of
inflammatory airway disorders.
While most studies have focused on the promotion of
the development of various cells or BALF exosome-induced airway
inflammatory disorders, few have paid attention to serum-derived
exosomes facilitating the pathogenesis of inflammatory airway
diseases. Thus, the present study investigated whether
serum-derived exosomes from House dust mite (HDM)-sensitized guinea
pigs could alter the phenotypic appearances of bronchial epithelial
cells and, if so, explore the underlying molecular mechanisms.
Materials and methods
Animals
Female guinea pigs (6–8 weeks old, n=20) were
obtained from Hunan Changsha Tianqin Biotechnology (http://cstqsw.com) and acclimated for a week before
the experiments. The animals were kept in a pathogen-free
environment and fed ad libitum. Our research protocol was
approved by the Animal Care Committee of Jiangxi Provincial
People's Hospital Affiliated with Nanchang University (approval no.
2021-052; Nanchang, China).
Cells, reagents and antibodies
The human bronchial epithelial cell line BEAS-2B was
obtained from The Cell Bank of Type Culture Collection of The
Chinese Academy of Sciences. HDM extract was purchased from
ALK-Abelló A/S, Dulbecco's modified Eagle's medium (DMEM) and
ExoQuick Exosome Isolation Reagent were acquired from Thermo Fisher
Scientific, Inc. Serum replacement was from Stemboscience, Inc.,
the PKH67 Green Fluorescent Cell Linker kit and DAPI were from
Sigma-Aldrich (Merck KGaA), while paraformaldehyde, Giemsa's stain
and Phosphotungstic acid hydrate were purchased from Beijing
Solarbio Science & Technology Co., Ltd. The BCA Protein
Quantitative kit was from CoWin Biosciences, polyvinylidene
difluoride (PVDF) membranes were from MilliporeSigma, interleukin
(IL)-4 and IL-13 ELISA kits were from Nanjing Jiancheng
Bioengineering Institute. The Immunoglobulin E (IgE; cat. no.
SFE40020) ELISA kit was purchased from Shanghai Shifeng, Inc.
(https://shfeng-edu.biomart.cn), and IL-6
(cat. no. EK 106/2-96) and nerve growth factor (NGF; cat. no. EK
1141-96) ELISA kits were from Hangzhou Multi Sciences (Lianke)
Biotech Co., Ltd. TAK-242 (cat. no. HY-11109) and BAY 11-7082 (cat.
no. HY-13453) were purchased from MedChemExpress, recombinant human
heat shock protein 70 (rHSP70; cat. no. 11660-H07H) was from Sino
Biological Technology Co., Ltd, and heat shock protein 70
(HSP70)-IN-1 (cat. no. M9273) was from AbMole Bioscience, Inc.
Primary antibodies against CD63 (cat. no. ab68418), IKKα/β (cat.
no. ab178870), phosphorylated (p)-IKKα/β (cat. no. ab194528) and
HSP70 (cat. no. ab31010) were procured from Abcam, anti-TLR4 (cat.
no. BA1717) was from Boster Biological Technology, and anti-p65
(cat. no. GB11997), anti-p-p65 (cat. no. GB11142-1), anti-β-actin
(cat. no. GB11001) and a HRP-conjugated goat anti-rabbit IgG
secondary antibody (cat. no. G1213) were purchased from Wuhan
Servicebio Technology Co., Ltd.
Cell culture
BEAS-2B cells were cultivated in DMEM supplemented
with 20% serum replacement (a cell culture supplement that replaces
fetal bovine serum to maintain cell growth and reproduction in
vitro), streptomycin (100 mg/ml) and penicillin (100 U/ml) at
37°C in a 5% CO2 atmosphere. The cells were passaged
every 2 days in a 1:3 ratio. In cellular co-culture with
serum-derived exosomes, serum replacement was used to prevent
interference from fetal bovine serum-derived exosomes. BEAS-2B
cells were co-cultured with serum-derived exosomes isolated from
the HDM group and PBS group at 37°C for 24 or 48 h separately.
BEAS-2B cells were also pretreated with or without 5 µg/ml
proteinase K (Beijing Solarbio Science & Technology Co., Ltd.),
5 µM BAY 11-7082, 5 µM HSP70-IN-1 and different concentrations of
TAK-242 (100 or 300 nM) at 37°C for 1 h, and then co-cultured with
serum-derived exosomes from the HDM group at 37°C for 24 h. BEAS-2B
cells were treated with or without rHSP70 (1 or 10 µg/ml) at 37°C
for 4 h.
Animal model
The guinea pigs (200–250 g, female) were housed in a
room maintained at moderate temperature (22±2°C) and humidity
(40–70%) with a 12 h light/dark cycle, and free access to food and
water for the duration of the present study. They were adaptively
fed for 7 days and randomly divided into two groups: Sham group
(PBS treatment group, n=10) and HDM group (n=10). HDM extracts
(100,000 U/ml) were diluted in a 0.1 mol/l PBS solution at
concentrations of 2,000 U/ml, 4,000 U/ml and 8,000 U/ml. A
HDM-induced asthma model was created as shown in Fig. 1. The guinea pigs in each group were
intraperitoneally injected with pentobarbital (35 mg/kg).
Subsequently, blood samples (5 ml per rodent) were withdrawn from
the heart via cardiac puncture. The guinea pigs were euthanized by
the immediate removal of the heart after exsanguination; death was
confirmed when the animals developed cardiac arrest, respiratory
arrest, corneal reflex arrest and rigor mortis. All procedures were
conducted strictly in accordance with the National Institutes of
Health Guide for the Care and Use of Laboratory Animals (33).
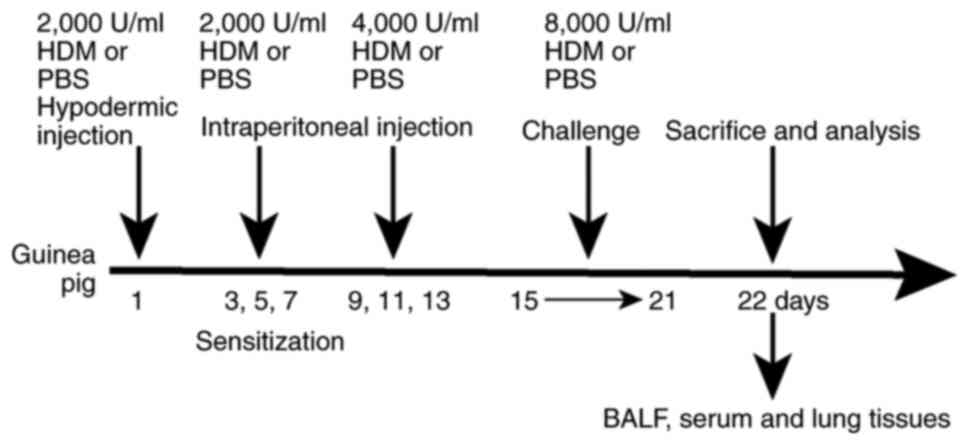 | Figure 1.Summary of the study protocol. The
HDM group guinea pigs were injected subcutaneously with 1,000 µl of
2,000 U/ml HDM on day 1, intraperitoneally injected with 1,000 µl
of 2,000 U/ml HDM on days 3, 5 and 7, and intraperitoneally with
500 µl of 4,000 U/ml HDM on days 9, 11 and 13. Guinea pigs in the
experimental group were then sensitized and challenged in atomized
boxes crafted for the present study with 8,000 U/ml HDM extract
from days 15 to 21, each time for 30 min. Sham group rodents were
sensitized and challenged with PBS, instead of HDM. Subsequently,
the animals were subjected to tracheotomy and washed with 5 ml
ice-cold PBS three times before BALF was collected. Serum from the
heart was collected using disposable needles, and lung tissues were
harvested. HDM, house dust mite; BALF, bronchoalveolar lavage
fluid. |
Lung histology
Harvested lung tissues from the guinea pigs were
fixed in 10% formalin solution for 24 h at 37°C and embedded in
paraffin. After deparaffinization, 5 µm sections of these tissues
were stained with hematoxylin for 5 min at 37°C and eosin for 5 min
at 37°C (H&E) to observe morphology, including pulmonary edema,
airway inflammation and airway epithelial injury under a light
microscope (ECLIPSE CI; Nikon Corporation).
