Introduction
Cardiovascular disease (CVD) is a general term for
numerous heart conditions. It can refer to several conditions:
Coronary heart disease (CHD), peripheral arterial disease,
cerebrovascular disease, rheumatic and congenital heart diseases
and venous thromboembolism. CVD is responsible for 31% of all
deaths and is the leading cause of mortality globally. Each year,
17.9 million people succumb to CVD; the majority of this is in the
form of CHD and cerebrovascular disease (1).
Over the past few years, there has been a growing
body of evidence that suggests that cancer and CVD share numerous
molecular pathways (2). This
interaction between different biological pathways indicates that
the same genes and proteins are often involved in developing both
diseases (2). Cancer and CVD share
a number of risk factors and pathogenic processes, including
chronic inflammation, oxidative stress and genetic instability
(3). There is evidence that DNA
damage and oxidative stress play a significant role in the
development of CVD (4). For
example, people with coronary artery disease have higher levels of
DNA damage, which is directly associated with atherosclerosis
severity (5).
Reactive oxygen species (ROS) are free radicals that
can damage DNA. ROS are produced as a byproduct of cellular
respiration and can also be produced by other processes, such as
inflammation (6). ROS can damage
DNA by causing single-strand breaks, double-strand breaks and base
modifications. These types of damage can lead to mutations, cancer
and other health problems (6). ROS
are essential for cellular functions at low levels, such as serving
as cellular messengers in redox signaling reactions (7). Cells have a variety of mechanisms to
regulate the levels of ROS. One such mechanism is the restriction
of respiration in the mitochondrial inner membrane to prevent the
production of ROS and protect other cellular components (7). Another manner by which to regulate
the levels of ROS is to protect DNA by complexing it with histones.
This prevents ROS from reaching the DNA and damaging it. Finally,
cells can also quench ROS by using antioxidant enzymes. These
enzymes neutralize ROS, rendering them harmless (7). ROS can damage DNA in two ways
(8). First, they can attack the
DNA bases, causing mutations. Second, they can also damage the DNA
backbone, causing breaks in the DNA strands (8). These breaks can be repaired by two
different pathways: The single-strand break repair (SSBR) pathway
and the double-strand break repair (DSBR) pathway (9).
The SSBR pathway repairs single-strand breaks in the
DNA backbone. The DSBR pathway repairs double-strand breaks in the
DNA backbone. Both of these pathways are important for maintaining
the integrity of DNA. Immediately following DNA damage,
lesion-specific proteins initiate the DNA damage response, a
collection of mechanisms that detect DNA damage, signal its
presence and promote DNA repair according to the type of damage
(10). Furthermore, cells can also
encounter DSBs, which are repaired by the homologous recombination
(HR) pathway (11).
From observation of the effects of ROS on DNA during
oxidative stress, it is considered that there is an organized and
sophisticated system to remove the effects of oxidative damage. In
human cells, the repair of oxidatively damaged DNA bases is
primarily carried out by the base excision repair mechanism
(BER).
In the first step of BER, a damage-specific DNA
glycosylase identifies the damaged base and removes it from the
DNA. In general, glycosylases are classified depending on their
function; they are either monofunctional or bifunctional (12).
An example of a DNA glycosylase in the first step of
BER is 8-oxoguanine DNA glycosylase (OGG1). It is located at
chromosome 3p25.3 and plays a significant role in the repair of
8-hydroxyguanine. The polymorphism rs1052133 in OGG1 leads to
substitution of the amino acid serine for cysteine at codon 326,
which shows a decrease in enzyme activity in OGG1-Ser326Cys
(13). Moreover, the polymorphism
rs1052133 is one of the common SNPs in the OGG1 gene and has been
linked to numerous different biological diseases such as breast
cancer (14–17), prostate cancer (18,19),
gastric cancer (20), colorectal
cancer (21), lung cancer
(22) and esophageal cancer
(23). Judging by the findings of
the previous studies, it is clear that polymorphism rs1052133 has a
considerable effect on enzyme activity, which might lead to more
oxidative damage (13).
In non-homologous end joining, the break is simply
joined together by ligation. The break is repaired in HR by copying
the homologous DNA sequence from a sister chromatid or another
homologous chromosome. In single-strand annealing (SSA), the break
is repaired by annealing the two DNA strands together (24). If the DSB occurs between direct
repeats, SSA is the only possible repair pathway. Otherwise, the
resected 3′ end of the broken DNA strand can invade the homologous
template to start the repair synthesis (24). There are two other mechanisms that
can occur after this step: Synthesis-dependent strand annealing
(SDSA) and break-induced repair (BIR) (24). SDSA is a type of HR repair in which
the invading 3′ end anneals to the homologous template and DNA
synthesis is used to fill in the gap. BIR is a type of HR repair in
which the invading 3′ end anneals to the homologous template, and
subsequently, a double Holliday junction is formed. The double
Holliday junction is then resolved, resulting in the repair of the
DSB (24).
HR is predominant in the G2 phase, and
BRCA1 initiates the ubiquitination of the downstream component.
