Introduction
The nucleus of human cells harbors 46 chromosomes
that are organized into smaller domains, which enable packaging of
different structural subunits devoted to different functions.
Chromosomes occupy specific locations within the nuclear space,
termed chromosome territories, which are further subdivided into
chromosomal compartments [topologically associated domains (TADs)]
that exhibit specific associations between promoter and enhancer
sites (1). It is now universally
accepted that location of co-regulated genes in different
chromosomes is not casual, as they occupy the same regions within
the nuclear space in different cells (2): This painting lies at the base of the
notion of ‘chromosome territories’ where the spatial correlation
between chromosomes changes throughout cell life (3). In this model, any gene exhibits
different neighbors depending on the specific phase of the cell
cycle and, in particular, on its activation state, indicating the
presence of a specific molecular apparatus that drives chromatin
mobility (4). As a corollary of
chromosome territories, it has been also demonstrated that gene
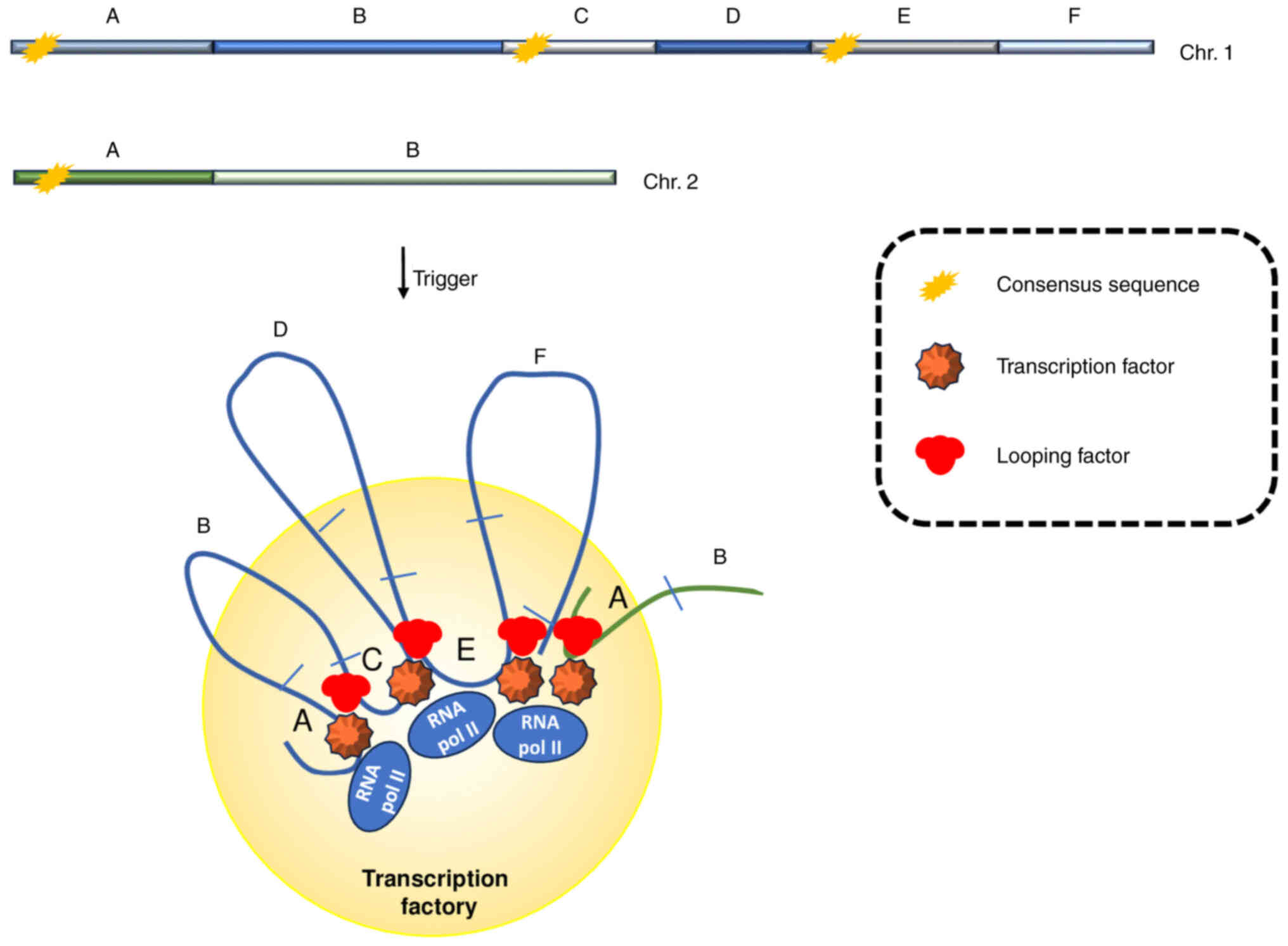
expression is grouped in several sites, called ‘transcription
factories’, where genes that share the activating stimuli are
concurrently transcribed (Fig. 1)
(5,6). Physically, transcription factories
can be viewed as nuclear granules with a diameter of roughly 50–100
nm reaching a number that varies between the different cell types
and ranges from few hundreds to ~30,000 (7,8).