Total cell counts
Precipitated cell suspension was conducted with 1 ml
PBS. A few droplets from the suspension were taken to the
cell-count boards to determine the total cell count in BALF per ml.
The remaining precipitated cells, including eosinophils,
neutrophils and lymphocytes, were fixed in 4% paraformaldehyde
solution for 30 min at 37°C and stained with Wright-Giemsa for 20
min at 37°C (at least 200 cells per sample) to deduce the
percentage of cells under a light microscope [ECLIPSE CI; Nikon
Corporation (magnification, ×40)].
ELISA
BALF IL-4 and IL-13 concentrations and serum IgE
levels from treated-guinea pigs and IL-6 and NGF contents in cell
supernatants were quantified using ELISA kits. In brief, the guinea
pigs were subjected to tracheotomy and washed with 5 ml ice-cold
PBS three times before BALF was collected. The obtained BALF
supernatants was centrifuged at 4°C (250 × g, 10 min). Different
concentrations of serum-derived exosomes (0, 50, 100, 200 µg/ml)
from the HDM group were added to BEAS-2B cells simultaneously and
incubated at 37°C for 24 and 48 h. After incubation, cell
supernatants were collected by centrifugation at 4°C (1,000 × g, 15
min).
Exosome isolation and
quantitation
Serum exosomes were isolated using the ExoQuick
Exosomes Isolation Reagent, according to the manufacturer's
recommended protocol. Briefly, serum from HDM-sensitized and
PBS-sensitized guinea pigs was differentially centrifuged at 4°C
(2,000 × g, 30 min) to remove cells and debris. The supernatants
were filtered through 0.22-µm filters to eliminate particles
>220 nm, and a reagent mixture was added to the well until the
solution was homogenous. The mixed suspension was then incubated at
4°C for 30 min and centrifuged at 10,000 × g for 10 min at room
temperature. Exosomes contained in the pellet at the bottom of the
tube were re-suspended in 200 µl PBS. Serum-produced exosomal
proteins were quantitated using the BCA Protein Assay kit, and
estimated by reference to a standard curve generated from proteins
[bovine serum albumin (BSA)] of known concentration.
Experimental groups
The samples were divided into three groups: Control
group (untreated cells), S-exo treatment group (exosomes from the
sham group) and H-exo treatment group (exosomes from the house dust
mite group).
Transmission electron microscopy
(TEM)
The ultrastructure of exosomes was observed using
TEM, referring to the methods described in a previous study
(34). A 20 µl drop of the exosomal
suspension was placed on parafilm and loaded to a carbon-coated
grid for 2 min. A 2% phosphotungstic acid solution prepared with
triple distilled water was used to stain the carbon-coated
grid-loaded suspension for 30 sec at room temperature. The sample
was then dried for 2 min under incandescent light, and the results
were observed and images captured using a transmission electron
microscope (JEM-1200EX; JEOL, Ltd.) at an acceleration voltage of
80 kV. The TEM images were cropped and scaled by Photoshop CS6
(Adobe Systems Incorporated).
Western blotting
After washing three times with precooled PBS and a
protease inhibitor cocktail on ice for 30 min, cells were harvested
in RIPA lysis buffer with 1 mM PMSF. The concentration of protein
was measured using a BCA protein assay kit. Total proteins (30 µg
per lane) were loaded, separated with 8–10% SDS-polyacrylamide
gels, and transferred onto PVDF membranes. The PVDF membranes were
blocked with 5% non-fat milk at room temperature for 2 h and then
incubated with a 1:1,000 dilution of the specific primary
antibodies at 4°C overnight. Followed by washing with TBS with 1%
Tween-20 three times (10 min each time), and incubation with
horseradish peroxidase-conjugated secondary antibodies (1:3,000) at
room temperature for 1 h. Then, the blots was visualized with
enhanced chemiluminescent solution (Beijing Solarbio Science &
Technology Co., Ltd.). Primary antibodies against the following
were used: CD63, HSP70, TLR4, IKKα/β, p-IKKα/β, p65, p-p65 and
β-actin. Band densities were analyzed using ImageJ software 6.0
(National Institutes of Health); the β-actin protein was used as an
internal reference.
Exosome labeling
Exosomes were labeled with PKH67 (a novel
fluorescent dye that labels living cells by binding to lipid
molecules in membrane structures) for general cell membrane
labelling according to the manufacturer's instructions, with minor
modifications. In brief, 100 µl serum-derived exosomes from the HDM
group were mixed with 1 ml Diluent C, and for control, 1 ml Diluent
C was mixed with PBS. The Diluent C added to the experiment and
control was prepared by mixing 1 µl PKH67 dye with 750 µl Diluent
C. Next, 1 ml of 1% BSA (Beijing Solarbio Science & Technology
Co., Ltd.) was added to the mixed experimental and control
solutions to stop the dyeing, and an ExoQuick Exosome Isolation
Reagent was used to precipitate the solutions and extract
exosomes.
Immunofluorescence staining
To detect the NF-κB subunit, p65, in nuclei, BEAS-2B
cells cultured in 6-well plates were treated with or without 100
µg/ml exosomes for 2 h at 37°C. Briefly, BEAS-2B cells were fixed
with 4.0% paraformaldehyde in PBS for 20 min at room temperature,
washed with PBS three times, and permeabilized in 0.25% Triton
X-100 at room temperature for 10 min. Non-specific binding was
blocked with 3% BSA in PBS for 30 min at room temperature, and the
cells were incubated with the p65 antibody at 4°C overnight,
followed by incubation with the horseradish peroxidase-conjugated
goat anti-rabbit IgG secondary antibody the next day at room
temperature for 1 h. Cells were then washed with PBS three times,
nuclei were stained with DAPI for 5 min at room temperature, and
detected with a fluorescence microscope (BX51; Olympus
Corporation).
Reverse transcription-quantitative
(RT-q)PCR
BEAS-2B cells were transferred into a tube
containing TRIzol reagent (Invitrogen; Thermo Fisher Scientific,
Inc.), and total RNA was extracted according to the manufacturer's
instructions. cDNA was synthesized from total RNA using RevertAid
First Strand cDNA Synthesis kit (Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol, and stored at −70°C until
further use. qPCR was performed using an SYBR Green Master Mix
(Roche Diagnostics) to verify the differential expression of the
genes. The thermocycling conditions for PCR were as follows:
Initial denaturation for 10 min at 95°C, followed by 40 cycles of
95°C for 15 sec and extension at 60°C for 1 min. RT-qPCR was
performed in duplicate and normalized to β-actin. Relative mRNA
expression was calculated using the 2−∆∆Cq method
(35). The primer sequences are
listed in Table I.
 | Table I.Sequences of primers used in reverse
transcription-quantitative PCR. |
Table I.
Sequences of primers used in reverse
transcription-quantitative PCR.
| Primer | Sequences
(5′→3′) |
|---|
| IL-6 | F:
GTAGTGAGGAACAAGCCAGAGC |
|
| R:
TACATTTGCCGAAGAGCCCT |
| NGF | F:
AGACATCAAGGGCAAGGAGGTG |
|
| R:
GCTGTCAACGGGATTTGGGTC |
| IL-8 | F:
CAGTTTTGCCAAGGAGTGCTAA |
|
| R:
AAACTTCTCCACAACCCTCTGC |
| TNF-α | F:
GCTGCACTTTGGAGTGATCG |
|
| R:
ATGAGGTACAGGCCCTCTGA |
| IL-1β | F:
GCGGCATCCAGCTACGAAT |
|
| R:
AAGCCTCGTTATCCCATGTGTC |
| β-actin | F:
CACCCAGCACAATGAAGATCAAGAT |
|
| R:
CCAGTTTTTAAATCCTGAGTCAAGC |
Statistical analysis
GraphPad Prism 6.0 (GraphPad Software, Inc.) was
used for all statistical analyses. Data are expressed as the mean ±
SD. An unpaired Student's t-test was used for comparisons between
two groups, and one-way ANOVA followed by the Bonferroni post hoc
test were employed for multiple comparisons. P<0.05 was
considered to indicate a statistically significant difference. Each
experiment was repeated three times.