Once the DSB is detected, other proteins called replication protein
A (RPA) and RAD51 recombinase (RAD51) bind to the DNA. RPA coats
the 3′ overhang of the broken DNA strand, while RAD51 forms a
nucleoprotein filament. This filament then invades the homologous
DNA strand, the other copy of the damaged gene. Several other
proteins, such as CtBP-interacting protein, breast cancer
susceptibility protein 2 and RAD52 homolog, help to facilitate this
process. Once the invading strand is in place, polymerases can add
new DNA nucleotides to fill in the gap, repairing the DSB (7,24).
As aforementioned, RAD51 has a crucial role in DSBR,
specifically in HR. RAD51, in eukaryotes, is a homolog of the RecA
protein, and it contains 339 amino acids. It is located at human
chromosome 15q15.1 and is highly polymorphic (25). There are five RAD51 paralogs,
RAD51B, RAD51C, RAD51D, X-ray repair cross complementing 2 (XRCC2)
and XRCC3, in the human genome. They play an essential role in HR,
and any loss of function would result in genomic instability
(25). RAD51 polymorphism
rs1801321; 172G>T is one of the most common polymorphisms
located at the 5′UTR (26). In
terms of clinical significance, rs1801321 has been linked with
endometrial cancer (27) and there
is a significant association with breast cancer (28,29).
At the same time, a number of studies have suggested no association
with ovarian cancer (30,31).
The present study aimed to provide data and evaluate
the significant association between CVD and the SNPs in OGG1
rs1052133 and RAD51 rs1801321 in a Saudi population.
Materials and methods
Patient population and ethics
statement
The present study included 240 individuals from King
Khalid University Hospital (KKUH; Riyadh, Kingdom of Saudi Arabia),
of which 120 were clinically hospitalized for CVD and 120 were
healthy, age- and sex-matched blood donors considered the control
group. Samples were collected from March to December 2021. Their
ethnicities were verified since the parents and grandparents of the
patients and controls were born in Saudi Arabia. The study was
reviewed and approved by the local committee from King Khaled
University Hospital and all patients provided written informed
consent. The present study was conducted according to the
guidelines of the Declaration of Helsinki and approved by the
institutional review board (approval no. IRB-HAPO-01-R-011) of
Al-Imam Muhammad Ibn Saud Islamic University, Riyadh, Saudi Arabia.
A questionnaire was administered to gather information on age,
demographics, sex, clinical features (such as disease duration) and
behavioral (such as smoking) factors. Data pertaining to
hypotension and hypertension events and pharmacological treatments
were also collected in the questionnaire and were then confirmed
using medical records. The clinical data are summarized in Table I.
 | Table I.Clinical data and characteristics of
the patients with CVD. |
Table I.
Clinical data and characteristics of
the patients with CVD.
|
Characteristics | CVD | Controls |
|---|
| N | 120 (50.0%) | 120 (50.0%) |
| Age, years | 62.0±12.0 | 54.8±15.3 |
| Sex |
|
|
|
Male | 85 (70.8%) | 80 (66.0%) |
|
Female | 35 (29.2%) | 40 (33.3%) |
| FBS, mmol/l | 8.2±3.9 | - |
| TG, mmol/l | 1.5±0.7 | - |
| TC, mmol/l | 4.2±1.1 | - |
| HDL-c, mmol/l | 1.1±0.9 | - |
| LDL-c, mmol/l | 2.6±0.8 | - |
| Smokers | 54 (45.0%) | 59 (49,2%) |
| Non-smokers | 66 (55.0%) | 61 (50.8%) |
| Hyper and
hypotension events | 89 (74.2%) | - |
|
| 110 (91.7%) |
|
| Pharmacological
treatments |
| No treatment |
|
Yes | 82 (68.3%) |
|
| No | 38 (31.7%) |
|
DNA extraction
DNA was extracted from 200 µl of EDTA anticoagulated
peripheral blood. The extraction was carried out following the
standards and procedures provided by the DNeasy® Blood
& Tissue Kit (cat. no. 69504; Qiagen GmbH). The purity and
concentration of the DNA were measured using a NanoDrop™ 8000
Spectrophotometer (Thermo Fisher Scientific, Inc.) using the ratio
A260/A280. The DNA samples were subsequently diluted, with each
sample diluted according to its concentration and DNA samples were
then stored at −20°C for genotyping.
Choice of SNPs
The two SNPs, rs1052133 in OGG1 and rs1801321 in
RAD51, were selected based on their functional properties. The
RAD51 polymorphism rs1801321; 172G>T is one of the most common
polymorphisms and is located at the 5′UTR (26). The OGG1 polymorphism rs1052133
leads to the substitution of the amino acid serine to cysteine at
codon 326 and shows a decrease in enzyme activity in OGG1-Ser326Cys
(13).
TaqMan SNP genotyping
TaqMan genotyping assay was used to determine
whether the SNPs, OGG1 rs1052133 and RAD51 rs1801321, were
prevalent in the test samples compared with the control samples.