They behave as chromatin hubs where at least two different active
RNA polymerases synthesize RNA on two different targets (9,10),
and harbor multi-protein complexes that reach local concentrations
sufficient to complete ordered expression of co-regulated genes
(11). In particular, it has been
calculated that any factory contains 8–10 active genes on average
(12,13).
Chromatin looping and distribution of genes
in different territories and transcription factories
The molecular mechanism supporting co-localization
of genes within the same factory needs to be investigated in deeper
detail; however, it is essentially based on the achievement of a
specific flexibility by chromatin that allows creation of several
loops able to change location of genes with respect to each other
along a chromatin fiber. On this regard, it could be hypothesized
that large-scale random movements of chromatin may allow
co-activated genes to meet the factories, triggering the multistep
process that drives their ordered recruitment to the factory
(14,15). This process implies the involvement
of different classes of proteins such as chromatin modifiers,
transcription factors and, notably, looping factors. Gene looping
calls for particular consideration as it governs the precise
apposition of enhancers and target genes mainly through different
mechanisms where architectural DNA binding proteins contribute to
looping by interacting with general or cell-specific factors
(16,17). Moreover, occasionally the process
involves the direct activity of general transcription factors
including the mediator complex and cohesin, that help the looping
between enhancers and promoters (18), whereas some other transcription
factors, such as ZNF143 and YY1 have been evidenced to enhance
chromatin loop formation in order to drive localization of specific
loci within the nucleus (19,20).
In addition, enhancer and non-coding RNA molecules have been deeply
involved in looping between enhancer and promoter sites of specific
genes, bringing these two elements into physical proximity in order
to allow correct transcriptional processing and indicating
chromatin looping as one of the features subjected to enhancer RNA
control (21–23). Finally, gene looping also concerns
in most cases the 3′-end polyadenylation sites of genes that bridge
with promoters and has been revealed to be essential for a correct
transcriptional output (24).
Interestingly, looping factors make a direct contribution to the
recruitment of a specific gene to the factory through the
establishment of protein bridges with the transcription factors
already located in shared factories (25), governing, in this way, the unequal
distribution of transcription throughout the nucleus in order to
gain optimized gene expression (Fig.
1) (26).
The separation of nuclear space in chromosome
territories is not inelastic and changes in response to
environmental stimuli that shape compartmentalization of chromatin
in parallel with restriction of the developmental progression
(27). In particular, the genome
appears widely dispersed within the nucleus in pluripotent stem
cells, probably due to the hyper-dynamic nature of looping of
chromatin fibers present in these cells (28). However, during lineage
specification, a dynamic equilibrium between chromatin mobility and
the transcriptional noise underlying specific gene silencing must
be reached (29,30). As the specialization process
progresses, chromatin compaction increases with large zones of the
nucleus devoid of DNA that accumulates along the nuclear envelope
and near the nucleolus: An image that correlates with the majority
of genes transcriptionally repressed and consequent appearance of
several Lamina-Associated-Domains where inhibitory markers prevail
(31). These states of chromatin
folding are acquired during cell type specialization and are
usually reversible depending upon the environmental signal: In this
regard an original example that underlines the role played by a
specific cell function, is represented by the rod photoreceptor
cells where compaction of chromosomes can be observed at the center
of the nuclear space instead of periphery. In fact, in these cells,
chromatin functions as a physical barrier to the scattering of
light when passing through, and its nuclear distribution appears to
be in accordance with the regulation of gene expression (32). The development of the concept of
Transcription Factory and the characterization of some of such
factories shed light on how the transcription factors drive the
topological genome reorganization in the context of the control of
gene expression. The authors' experience has been focused
essentially on steroid receptors, in particular estrogen and
retinoic acid receptors, but other examples of TFs mediating the
assembly of transcription factories include Nanog (33) and Sox2 (34) in differentiation of pluripotent
stem cells, Klf1 (35) in
erythrocytes differentiation, Pax3 (36) and MyoD (37) during myogenesis, Pax5 (38) in B cell differentiation.