Results
Successful establishment of allergic
asthma model in guinea pigs via HDM sensitization and
challenge
Bronchial and alveolar septum structures in the sham
group were normal, with fewer inflammatory cells infiltrating the
lung tissues, but in the HDM group, bronchial mucosal edema,
increased mucus secretion and observable inflammatory cell
infiltration were observed (Fig.
2A). Serum IgE levels increased significantly in the HDM group
compared with those in the sham group (Fig. 2B), as did IL-4 and IL-13 BALF levels
(Fig. 2C) and total BALF cell
numbers, with increases in inflammatory cells, such as lymphocytes,
neutrophils and eosinophils (Fig.
2D). A model of allergic asthma was successfully established in
guinea pigs via HDM sensitization and challenge.
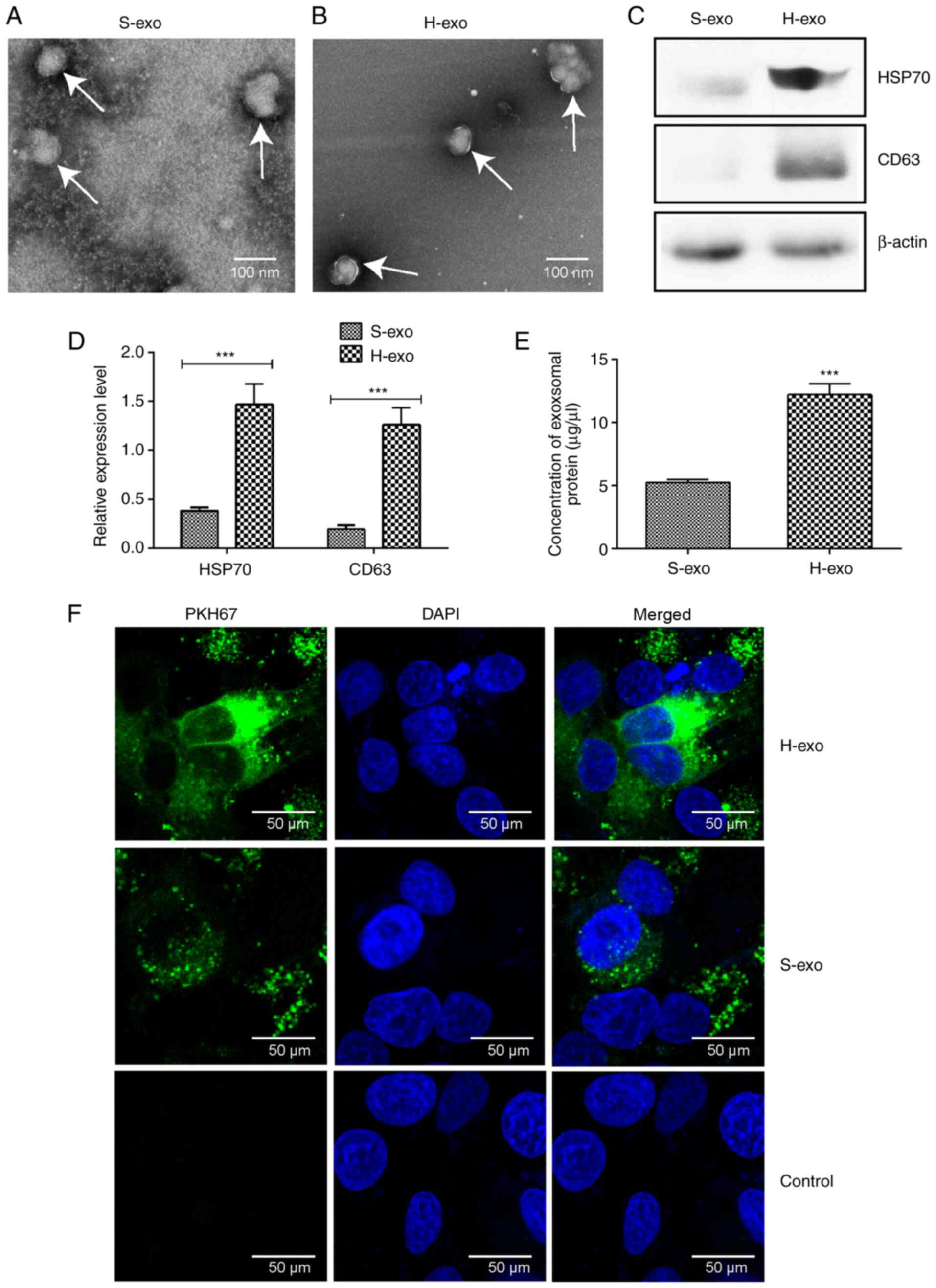
BEAS-2B cells efficiently incorporated
serum-derived exosomes
TEM revealed that exosome sizes in the two groups
were in the range of 30–120 nm in diameter, and the vesicles
appeared as double-layer circles (Fig.
3A and B). According to western blot analyses, the expression
levels of exosome marker proteins, HSP70 and CD63, in the H-exo
group were significantly higher than those in the S-exo group
(Fig. 3C and D), as were serum
exosomal protein amounts (Fig. 3E).
The uptake of serum-derived exosomes by BEAS-2B cells was observed
24 h later; green fluorescence was perceived in BEAS-2B cell
cytoplasm (Fig. 3F). The data
demonstrated that BEAS-2B cells effectively absorbed serum-derived
exosomes from the H-exo and S-exo groups.
Incubating BEAS-2B cells with
serum-derived exosomes induces changes in gene expression
profiles
The ability of serum-derived exosomes to stimulate a
proinflammatory response in BEAS-2B cells was explored. The cells
were cultured separately with or without 100 mg/ml serum-derived
exosomes obtained from the H-exo and S-exo groups for 24 h, and
cytokine expression was determined using RT-qPCR. The results
showed that IL-6 and NGF secretions were higher in exosomes from
the H-exo group than in those from the control group (Fig. 4A). To determine whether IL-6 and NGF
levels in exosome-treated BEAS-2B cells were
concentration-dependent; BEAS-2B cells were co-cultured with the
concentration gradient of exosomes from the HDM group at different
times (24 and 48 h). In the exosome-treated BEAS-2B cells, the
increase in mRNA expression levels and secretion of IL-6 and NGF
was only dose-dependent for 50 and 100 µg/ml. There was a decrease
at 200 µg/ml (Fig. 4B-E).
Therefore, serum-derived exosomes from the HDM group modulated
BEAS-2B cell phenotypic appearances and enhanced the inflammatory
responses.