TaqMan genotyping assay (cat. no. 4351379; Applied biosystem, USA)
was used for OGG1 rs1052133
[VIC/FAM]5′-CTGTTCAGTGCCGACCTGCGCCAAT[C/G]CCGCCATGCTCAGGAGCCACCAGCA-3′)
and RAD51 rs1801321 ([VIC/FAM]
5′-GCCGTGCGGGTCGGGCGCGTGCCAC[GT]CCCGCGGGGTGAAGTCGGAGCGCGG-3′). The
genotyping was performed in a 96-well format by applying a
QuantStudio 7 Flex Real-Time PCR system (Applied Biosystems, Foster
City, CA, USA). To prepare the plates for genotyping, the following
were added to each well in a 0.1-ml plate: 5 µl TaqMan master mix,
0.25 µl SNP reagent (Applied Biosystems, USA), 2.75 µl RNase-free
water and 2 µl DNA from each sample. The total volume in each well
was 10 µl. Thermocycling conditions were as follows: 30-sec
pre-read phase at 60°C, a 10-min initial activation step at 95°C,
and then proceeded to 45 cycles of PCR amplification. Each cycle
encompassed a 15-sec denaturation period at 95°C, followed by a
1-min annealing phase at 60°C, and concluded with a 30-sec
extension phase also at 60°C.
Protein-protein interaction (PPI)
network
The PPI network was generated using GENEMANIA
(http://www.genemania.org/), which is a
database and a tool that predicts gene functions based on different
sources and databases (32). The
network showed physical interaction if two gene products were found
to interact in a PPI study, co-expression if two genes were linked
and their expression levels were similar in a gene expression
study, predictions if there were functional relationships between
genes (often protein interactions) and co-localization if genes
were expressed in the same tissue or if their products were found
in the same cellular location. The network also revealed pathway
data if two gene products participated in the same reaction in a
pathway, genetic interaction if two genes were functionally
associated (for example, if the effects of perturbing one gene were
found to be modified by perturbations to a second gene) and shared
protein domains if two gene products had the same protein domain.
In total, the network demonstrated seven categories to detect
related genes: Physical interaction, co-expression, predicted,
co-localization, pathway, genetic interactions and shared protein
domains.
Statistical analysis
The genotype frequencies were tested using the
Hardy-Weinberg exact test. The significant differences between the
controls were calculated using the chi-square test. The odds ratio
(OR) and 95% confidence interval were calculated using Fisher's
exact test and SPSS version 16.0 statistical package (SPSS, Inc.).
P<0.05 was considered to indicate a statistically significant
difference.
Results
Analysis of clinical characteristics
as well as identification of the risk for CVD related to RAD51 SNP
and OGG1 SNP in patients from Saudi Arabia
The clinical characteristics of the 120 cases of CVD
and matched controls, with the same age, sex, ethnicity, smoking
habits and pharmacological treatments are presented in Table I.
A total of 70.8% of the patients with CVD were male
and 29.2% were female patients, whereas, in the control group, 66%
were men and 33.3% were women. Most of the patients were men due to
the availability of blood samples. Thus, the authors tried to match
the ratio of CVD cases and the control group. It is worth stating
that the ethnic distribution was 100% Saudi Arabian for all of the
patients of the study. The mean age of the study population was