It is also important to clarify that the
transcription factories represent by themselves well-defined
entities, but more insight into three-dimensional chromatin changes
associated with the activation of transcription could arise from
the deeper knowledge about nuclear structures such as the
membrane-less organelle (MLOs) (39). Evidence exists suggesting that
nuclear condensate formation could be involved in the regulation of
various aspects of gene expression, as numerous transcription
factors, such as the steroid receptors, undergo Liquid-Liquid Phase
Separation, the process leading to MLOs formation. Androgen
receptor (AR), estrogen receptor and glucocorticoid receptor (GR)
condensates were observed in the presence of the Mediator Complex
subunit 1. The formation of condensates is subordinated to the
interaction of the receptor with specific chromatin regions in the
nucleus. The ligand-bound steroid receptors are translocated to the
nucleus, where they form transcriptionally active foci. It has been
reported that AR and GR foci are endowed with properties of MLOs
(40). Thus, it is conceivable
that formation of MLOs including steroid receptor-chromatin
complexes contributes to stabilization of the transcription
factories architecture and an integrated analysis of both processes
could provide comprehensive insight into the regulation of gene
expression.
On one extreme side of cellular differentiation and
growth stands cancer, where altered nuclear architecture shows an
irregular occupancy of the nucleus by chromosomes with altered
compaction of chromatin and its DNA content. In particular, cancer
cells display changes in the appearance and number of the nucleoli,
the first described transcription factories assembled in proximity
of the nucleolar organizing regions (41). In fact, an increase in the number
of nucleoli parallels a decrease in the stability of contacts
between chromosomes carrying rRNA loci, and consequent changes in
nuclear organization that may impact adaptive responses. In this
regard, cells have evolved a complex machinery to preserve the
organization of nuclear space in which chromatin mobility is
balanced by factors that restrain its amplitude; however, in
transformed cells alterations in bridging between regulatory
elements such as enhancers or insulators with promoters are well
documented (42). An increase in
mobility of cancer genome may also be presumably responsible for
the increased gene expression with consequent phenotypic
variability among cancer cells, functioning as the base of tumor
progression due to altered contacts with the transcription factors
that normally govern gene expression (43). As an example, translocations of
Myc and Igh genes, most frequently found in
plasmacytoma cells (44), share
the same factories presumably because they require similar
transcription factors that start the process of translocation
(45,46). The observed increase in genome
mobility may be due, at least in some cases, to changes in the
activity of genome organizer proteins such as SATB1 that has been
involved in the emergence of aggressive tumor phenotypes (47). Finally, a spatial relation between
the three-dimensional architecture of the genome and chromosomal
alterations has been reported in cancer cells where proximity of
regions transcribed at the same time may be used to predict
variations in gene-copy number observed frequently in malignant
cells (48,49).
Experimental strategies to highlight
transcriptional looping
Chromosome conformation capture
(3C)
It is well established that transcription is
governed by post-translational modifications at the N-terminal
tails of the histone octamer which represents the major protein
component of nucleosomes, the fundamental subunit of chromatin.
Such modifications follow a precise code (50), and are especially aimed at
governing the interaction of transcription factors with the DNA
located at the promoter regions of genes to be activated and/or
repressed (51,52). Among these modifications,
methylation of lysine 4 (K4) and demethylation of lysine 9 (K9) in
histone H3, the most protruding tail from the octamer histone disc,
are mostly involved in gene activation (53,54).
In particular, demethylation of H3K9 is catalyzed by specific
demethylases recruited to the regulatory sites of activated genes
(55), and produces nuclear
reactive oxygen species (ROS) that induce single-strand DNA breaks
that rule chromatin plasticity, easing the productive
transcriptional output (56,57).