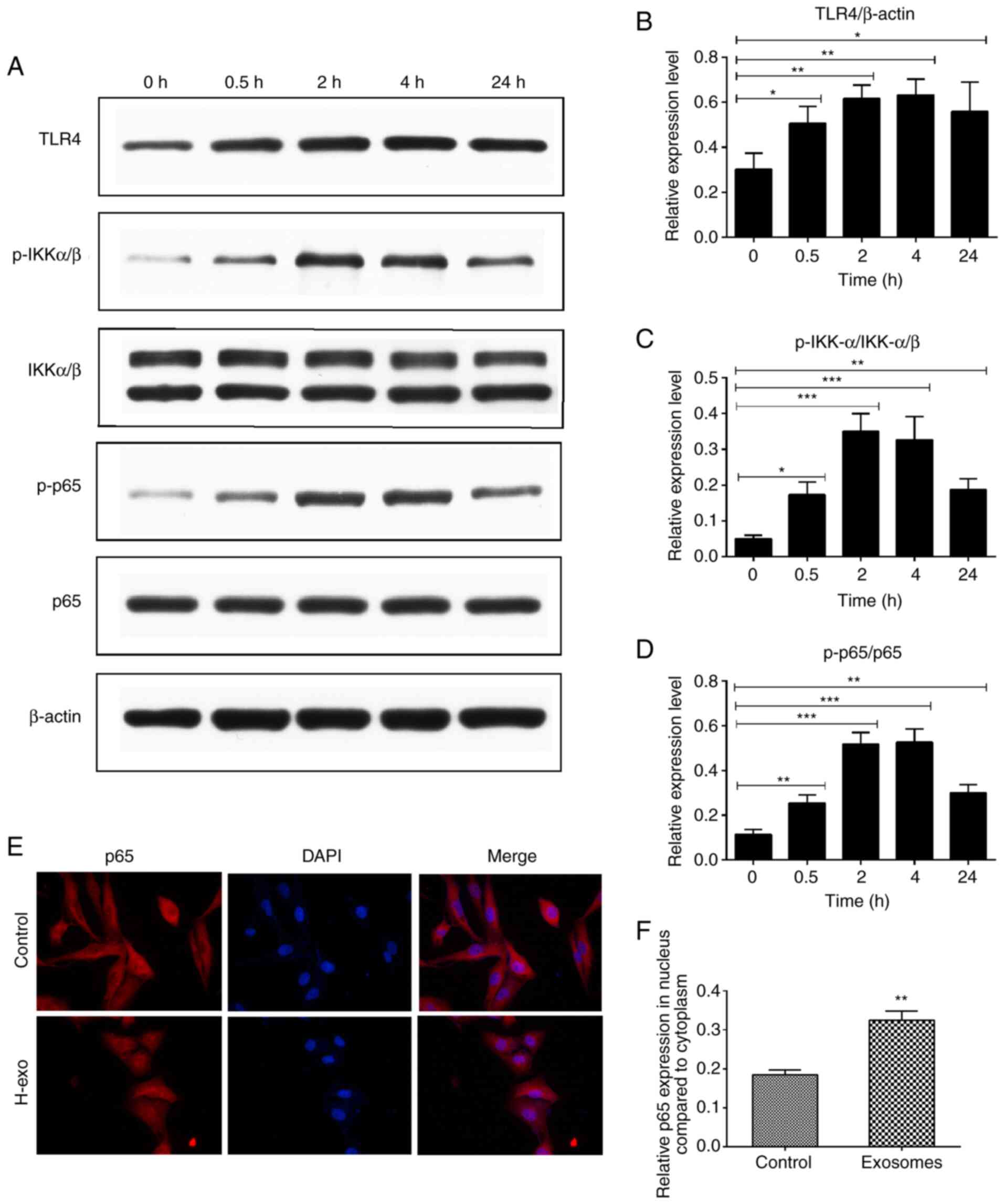
Serum-derived exosomes from the HDM
group induces TLR4-NF-κB pathway activation in BEAS-2B cells
To determine the signaling pathway through which
serum-derived exosomes from the HDM group promoted the inflammatory
response in BEAS-2B cells, TLR4-NF-κB pathway protein levels were
evaluated using western blot analysis, and the signaling events
triggered by exosomes were observed. As presented in Fig. 5A, IKKα/β and p65 phosphorylation
resulted in a rapid NF-κB activation 30 min after exosome addition.
As expected, stimulation with exosomes significantly increased
TLR4, p-IKKα/β and p-p65 expression levels, with maximum elevations
noted at 2 or 4 h (Fig. 5A-D).
Generally, NF-κB signaling activation is linked to p65 protein
phosphorylation and translocation to the cell nucleus.
Serum-derived exosomes from the HDM group stimulation resulted in
the translocation of p65 from the cytoplasm to the nucleus
(Fig. 5E and F).
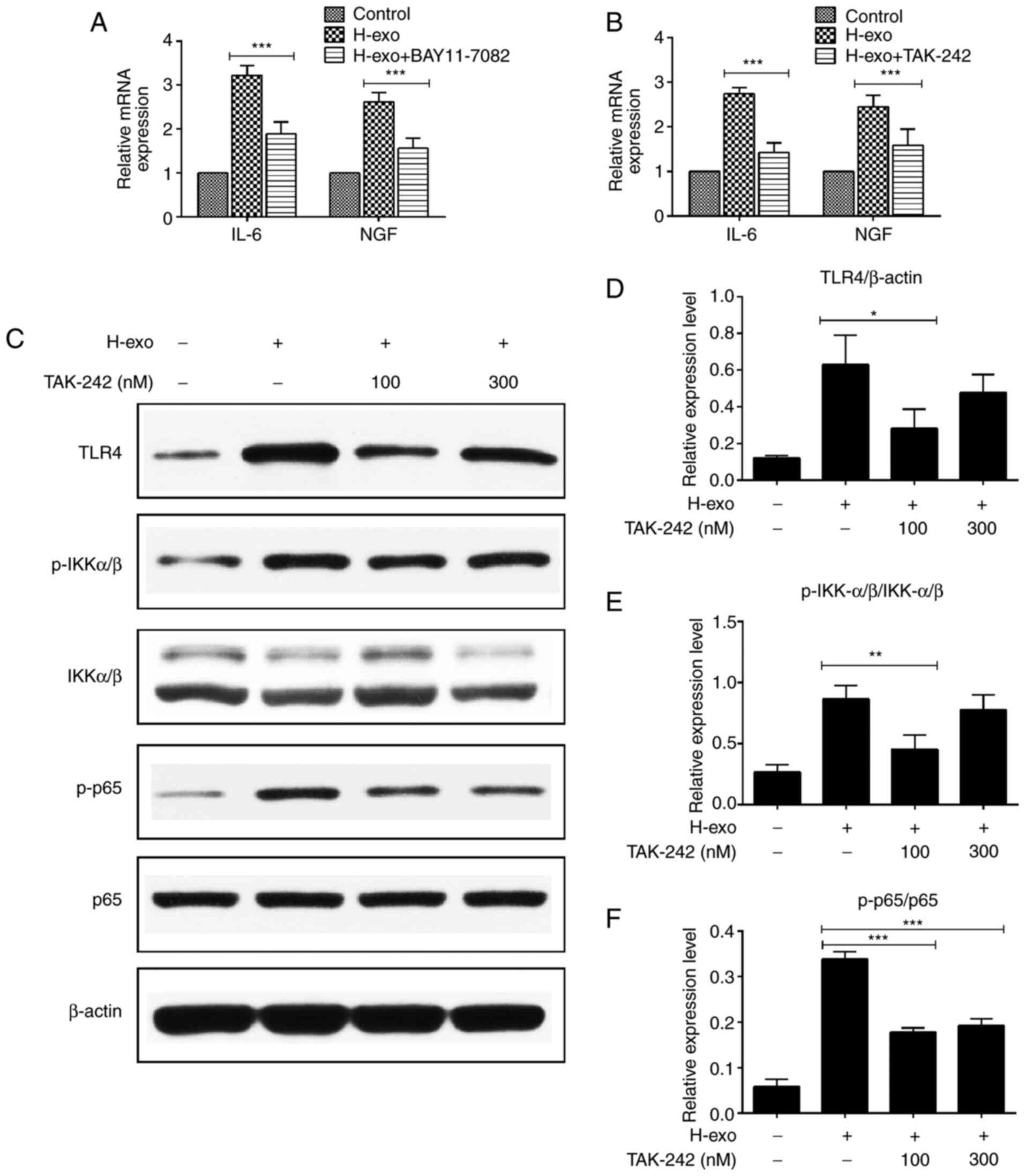
TAK-242 reduces the expression of
NF-κB phosphorylation in BEAS-2B cells
IL-6 and NGF mRNA levels in BEAS-2B cells were
evaluated via RT-qPCR after blocking the TLR4-NF-κB pathway. It was
demonstrated that IL-6 and NGF expression levels decreased when
BEAS-2B cells were treated with serum-derived exosomes from the HDM
group in the presence of the NF-κB inhibitor (BAY 11–7082) and TLR4
inhibitor (TAK-242) (Fig. 6A and
B). TLR4, p-IKKα/β and p-p65 expression levels increased after
BEAS-2B cell stimulation by serum-derived exosomes from the HDM
group. To verify the TLR4 pathway protein levels obtained above,
various concentrations of TAK-242 were added to BEAS-2B cells.
TLR4, p-IKKα/β and p-p65 protein levels were partially inhibited in
BEAS-2B cells by TAK-242 (Fig.
6C-F). These findings indicated that blocking TLR4 suppressed
the exosome-mediated signaling pathway in BEAS-2B cells.