62±12 and 54.8±15.3 years for patients with CVD and healthy
subjects, respectively.
Genotype frequencies of OGG1 gene
rs1052133 and RAD51 gene rs1801321 in CVD and control groups
To analyze the role of the two polymorphisms of OGG1
and RAD51 in the pathophysiology of CVD, the association between
the patients with CVD and healthy controls were investigated by
comparing the allele frequencies of the two SNPs. The distributions
of the alleles and genotypes with significances and OR are reported
in Table II. Hardy-Weinberg
equilibrium was used to measure the genotype frequencies of all
SNPs.
 | Table II.Genotype frequencies of OGG1 gene
polymorphism and RAD51 gene polymorphism in patients with CVD and
controls. |
Table II.
Genotype frequencies of OGG1 gene
polymorphism and RAD51 gene polymorphism in patients with CVD and
controls.
| Gene | SNP ID | Genotype | CVD (%) | Control (%) | OR | 95% CI |
χ2-value | P-value |
|---|
| OGG1 | rs1052133 | CC | 48 (40%) | 50 (42%) | Ref |
|
|
|
|
|
| CG | 57 (47%) | 58 (48%) | 1.02 | 0.5972–1.7549 | 0.01 | 0.920344 |
|
|
| GG | 15 (12.5%) | 10 (8%) | 1.56 | 0.6398–3.8156 | 0.97 | 0.325 |
|
|
| CG + GG | 72 (60%) | 68 (57%) | 0.91 | 0.5410–1.5196 | 0.14 | 0.709 |
|
|
| C | 153 (64%) | 158 (66%) | Ref |
|
|
|
|
|
| G | 87 (36%) | 78 (32%) | 0.87 | 0.5949–1.2669 | 0.54 | 0.463 |
| RAD51 | rs1801321 | GG | 15(12.5%) | 59 (49%) | Ref |
|
|
|
|
|
| GT | 2 (2%) | 0 | 0.14 | 0.0115–1.5923 | 3.34 | 0.0076a |
|
|
| TT | 103 (86%) | 60 (50%) | 0.16 | 0.0835–0.2989 | 36.0 |
1.98×10−9a |
|
|
| GT + TT | 105 (87%) | 60 (50%) | 0.16 | 0.0834–0.2977 | 36.3 |
1.24×10−9a |
|
|
| G | 32 (26.6%) | 118 (98%) | Ref |
|
|
|
|
|
| T | 208 (87%) | 120 (100%) | 0.17 | 0.1068–0.2586 | 70.22 |
5.92×10−17a |
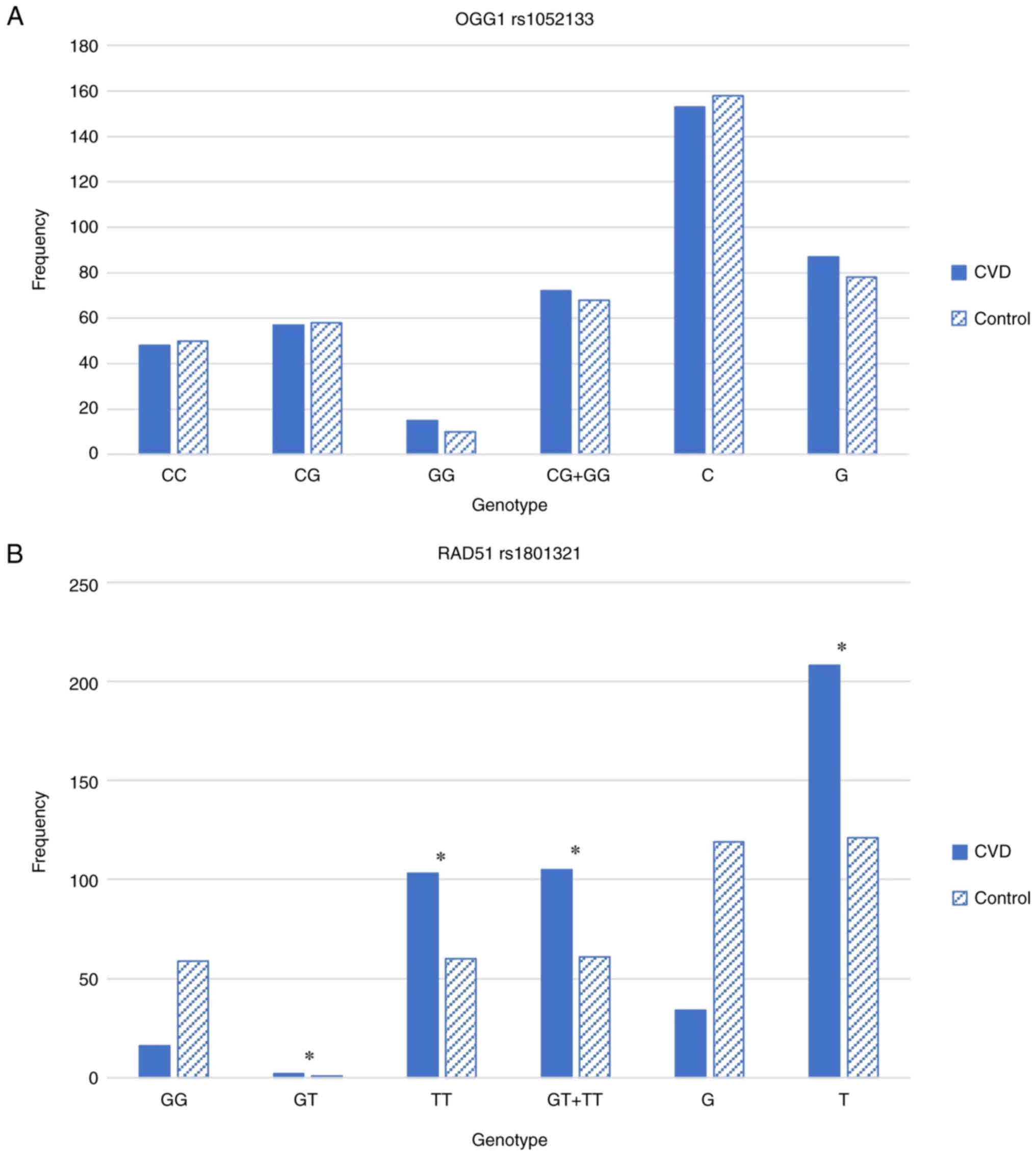
For OGG1 rs1052133C>G, the genotype frequencies
of the OGG1 gene in the patients with CVD and controls are shown in
Fig. 1A. No significant
differences were observed in the genotype and allele frequencies
for the C and G alleles between the patients with CVD and normal
controls. For RAD51 rs1801321G>T, the genotype frequencies of
the RAD51 gene in the patients with CVD and controls are
illustrated in Fig. 1B. The
controls were homozygous for either the G allele or T allele, while
the CVD samples revealed a strong association with rs1801321G>T
polymorphism. The TT genotype was strongly associated with CVD in
the Saudi population with P<1.98×10−9. It appears
that heterogenicity is not common in RAD51, as there were only
three samples (two CVD and one control) that showed a GT genotype.
The frequency of the wild-type GG genotype was higher in the
controls than in the CVD samples. By contrast, the TT genotype was
found to be more frequent in the CVD samples than in the
controls.
The samples were categorized into two groups based
on patient age: <60 and >60 years of age. No significant
differences in terms of the genotype and allele frequencies of OGG1
rs1052133 and RAD1 rs1801321 were revealed between patients <60
and >60 years old (Table
III). Conversely, the association between sex, OGG1 and RAD51
variants and the risk of developing CVD was also examined (Table IV). It was revealed that only the
rs1801321G>T polymorphism in male patients had a significant
association with CVD development.
 | Table III.Genotype frequencies of OGG1 gene
polymorphism and RAD51 gene polymorphism in patients with CVD and
controls based on age. |
Table III.
Genotype frequencies of OGG1 gene
polymorphism and RAD51 gene polymorphism in patients with CVD and
controls based on age.
| Gene | SNP ID | Genotype | CVD <60 | CVD >60 | OR | 95% CI |
χ2-value | P-value |
|---|
| OGG1 | rs1052133 | CC | 24 | 37 | Ref | | | |
|
|
| CG | 26 | 31 | 1.19 | 0.5524–2.5733 | 0.20096 | 0.65395 |
|
|
| GG | 7 | 8 | 1.14 | 0.3577–3.6512 | 0.05081 | 0.82167 |
|
|
| CG + GG | 33 | 39 | 1.18 | 0.5687–2.4560 | 0.2005 | 0.65432 |
|
|
| C | 74 | 79 | Ref | | | |
|
|
| G | 40 | 47 | 1.1 | 0.6494–1.8653 | 0.12693 | 0.72163 |
| RAD51 | rs1801321 | GG | 9 | 7 | Ref |
|
|
|
|
|
| GT | 1 | 1 | 1.29 |
(0.0678–24.3835) | 0.0281 | 0.8668 |
|
|
| TT | 45 | 58 | 1.66 |
(0.5731–4.7914) | 0.8815 | 0.34779 |
|
|
| GT + TT | 46 | 59 | 0.5711 |
(0.5711–4.7613) | 0.8667 | 0.35187 |
|
|
| G | 19 | 15 | Ref | | | |
|
|
| T | 91 | 117 | 1.63 |
(0.7846–3.3804) | 1.7349 | 0.18778 |
 | Table IV.Genotype frequencies of OGG1 gene
polymorphism and RAD51 gene polymorphism in patients with CVD and
controls based on sex. |
Table IV.
Genotype frequencies of OGG1 gene
polymorphism and RAD51 gene polymorphism in patients with CVD and
controls based on sex.
| Gene | SNP ID | Genotype | Male patients with
CVD | Female patients
with CVD | OR | 95% CI |
χ2-value | P-value |
|---|
| OGG1 | rs1052133 | CC | 38 | 10 | Ref | | | |
|
|
| CG | 37 | 20 | 2.05 |
(0.8487–4.9711) | 2.5943 | 0.10725 |
|
|
| GG | 9 | 6 | 2.53 |
(0.7288–8.8064) | 2.2159 | 0.1366 |
|
|
| CG + GG | 46 | 26 | 2.15 |
(0.9213–5.0075) | 3.2011 | 0.07359 |
|
|
| C | 113 | 40 | Ref | | | |
|
|
| G | 55 | 32 | 1.64 |
(0.9336–2.8937) | 2.9887 | 0.08385 |
| RAD51 | rs1801321 | GG | 14 | 2 | Ref | | | |
|
|
| GT | 1 | 0 | - | - | 0.46667 | 0.49452 |
|
|
| TT | 70 | 33 | 0.94 | 0.1643–5.4103 | 0.00436 | 0.94736 |
|
|
| GT + TT | 71 | 33 | 0.93 | 0.1620–5.3329 | 0.00672 | 0.93469 |
|
|
| G | 29 | 4 | Ref | | | |
|
|
| T | 141 | 66 | 3.39 | 1.1461–10.0483 | 5.902 |
0.01512a |
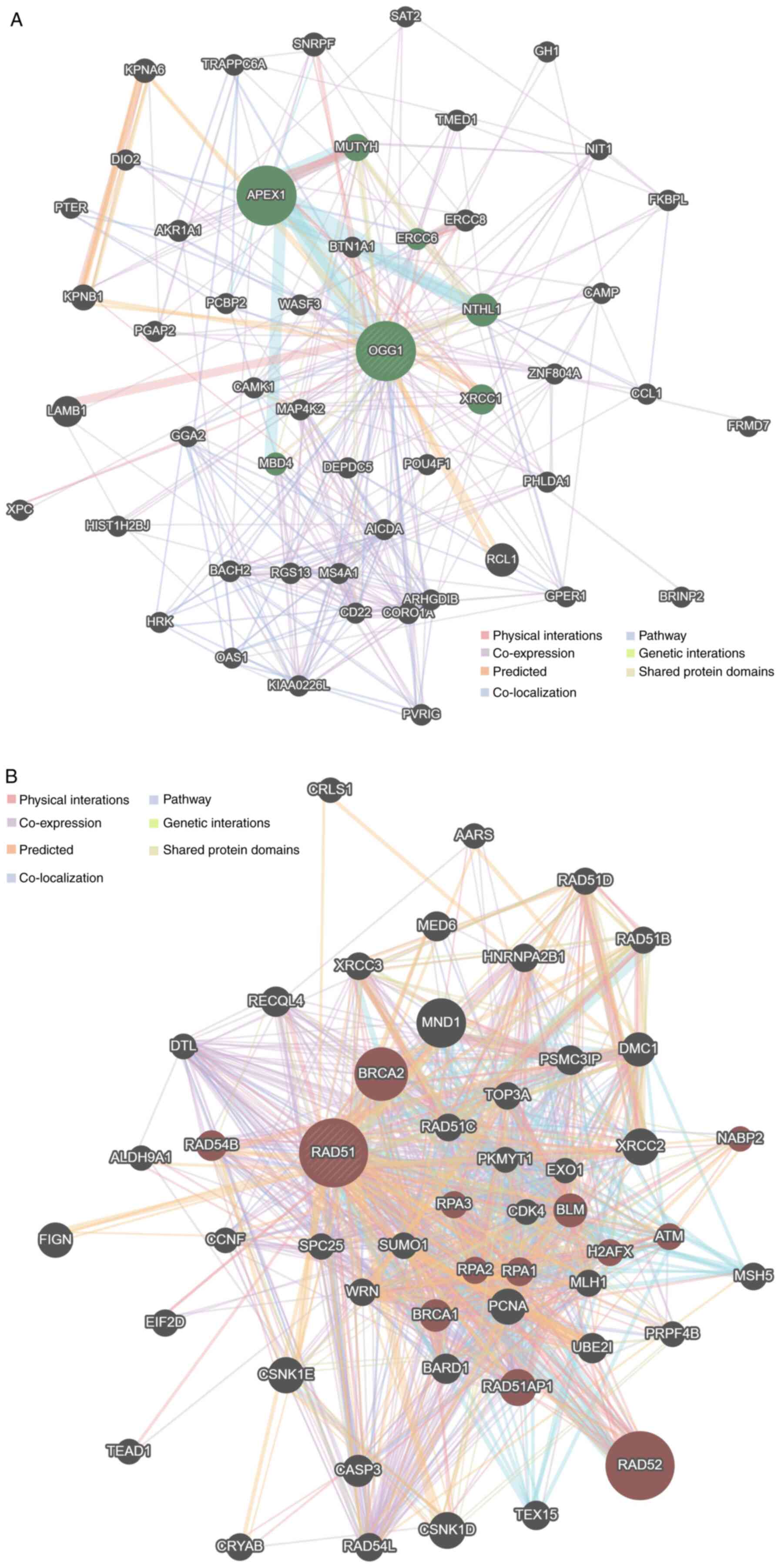
PPI network
Two networks were generated for the OGG1 gene and
the RAD51 gene. For each gene, the PPI network revealed a total of
50 interacting genes for the OGG1 gene (Fig. 2A) and the RAD51 gene (Fig. 2B).
Discussion
DNA damage can be caused by endogenous or exogenous
agents. Cells have different mechanisms to repair DNA and
continuously monitor chromosomes in an attempt to correct any
damage. Thus, DNA repair genes play a critical role in protecting
cells from the consequences of genetic mutations. Any damage in DNA
repair genes may lead to a failure to maintain the integrity of the
genome and the normal function of cells. Several studies have
evaluated the association between single-nucleotide polymorphisms