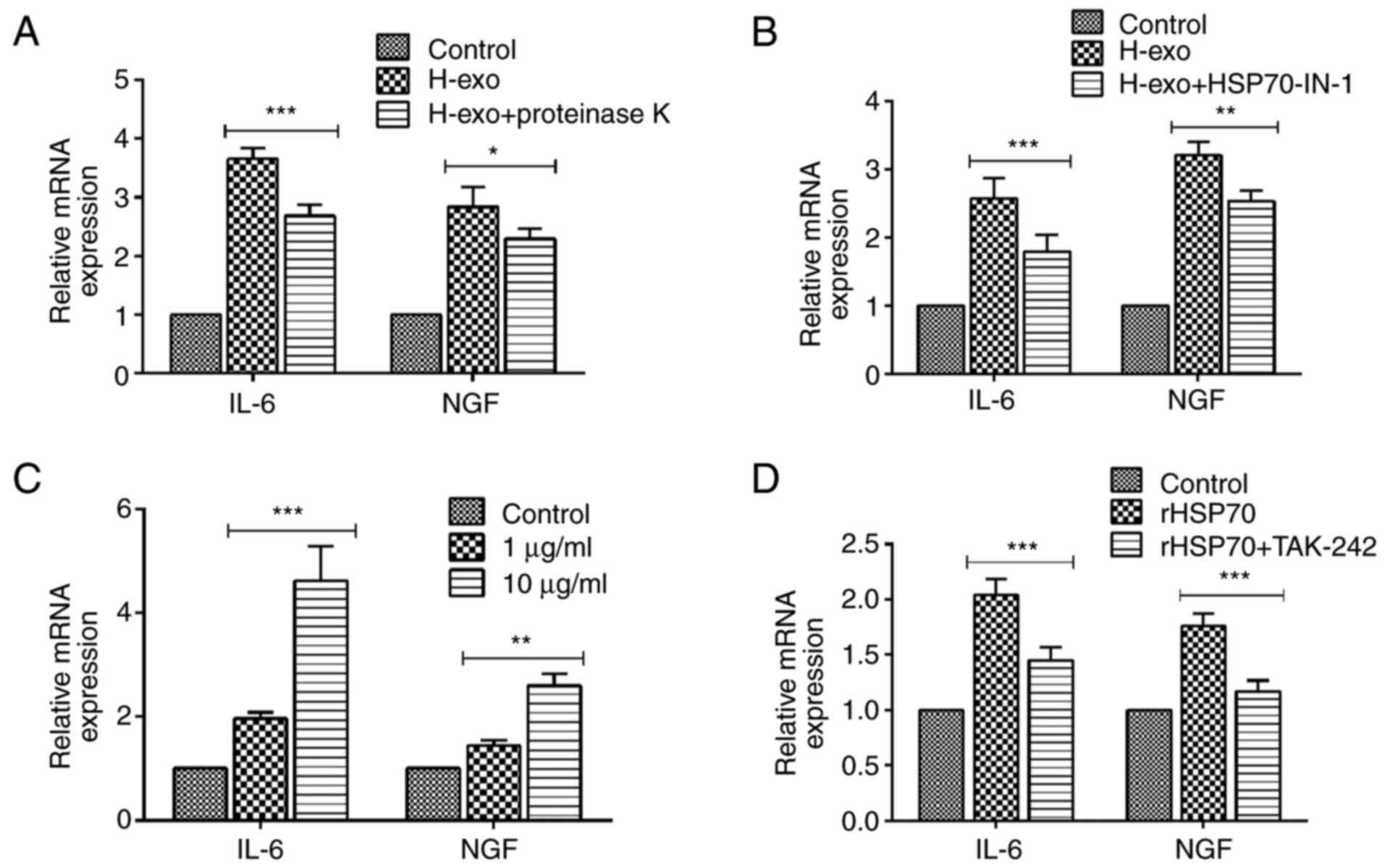
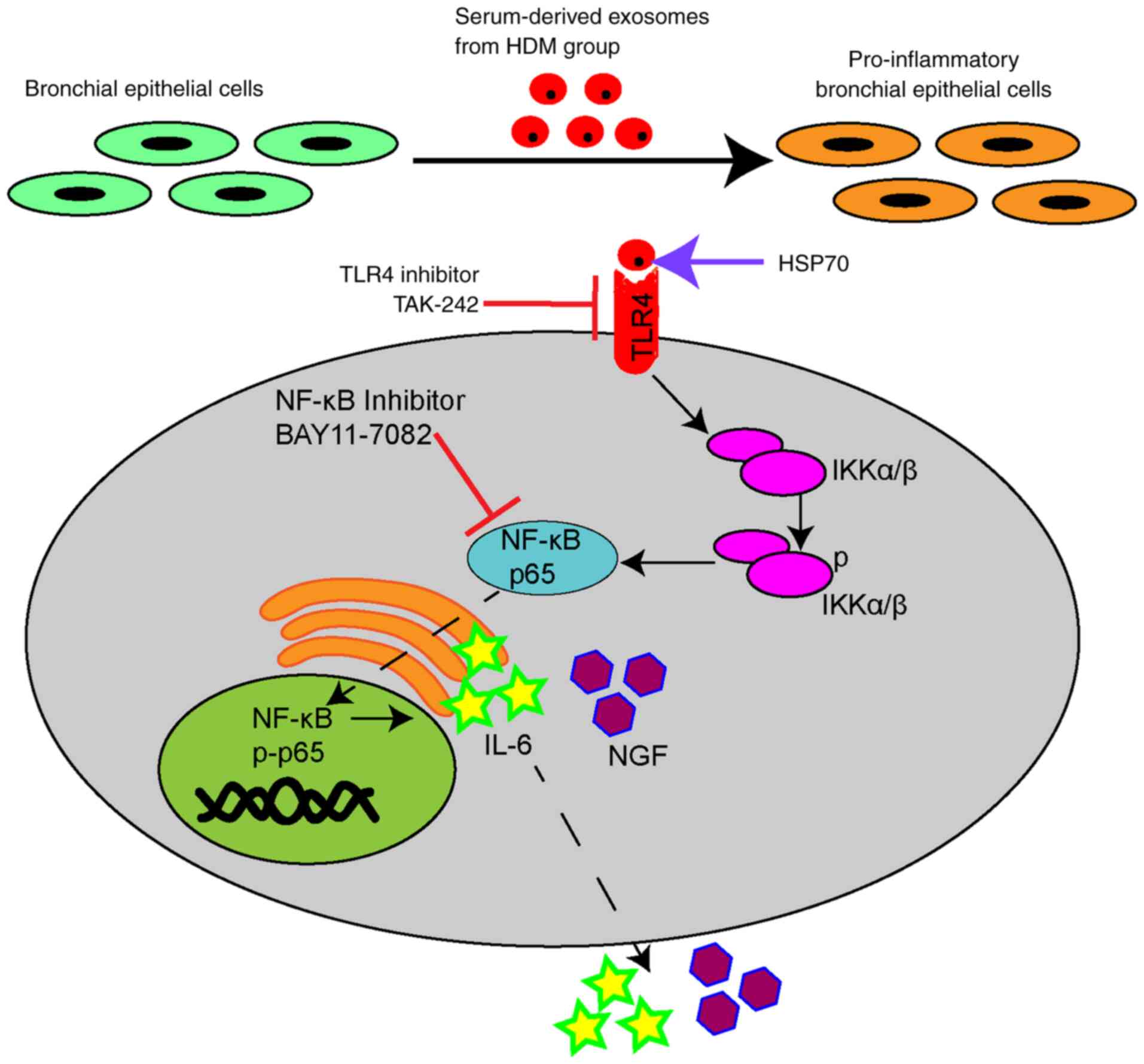
Serum-derived exosomes from the HDM
group enable the transfer of HSP70 into BEAS-2B cells and
contribute to cytokine secretion via the TLR4-NF-κB pathway
A variety of biological molecules, including nucleic
acids (DNA, miRNA, RNA), proteins and lipids, have been classified
as exosome contents (36). To
investigate whether exosomal surface proteins have an effect on the
production of cytokines, exosomes were treated with proteinase K
before they were added to BEAS-2B cells. Cytokine upregulation was
partially suppressed by proteinase K compared with the levels in
the exosome group not treated with the enzyme (Fig. 7A), indicating that exosomal surface
proteins prompted the increased expression of cytokines. Based on a
previous finding that exosomal HSP70 from mycobacteria-infected
macrophage cells induced an inflammatory response in uninfected
macrophages (37), it was further
examined whether HSP70 with serum-derived exosomes from the HDM
group could also exert a pro-inflammatory effect in recipient
cells. HSP70-IN-1, an inhibitor of HSP70, suppressed the increased
expression of IL-6 and NGF induced by serum-derived exosomes in
BEAS-2B cells (Fig. 7B). Various
concentrations of rHSP70 were added to BEAS-2B cell cultures to
mimic the role of exosomal proteins, and they promoted IL-6 and NGF
mRNA levels in a dose-dependent manner (Fig. 7C); however, IL-6 and NGF expression
levels were diminished after treatment with TAK-242 (Fig. 7D). Collectively, these data showed
that exosomes-containing HSP70 from serum in the HDM group
activated the TLR4-NF-κB signaling pathway in BEAS-2B cells.
Discussion
Previous investigations have shown that an
alteration in the airway microenvironment can further aggravate
inflammatory airway disorders, including chronic bronchitis,
bronchiectasis and asthma (38,39).
Airway epithelial cells, which are the first line of defense
against different stimuli, play an essential role in maintaining
the normal functioning of the airway microenvironment (3). Cell-to-cell communication is key to
regulating the underlying mechanism of inflammatory lung diseases
(40). Secreted soluble molecules,
such as chemokines, cytokines and cell surface receptors, are also
involved in this regulatory process (41). Exosomes, as extracellular functional
units, have recently attracted research interest due to their
crucial role in the pathogenesis of various diseases (18). The present study focused on the role
of exosomes on the bronchial epithelium to mimic the effect of
external stimuli on the airway microenvironment. In general,
exosomes measure between 30 and 120 nm in size, but those from the
H-exo group appeared smaller than the ones from the S-exo group
(Fig. 3A and B), which was
speculated to be possibly caused by partial exosomal fusion.
However, the absence of a blank control group proved detrimental in
the ability to make solid comparisons and inferences. Emerging
evidence indicates that the secretion of exosomes from eosinophils
in asthmatic subjects is higher than in healthy controls (16). Similarly, the protein levels of
airway exosomal surface markers, including CD81, CD36 and HLA-DR,
are significantly elevated in asthmatic subjects relative to
healthy controls (42). As
expected, the present study found that serum-derived exosomal
surface molecules, such as CD63 and HSP70, increased in the H-exo
group, but were notably attenuated in the S-exo group (Fig. 3C and D), possibly because the number
of exosomes released from body fluids are not the same under
different conditions, and the effects of these exosomes may also
differ in vivo and in vitro.
In the past decade, exosomes, as mediators of
intercellular crosstalk, have been shown to have the capacity to
transfer their cargos to influence the physiological and
pathological functions of receptor cells or parent cells. Valadi
et al (43) demonstrated
that exosomal RNA from mast cells in mice transferred to human mast
cells and eventually translated into proteins in recipient cells.
Exosomes from neutrophils are also reportedly rapidly up-taken by
airway smooth muscle cells, and their potential proliferative
ability increases after contact with lipopolysaccharides (44). Additionally, exosomes exist in
biological fluids upon their release from cells and can be
internalized by various cell types to alter their phenotypic
appearances and functions (17,45). A
previous study suggested that exosomes isolated from different body
fluids enhance the production of inflammatory cytokines, such as
IL-1β, TNF-α and IL-6, in monocytic cells (46). In the current study, BEAS-2B cells
efficiently incorporated serum-derived exosomes from the HDM and
sham groups (Fig. 3F). Because this
part of the study focused on whether BEAS-2B cells could take up
serum-derived exosomes, cell morphology images were not taken.
Serum-derived exosomes from the HDM group interacted with BEAS-2B
cells to induce increased IL-6 and NGF expression levels, but the
increase was only concentration-dependent for 50 and 100 µg/ml
exosome-treated cells (Fig. 4A-E).
The exosomal treatments were normalized to the respective time
controls, so the concentrations of cytokines for the different
concentrations of the exosomes between the 24 and 48 h groups were
not compared. IL-6 is a member of the interleukin family that plays
a significant role in the occurrence and development of
inflammatory airway disorders (47), while NGF, a member of growth
factors, mediates the generation, proliferation, differentiation
and maturation of inflammatory cells, and is involved in the
mechanism of the airway neurogenic inflammatory response (48). No gene-expression changes were
observed when serum-derived exosomes from the sham group were
co-cultured with BEAS-2B cells (Fig.
4A), suggesting that these exosomes played a crucial role in
maintaining the normal physiological function of the guinea pigs,
possibly having no ability to alter the phenotypic appearances and
functions of nearby or distant target cells.
Previous studies have also revealed that exosomes
secreted by various cells or biological fluids promote local or
systematic inflammatory processes to modulate the pathogeneses of
various diseases (18,49). However, the underlying molecular
mechanisms of the processes in the various diseases are different.
According to a previous inquiry, airway epithelial cell apoptosis
is prompted by exosomes from eosinophils in patients with asthma,
and these exosomes can enhance the proliferation of bronchial
smooth muscle cells via ERK1/2 activation (50). Mature dendritic cell-released
exosomes can increase an inflammatory phenotype in the endothelium
through membrane TNF-α, activating the NF-κB signaling pathway
(51). Exosomes released from
various body fluids allegedly also stimulate pro-inflammatory
cytokine secretion via the NF-κB- and STAT3-mediated signaling
pathway in a TLR-dependent manner (46), and plasma-derived exosomal
mitochondrial DNA in patients with chronic heart failure induces
IL-8 and IL-1β secretion via the TLR9-NF-κB pathway (52). These findings suggest that exosomes
from different sources mediate cell-to-cell communication in a
variety of ways.
We hypothesized that serum-derived exosomes in the
HDM group could trigger BEAS-2B cell inflammation by activating the
NF-κB pathway through TLR4. IKKα and IKKβ, subunits of the IIKK,
are essential for IκB phosphorylation and NF-κB activation
(53). The NF-κB family, including
RelA/p65, RelB, Rel/c-Rel, p50 (p105/NF-κB1) and p52 (p100/NF-κB2),
exert a crucial role in the pathology of the airway by modulating
cytokine and chemokine secretion (54). In the present study, it was observed
that these exosomes stimulated the increased expression of TLR4,
p-IKKα/β and p-p65, and the NF-κB subunit, p65, translocated into
the nuclei of BEAS-2B cells to induce cytokine secretion (Fig. 5A-F). However, after treatment with
exosomes for 24 h, TLR-4, p-IKK-α/β and p-p65 expression levels
were lower than those obtained from treatment with exosomes for 2
and 4 h, possibly because the proinflammatory effect of exosomes
co-cultured with cells decreases over time, and the longer the
reaction time, the more likely it is that exosomal content loses
its activity. Blocking NF-κB and TLR4 downregulated cytokine
expression levels (Fig. 6A and B).
TAK-242 partially inhibited the TLR4-NF-κB signaling pathway and
attenuated the protein expression levels in this pathway (Fig. 6C-F). However, 100 nM TAK-242
partially inhibited the activation of TLR4 and p-IKK-α/β more than
300 nM TAK-242 did, perhaps because volumes of TAK-242 below 100 nM
produced a concentration-dependent effect, and volumes above 100 nM
were in the plateau phase when interacting on BEAS-2B cells. On the
other hand, the amount of TLR4 receptors in bronchial epithelial
cells might have been insufficient, and its inhibitory effect may
have been attenuated because of the high concentration of TAK-242.
Therefore, exosomes from the HDM group could activate the
TLR4-NF-κB pathway in BEAS-2B cells and contribute to the
inflammatory response.
This research was significantly limited by the lack
of exosome samples from the serum of patients with asthma, mainly
because it is difficult to obtain serum samples from patients with
moderate to severe asthma. Secondly, there is an ethical issue that
the consent of patients with asthma is required before the obtained
serum-derived exosomal samples are used for research. Thirdly, the
extraction of serum-derived exosomes from animal models of asthma
and co-culture with BEAS-2B cells are the preliminary experiments
of our team. However, research on the involvement of serum-derived
exosomes in the pathogenesis of asthma is far from complete.