(SNPs) and developing several types of cancers and chronic
diseases. OGG1 has been revealed to prevent the accumulation of
mutations that occur as a result of exposure to reactive oxygen and
regulates the transcription of various oxidative stress response
genes (33). RADA51 has been
demonstrated to be responsible for repairing double-strand DNA
breaks (34). Polymorphisms in
OGG1 and RADA51 have been linked to the development of several
types of cancers, as this mutation alters the functions of proteins
(35,36).
In the present study, the association between
previously identified SNPs in OGG1 and RAD51 and susceptibility to
CVD in individuals from Saudi Arabia was evaluated. The results
revealed that for OGG1, there was no significant association
between the rs1052133 SNP and the development of CVD, although OGG1
rs1052133 has a carcinogenic role (37). Conversely, the results showed that
there was a significant association between RAD51 rs1801321 and
CVD. The polymorphism rs1801321 plays a substantial role in
promoter activity by substituting G to T on position 172+ (26). Sorting the samples based on age did
not reveal a significant association between carrying the RAD51
rs1801321 polymorphism and developing CVD, but the RAD51
rs1801321G>T polymorphism in male patients exhibited a
significant association with CVD development.
In terms of the protein-protein network, cooperation
between RAD51 and the breast cancer susceptibility protein BRCA2 is
crucial for the repair of double-strand DNA breaks by HR (2). BRCA2 plays a role in HR by
interacting with RAD51. It captures RAD51, transports it to the
damaged site, and subsequently promotes the creation of helical
RAD51-single-stranded DNA nucleoprotein filaments. These filaments
actively search for a homologous DNA template (38). It has been revealed that promoter
activity of RAD51 is significantly enhanced by substituting G at
the polymorphic position +172 for T (26). The elevated activity of HR could
paradoxically result in genomic instability by causing
inappropriate recombination (26).
Consequently, accumulation of DNA damage leads to unaffordable
sequelae such as senescence, apoptosis and inflammation which in
turn could lead to CVD (39).
In conclusion, analyzing the frequencies of RAD51
gene polymorphisms revealed that there was a significant
association between CVD and RAD51 rs1801321 for both male and
female patients regardless of age, while OGG1 rs1052133 exhibited
no significant association with development of CVD in a Saudi
population. To the best of our knowledge, no previous research has
evaluated the association between CVD and RAD51 rs1801321., thus,
more studies with larger sample sizes should be conducted to
confirm the findings of the present study.