Previous analysis of the expression of miR-125b in the serum
exosomes of patients with different severities of asthma compared
with healthy subjects showed an altered miR-125b content, and thus
may have potential as a diagnostic marker for asthma (55). Therefore, further research must be
conducted to determine whether serum-derived exosomes from
asthmatic patients of different severity play a pro-inflammatory
role in bronchial epithelial cells or other receptor cells, and
this is our next research focus.
Exosomal surface molecules can mediate intracellular
signaling pathways through direct contact with receptors on target
cells. Anand et al (37)
demonstrated that exosomal surface HSP70 levels from macrophages
infected with mycobacteria are expressed higher than in controls,
and HSP70 in exosome-treated macrophages activates NF-κB signaling
to stimulate the release of TNF-α in uninfected macrophages.
Circulating HSP70 levels from patients with asthma are relevant to
the severity of disorders and the symptom of asthma and, therefore,
may contribute to the pathogenesis of the disease (56). The present study showed that
serum-derived exosomal HSP70 in the HDM group was higher than in
the sham group, suggesting that exosomal surface HSP70 may be
involved in the pathogenesis process. HSP70 on the surface of
serum-derived exosomes from the HDM group could, therefore, alter
BEAS-2B cell phenotypic appearances by regulating the TLR4-NF-κB
signaling pathway (Fig. 7A-D). A
previous study has shown that treatment with proteinase K-digested
HSP70 in bone marrow-derived dendritic cells results in a reduction
in HSP70-dependent cytokines (57).
A previous study reported that the increased secretion of
inflammatory factors could be markedly suppressed following the
pretreatment of mesenchymal stem cells with proteinase K compared
with a A549 exosome-treated group (58). As expected, the results of the
current study indicated that the increases in cytokine
concentrations were partially repressed by proteinase K and the
HSP70 blocker compared with the exosome-treated group without the
enzyme. Therefore, it should be considered that other exosomal
components, including nucleic acids and other proteins, possibly
participate in this inflammatory response and this should be
further explored.
In conclusion, it was demonstrated that
serum-derived exosomes interacted with BEAS-2B cells and could
alter their phenotypic appearance. Additionally, the
HSP70-modulated inflammatory effect on the surface of serum-derived
exosomes from the HDM group upregulated IL-6 and NGF expression
levels by activating the TLR4-NF-κB pathway (Fig. 8). Overall, exosomal presence in
HDM-sensitized guinea pigs could be influential in the underlying
mechanism of inflammatory airway diseases; however, blocking
exosome-mediated communication between cells would attenuate the
inflammation, potentially partially relieving symptoms of
inflammatory airway diseases in guinea pigs.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Natural
Science Foundation of China (grant nos. 81460004 and 82160006); the
Jiangxi Provincial Cultivation Program for Academic and Technical
Leaders of Major Subjects (grant no. 20172BCB22025); and the
Jiangxi Provincial Natural Science Foundation General Project
(grant no. 20202BAB206003).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JW and CL conceived and designed the present study.
CL, XLH, XZ, ZFW and LXD performed the experiments. CL, JPL and XLH
analyzed the experimental data. CL wrote the manuscript. CL and JW
confirm the authenticity of all the raw data. All authors have read
and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Animal Care
and Committee of Jiangxi Provincial People's Hospital Affiliated to
Nanchang University (approval no. 2021-052; Nanchang, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Zanini A, Cherubino F, Zampogna E, Croce
S, Pignatti P and Spanevello A: Bronchial hyperresponsiveness,
airway inflammation, and reversibility in patients with chronic
obstructive pulmonary disease. Int J Chron Obstruct Pulmon Dis.
10:1155–1161. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Alashkar AB, Miethe S, Pogge VSE, Potaczek
DP and Garn H: Epigenetic regulation of airway epithelium immune
functions in asthma. Front Immunol. 11:17472020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fujita Y, Kosaka N, Araya J, Kuwano K and
Ochiya T: Extracellular vesicles in lung microenvironment and
pathogenesis. Trends Mol Med. 21:533–542. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Weitnauer M, Mijošek V and Dalpke AH:
Control of local immunity by airway epithelial cells. Mucosal
Immunol. 9:287–298. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Holgate ST: Epithelium dysfunction in
asthma. J Allergy Clin Immunol. 120:1233–1246. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gon Y and Hashimoto S: Role of airway
epithelial barrier dysfunction in pathogenesis of asthma. Allergol
Int. 67:12–17. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mitchell PD and O'Byrne PM:
Epithelial-derived cytokines in asthma. Chest. 151:1338–1344. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mitchell PD and O'Byrne PM: Biologics and
the lung: TSLP and other epithelial cell-derived cytokines in
asthma. Pharmacol Ther. 169:104–112. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sun Z, Ji N, Ma Q, Zhu R, Chen Z, Wang Z,
Qian Y, Wu C, Hu F, Huang M and Zhang M: Epithelial-mesenchymal
transition in asthma airway remodeling is regulated by the
IL-33/CD146 axis. Front Immunol. 11:15982020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kato A and Schleimer RP: Beyond
inflammation: Airway epithelial cells are at the interface of
innate and adaptive immunity. Curr Opin Immunol. 19:711–720. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Videira RF and da Costa Martins PA:
Non-coding RNAs in cardiac intercellular communication. Front
Physiol. 11:7382020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Barros FM, Carneiro F, Machado JC and Melo
SA: Exosomes and immune response in cancer: Friends or foes? Front
Immunol. 9:7302018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Smith VL, Cheng Y, Bryant BR and Schorey
JS: Exosomes function in antigen presentation during an in vivo
mycobacterium tuberculosis infection. Sci Rep. 7:435782017.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chan BD, Wong WY, Lee MM, Cho WC, Yee BK,
Kwan YW and Tai WC: Exosomes in inflammation and inflammatory
disease. Proteomics. 19:e18001492019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhao B, Li X, Shi X, Shi X, Zhang W, Wu G,
Wang X, Su L and Hu D: Exosomal microRNAs derived from human
amniotic epithelial cells accelerate wound healing by promoting the
proliferation and migration of fibroblasts. Stem Cells Int.
2018:54204632018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mazzeo C, Cañas JA, Zafra MP, Rojas Marco
A, Fernández-Nieto M, Sanz V, Mittelbrunn M, Izquierdo M, Baixaulli
F, Sastre J and Del Pozo V: Exosome secretion by eosinophils: A
possible role in asthma pathogenesis. J Allergy Clin Immun.
135:1603–1613. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhao L, Yu J, Wang J, Li H, Che J and Cao
B: Isolation and identification of miRNAs in exosomes derived from
serum of colon cancer patients. J Cancer. 8:1145–1152. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Alipoor SD, Mortaz E, Garssen J,
Movassaghi M, Mirsaeidi M and Adcock IM: Exosomes and exosomal
miRNA in respiratory diseases. Mediat Inflamm. 2016:1–11. 2016.
View Article : Google Scholar
|
|
19
|
Mathivanan S, Ji H and Simpson RJ:
Exosomes: Extracellular organelles important in intercellular
communication. J Proteomics. 73:1907–1920. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Turturici G, Tinnirello R, Sconzo G and
Geraci F: Extracellular membrane vesicles as a mechanism of
Cell-to-Cell communication: Advantages and disadvantages. Am J
Physiol Cell Physiol. 306:C621–C633. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hough KP and Deshane JS: Exosomes in
allergic airway diseases. Curr Allergy Asthma Rep. 19:262019.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kadota T, Fujita Y, Yoshioka Y, Araya J,
Kuwano K and Ochiya T: Extracellular vesicles in chronic
obstructive pulmonary disease. Int J Mol Sci. 17:18012016.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mortaz E, Alipoor SD, Varahram M, Jamaati
H, Garssen J, Mumby SE and Adcock IM: Exosomes in severe asthma:
Update in their roles and potential in therapy. Biomed Res Int.