Acknowledgements
The authors extend their appreciation to the
Deputyship for Research & Innovation, Ministry of Education in
Saudi Arabia for funding this research (grant no.
IFKSURC-1-3401).
Funding
The present study was supported by the Deputyship for Research
and Innovation, Ministry of Education in Saudi Arabia (grant no.
IFKSURC-1-3401).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
AmA, AbA and MA designed the study. AmA, AbA, JS and
SA conducted the experiments. SA contributed new reagents/analytic
tools. AmA, AbA, MBA, JS and MA analyzed the data and wrote the
paper. MBA and MA confirm the authenticity of all the raw data. All
authors have read and approved the final manuscript.
Ethics approval and consent to
participate
The protocol of the study was reviewed and approved
by the local committee from King Khaled University Hospital and all
patients provided written informed consent. The present study was
conducted according to the guidelines of the Declaration of
Helsinki and approved by the institutional review board of Al-Imam
Muhammad Ibn Saud Islamic University (approval no.
IRB-HAPO-01-R-011). The samples were originally collected from King
Khalid University Hospital (Riyadh, Saudi Arabia) by Dr Mikhlid M.
Almutairi, who obtained Institutional Review Board approval from
Al-Imam Muhammed Ibn Saud Islamic University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
World Health Organization (WHO), .
Cardiovascular diseases (CVDs). WHO; Geneva: http://www.who.int/mediacentre/factsheets/fs317/en/December
21–2020
|
|
2
|
Ramos KS and Partridge CR: Atherosclerosis
and cancer: Flip sides of the neoplastic response in mammalian
cells? Cardiovasc Toxicol. 5:245–255. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hansen ES: International commission for
protection against environmental mutagens and carcinogens. ICPEMC
working paper 7/1/2. Shared risk factors for cancer and
atherosclerosis-a review of the epidemiological evidence. Mutat
Res. 239:163–179. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
De Flora S and Izzotti A: Mutagenesis and
cardiovascular diseases Molecular mechanisms, risk factors, and
protective factors. Mutat Res. 621:5–17. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Botto N, Rizza A, Colombo MG, Mazzone AM,
Manfredi S, Masetti S, Clerico A, Biagini A and Andreassi MG:
Evidence for DNA damage in patients with coronary artery disease.
Mutat Res. 493:23–30. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Henle ES and Linn S: Formation,
prevention, and repair of DNA damage by iron/hydrogen peroxide. J
Biol Chem. 272:19095–19098. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chatterjee N and Walker GC: Mechanisms of
DNA damage, repair, and mutagenesis. Environ Mol Mutagen.
58:235–263. 2017. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Teebor GW, Boorstein RJ and Cadet J: The
repairability of oxidative free radical mediated damage to DNA: A
review. Int J Radiat Biol. 54:131–150. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Demple B and DeMott MS: Dynamics and
diversions in base excision DNA repair of oxidized abasic lesions.
Oncogene. 21:8926–8934. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Harper JW and Elledge SJ: The DNA damage
response: Ten years after. Mol Cell. 28:739–745. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Minten EV and Yu DS: DNA repair:
translation to the clinic. Clin Oncol. 31:303–310. 2019. View Article : Google Scholar
|
|
12
|
Whitaker AM, Schaich MA, Smith MS, Flynn
TS and Freudenthal BD: Base excision repair of oxidative DNA
damage: From mechanism to disease. Front Biosci (Landmark Ed).
22:14932017. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Okasaka T, Matsuo K, Suzuki T, Ito H,
Hosono S, Kawase T, Watanabe M, Yatabe Y, Hida T, Mitsudomi T, et
al: hOGG1 Ser326Cys polymorphism and risk of lung cancer by
histological type. J Hum Genet. 54:739–745. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Rodrigues P, de Marco G, Furriol J,
Mansego ML, Pineda-Alonso M, Gonzalez-Neira A, Martin-Escudero JC,
Benitez J, Lluch A, Chaves FJ and Eroles P: Oxidative stress in
susceptibility to breast cancer: study in Spanish population. BMC
Cancer. 14:1–5. 2014. View Article : Google Scholar
|
|
15
|
Ming-Shiean H, Yu JC, Wang HW, Chen ST,
Hsiung CN, Ding SL, Wu PE, Shen CY and Cheng CW: Synergistic
effects of polymorphisms in DNA repair genes and endogenous
estrogen exposure on female breast cancer risk. Ann Surg Oncol.
17:760–771. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sanjari Moghaddam A, Nazarzadeh M, Bidel
Z, Karamatinia A, Darvish H and Mosavi Jarrahi A: hOGG 1 gene
polymorphism and breast cancer risk: A systematic review and
meta-analysis study. Breast J. 24:70–73. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Romanowicz H, Pyziak Ł, Jabłoński F, Bryś
M, Forma E and Smolarz B: Analysis of DNA repair genes
polymorphisms in breast cancer. Pathol Oncol Res. 23:117–123. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen Y, Li J, Li T and Mo Z: hOGG1 C1245G
gene polymorphism associated with prostate cancer: A meta-analysis.
Int J Biol Markers. 30:e161–e168. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dhillon VS, Yeoh E and Fenech M: DNA
repair gene polymorphisms and prostate cancer risk in South
Australia-results of a pilot study. Urol Oncol. 29:641–646. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Takezaki T, Gao CM, Wu JZ, Li ZY, Wang JD,
Ding JH, Liu YT, Hu X, Xu TL, Tajima K and Sugimura H: hOGG1
Ser326Cys polymorphism and modification by environmental factors of
stomach cancer risk in Chinese. Int J Cancer. 99:624–627. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Su Y, Xu A and Zhu J: The effect of
oxoguanine glycosylase 1 rs1052133 polymorphism on colorectal
cancer risk in Caucasian population. Tumor Biol. 35:513–517. 2014.
View Article : Google Scholar
|
|
22
|
Li Z, Guan W, Li MX, Zhong ZY, Qian CY,
Yang XQ, Liao L, Li ZP and Wang D: Genetic polymorphism of DNA
base-excision repair genes (APE1, OGG1 and XRCC1) and their
correlation with risk of lung cancer in a Chinese population. Arch
Med Res. 42:226–234. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tse D, Zhai R, Zhou W, Heist RS, Asomaning
K, Su L, Lynch TJ, Wain JC, Christiani DC and Liu G: Polymorphisms
of the NER pathway genes, ERCC1 and XPD are associated with
esophageal adenocarcinoma risk. Cancer Causes Control.
19:1077–1083. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Do AT, Brooks JT, Le Neveu MK and LaRocque
JR: Double-strand break repair assays determine pathway choice and
structure of gene conversion events in Drosophila melanogaster. G3
(Bethesda). 4:425–432. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nowacka-Zawisza M, Wiśnik E, Wasilewski A,
Skowrońska M, Forma E, Bryś M, Różański W and Krajewska WM:
Polymorphisms of homologous recombination RAD51, RAD51B, XRCC2, and
XRCC3 genes and the risk of prostate cancer. Anal Cell Pathol
(Amst). 2015:8286462015.PubMed/NCBI
|
|
26
|
Hasselbach L, Haase S, Fischer D, Kolberg
HC and Stürzbecher HW: Characterisation of the promoter region of
the human DNA-repair gene Rad51. Eur J Gynaecol Oncol. 26:589–598.
2005.PubMed/NCBI
|
|
27
|
Michalska MM, Samulak D, Romanowicz H and
Smolarz B: Association of polymorphisms in the 5′ untranslated
region of RAD51 gene with risk of endometrial cancer in the Polish
population. Arch Gynecol Obstet. 290:985–991. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Al Zoubi MS, Zavaglia K, Mazanti C, Al
Hamad M, Al Batayneh K, Aljabali AAA and Bevilacqua G:
Polymorphisms and mutations in GSTP1, RAD51, XRCC1 and XRCC3 genes
in breast cancer patients. Int J Biol Markers. 32:e337–e343. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tulbah S, Alabdulkarim H, Alanazi M,
Parine NR, Shaik J, Pathan AA, Al-Amri A, Khan W and Warsy A:
Polymorphisms in RAD51 and their relation with breast cancer in
Saudi females. Onco Targets Ther. 9:269–277. 2016.PubMed/NCBI
|
|
30
|
Smolarz B, Makowska M, Samulak D,
Michalska MM, Mojs E, Romanowicz H and Wilczak M: Association
between polymorphisms of the DNA repair gene RAD51 and ovarian
cancer. Pol J Pathol. 64:290–295. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Romanowicz-Makowska H, Smolarz B, Samulak
D, Michalska M, Lewy J, Burzyński M and Kokołaszwili G: A single
nucleotide polymorphism in the 5′ untranslated region of RAD51 and
ovarian cancer risk in Polish women. Eur J Gynaecol Oncol.
33:406–410. 2012.PubMed/NCBI
|
|
32
|
Warde-Farley D, Donaldson SL, Comes O,
Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT,
et al: The GeneMANIA prediction server: Biological network
integration for gene prioritization and predicting gene function.
Nucleic Acids Res. 38:214–220. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang K, Maayah M, Sweasy JB and Alnajjar
KS: The role of cysteines in the structure and function of OGG1. J
Biol Chem. 296:1000932021. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Daboussi F, Dumay A, Delacôte F and Lopez
BS: DNA double-strand break repair signalling: The case of RAD51
post-translational regulation. Cell Signal. 14:969–975. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Duan WX, Hua RX, Yi W, Shen LJ, Jin ZX,
Zhao YH, Yi DH, Chen WS and Yu SQ: The association between OGG1
Ser326Cys polymorphism and lung cancer susceptibility: A
meta-analysis of 27 studies. PLoS One. 7:e359702012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Thacker J: The RAD51 gene family, genetic
instability and cancer. Cancer Lett. 219:125–135. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hassan FM: OGG1 rs1052133 polymorphism and
genetic susceptibility to chronic myelogenous leukaemia. Asian Pac
J Cancer Prev. 20:925–928. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lord CJ and Ashworth A: RAD51, BRCA2 and
DNA repair: A partial resolution. Nat Struct Mol Biol. 14:461–462.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wu L, Sowers JR, Zhang Y and Ren J:
Targeting DNA damage response in cardiovascular diseases: From
pathophysiology to therapeutic implications. Cardiovasc Res.
119:691–709. 2023. View Article : Google Scholar : PubMed/NCBI
|