2018:28621872018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cañas JA, Sastre B, Rodrigo-Muñoz JM and
Del PV: Exosomes: A new approach to asthma pathology. Clin Chim
Acta. 495:139–147. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Huang F, Jia H, Zou Y, Yao Y and Deng Z:
Exosomes: An important messenger in the asthma inflammatory
microenvironment. J Int Med Res. 48:12207027722020.
|
|
26
|
Levanen B, Bhakta NR, Torregrosa PP,
Barbeau R, Hiltbrunner S, Pollack JL, Skold CM, Svartengren M,
Grunewald J, Gabrielsson S, et al: Altered microRNA profiles in
bronchoalveolar lavage fluid exosomes in asthmatic patients. J
Allergy Clin Immunol. 131:894–903. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bartel S, La Grutta S, Cilluffo G,
Perconti G, Bongiovanni A, Giallongo A, Behrends J, Kruppa J,
Hermann S, Chiang D, et al: Human airway epithelial extracellular
vesicle miRNA signature is altered upon asthma development.
Allergy. 75:346–356. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kulshreshtha A, Ahmad T, Agrawal A and
Ghosh B: Proinflammatory role of epithelial cell-derived exosomes
in allergic airway inflammation. J Allergy Clin Immun.
131:1194–1203. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Paredes PT, Esser J, Admyre C, Nord M,
Rahman QK, Lukic A, Radmark O, Gronneberg R, Grunewald J, Eklund A,
et al: Bronchoalveolar lavage fluid exosomes contribute to cytokine
and leukotriene production in allergic asthma. Allergy. 67:911–919.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hough KP, Chanda D, Duncan SR, Thannickal
VJ and Deshane JS: Exosomes in immunoregulation of chronic lung
diseases. Allergy. 72:534–544. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tan D, Armitage J, Teo TH, Ong NE, Shin H
and Moodley YP: Elevated levels of circulating exosome in COPD
patients are associated with systemic inflammation. Respir Med.
132:261–264. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xu H, Ling M, Xue J, Dai X, Sun Q, Chen C,
Liu Y, Zhou L, Liu J, Luo F, et al: Exosomal microRNA-21 derived
from bronchial epithelial cells is involved in aberrant
epithelium-fibroblast cross-talk in COPD induced by cigarette
smoking. Theranostics. 8:5419–5433. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Care NRCU and Animals AUOL: Guide for the
Care and Use of Laboratory Animals. National Academies Press US;
Washington, DC: 2011
|
|
34
|
Tang YT, Huang YY, Zheng L, Qin SH, Xu XP,
An TX, Xu Y, Wu YS, Hu XM, Ping BH and Wang Q: Comparison of
isolation methods of exosomes and exosomal RNA from cell culture
medium and serum. Int J Mol Med. 40:834–844. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang L and Yu D: Exosomes in cancer
development, metastasis, and immunity. Biochim Biophys Acta Rev
Cancer. 1871:455–468. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Anand PK, Anand E, Bleck CK, Anes E and
Griffiths G: Exosomal Hsp70 induces a pro-inflammatory response to
foreign particles including mycobacteria. PLoS One. 5:e101362010.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Dickson RP, Erb-Downward JR and Huffnagle
GB: Homeostasis and its disruption in the lung microbiome. Am J
Physiol Lung Cell Mol Physiol. 309:L1047–L1055. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Mendez R and Banerjee S, Bhattacharya SK
and Banerjee S: Lung Inflammation and disease: A perspective on
microbial homeostasis and metabolism. Iubmb Life. 71:152–165. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Paplinska-Goryca M, Misiukiewicz-Stepien
P, Nejman-Gryz P, Proboszcz M, Mlacki M, Gorska K and Krenke R:
Epithelial-macrophage-dendritic cell interactions impact alarmins
expression in asthma and COPD. Clin Immunol. 215:1084212020.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Cornwell WD, Kim V, Song C and Rogers TJ:
Pathogenesis of inflammation and repair in advanced COPD. Semin
Respir Crit Care Med. 31:257–266. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Admyre C, Bohle B, Johansson SM,
Focke-Tejkl M, Valenta R, Scheynius A and Gabrielsson S: B
cell-derived exosomes can present allergen peptides and activate
allergen-specific T cells to proliferate and produce TH2-like
cytokines. J Allergy Clin Immunol. 120:1418–1424. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Valadi H, Ekström K, Bossios A, Sjöstrand
M, Lee JJ and Lötvall JO: Exosome-mediated transfer of mRNAs and
microRNAs is a novel mechanism of genetic exchange between cells.
Nat Cell Biol. 9:654–659. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Vargas A, Roux-Dalvai F, Droit A and
Lavoie JP: Neutrophil-derived exosomes: A new mechanism
contributing to airway smooth muscle remodeling. Am J Respir Cell
Mol Biol. 55:450–461. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Sahoo S and Losordo DW: Exosomes and
cardiac repair after myocardial infarction. Circ Res. 114:333–344.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Bretz NP, Ridinger J, Rupp AK, Rimbach K,
Keller S, Rupp C, Marmé F, Umansky L, Umansky V, Eigenbrod T, et
al: Body fluid exosomes promote secretion of inflammatory cytokines
in monocytic cells via Toll-like receptor signaling. J Biol Chem.
288:36691–36702. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Rincon M and Irvin CG: Role of IL-6 in
asthma and other inflammatory pulmonary diseases. Int J Biol Sci.
8:1281–1290. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Abram M, Wegmann M, Fokuhl V, Sonar S,
Luger EO, Kerzel S, Radbruch A, Renz H and Zemlin M: Nerve growth
factor and neurotrophin-3 mediate survival of pulmonary plasma
cells during the allergic airway inflammation. J Immunol.
182:4705–4712. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Othman N, Jamal R and Abu N:
Cancer-derived exosomes as effectors of key inflammation-related
players. Front Immunol. 10:21032019. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Cañas JA, Sastre B, Rodrigo-Muñoz JM,
Fernández-Nieto M, Barranco P, Quirce S, Sastre J and Del PV:
Eosinophil-derived exosomes contribute to asthma remodelling by
activating structural lung cells. Clin Exp Allergy. 48:1173–1185.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Gao W, Liu H, Yuan J, Wu C, Huang D, Ma Y,
Zhu J, Ma L, Guo J, Shi H, et al: Exosomes derived from mature
dendritic cells increase endothelial inflammation and
atherosclerosis via membrane TNF-α mediated NF-κB pathway. J Cell
Mol Med. 20:2318–2327. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Ye W, Tang X, Yang Z, Liu C, Zhang X, Jin
J and Lyu J: Plasma-derived exosomes contribute to inflammation via
the TLR9-NF-κB pathway in chronic heart failure patients. Mol
Immunol. 87:114–121. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zandi E, Rothwarf DM, Delhase M, Hayakawa
M and Karin M: The IkappaB kinase complex (IKK) contains two kinase
subunits, IKKalpha and IKKbeta, necessary for IkappaB
phosphorylation and NF-kappaB activation. Cell. 91:243–252. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Schuliga M: NF-kappaB signaling in chronic
inflammatory airway disease. Biomolecules. 5:1266–1283. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Zhao M, Juanjuan L, Weijia F, Jing X,
Qiuhua H, Hua Z, Fuhe L and Hao P: Expression levels of
microRNA-125b in serum exosomes of patients with asthma of
different severity and its diagnostic significance. Curr Drug
Metab. 20:781–784. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Hou C, Zhao H, Li W, Liang Z, Zhang D, Liu
L, Tong W, Cai SX and Zou F: Increased heat shock protein 70 levels
in induced sputum and plasma correlate with severity of asthma
patients. Cell Stress Chaperones. 16:663–671. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Spiering R, van der Zee R, Wagenaar J, van
Eden W and Broere F: Mycobacterial and mouse HSP70 have
immuno-modulatory effects on dendritic cells. Cell Stress
Chaperones. 18:439–446. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Li X, Wang S, Zhu R, Li H, Han Q and Zhao
RC: Lung tumor exosomes induce a pro-inflammatory phenotype in
mesenchymal stem cells via NFκB-TLR signaling pathway. J Hematol
Oncol. 9:422016. View Article : Google Scholar : PubMed/NCBI
|