In order to analyze the involvement of gene folding
in the expression of responsive genes induced by estrogens, the
previously described strategy ‘Capturing chromosome conformation’,
also known as ‘3C’ was first followed (58). In synthesis, the experimental
design is based on the quantification of contact frequencies at any
time between two loci distant on linear DNA, as revealed by
quantitative polymerase chain reaction (qPCR) amplifications
yielding incomparably richer data with respect to previous
strategies based on light microscopy, and providing deeper insight
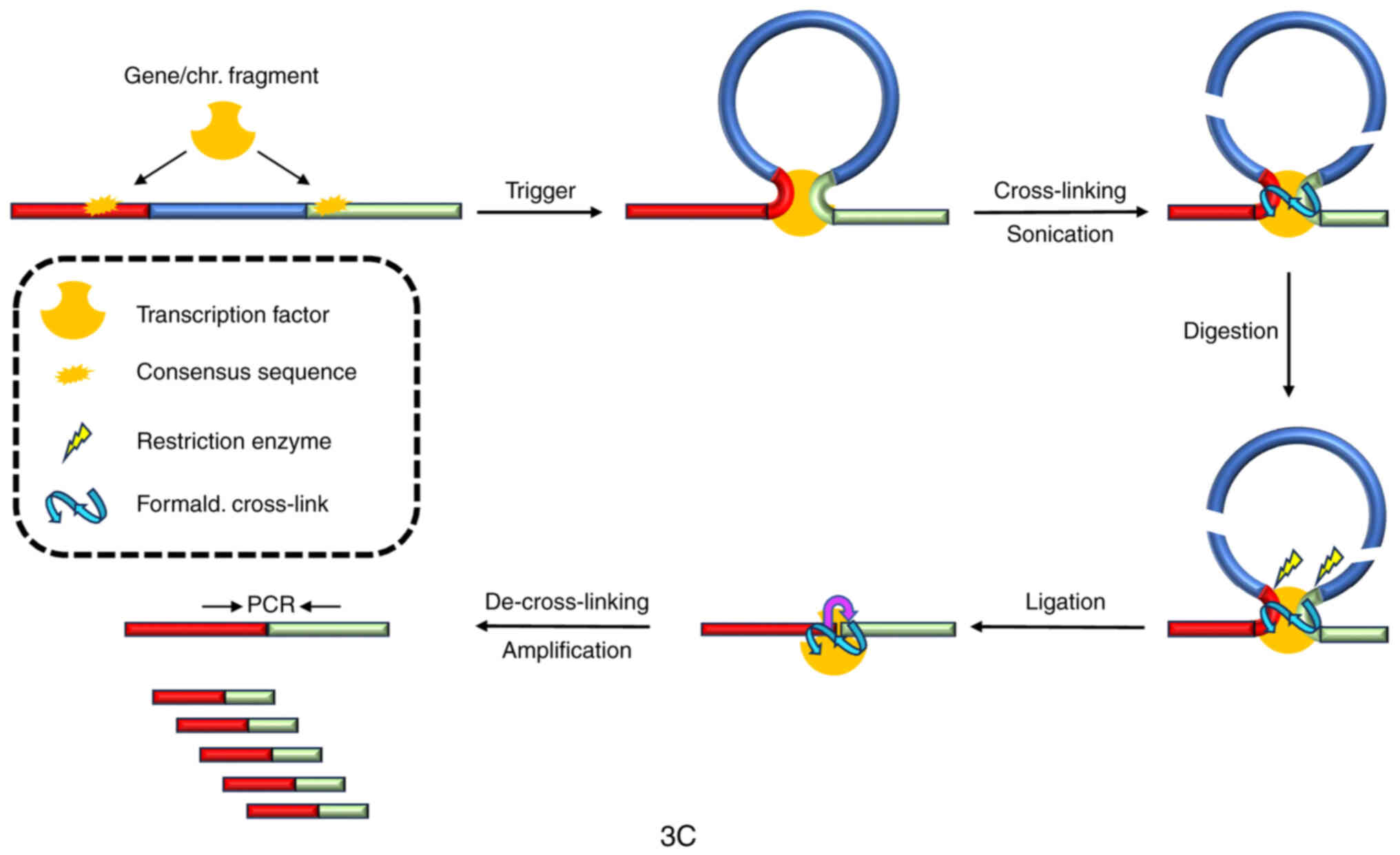
into the spatial organization of specific loci (Fig. 2).
The main steps of 3C experimental design may be
summarized as follows: i) cross-linking of chromatin using a
soluble fixative agent such as formaldehyde to generate covalent
bonds between the sites on DNA bridged by proteins; ii) isolation
and digestion of chromatin with selected restriction enzymes; iii)
ligation of sticky ends of digested fragments at low DNA
concentration to favor intramolecular over intermolecular
ligations; iv) reverse of cross-linking to obtain purified DNA in
order to interrogate the rearranged fragments by qPCR using
locus-specific primers encompassing fragments of the sites supposed
to be bridged under the investigated experimental conditions; and
v) comparison of the PCR results from cross-linked templates with
control templates without bridging, to discriminate non-specific
ligations. In principle, the cross-linking frequency between two
sites on DNA is inversely proportional to their relative distance
unless they are put in spatial contact by a particular protein
factor. Therefore, a picture of the three-dimensional (3D)
architecture of a particular locus may be deduced by comparing
linking frequencies from cross-linked chromatin to the control
(Fig. 2).
Historically, yeast chromosome III has been the
first to be analyzed, and its 3D conformation has revealed
proximity between the telomeres that conferred the chromosome the
form of a sort of ring (58).
Next, 3C experimental approach was adapted for analysis of mammary
cells and, even though it can be especially used to detect contacts
between sites spanning only few hundred kilobases (59), it confirmed the existence of
dynamic chromatin loops between promoters/enhancers and their
target genes, conferring chromatin a peculiar conformation
dependent on the transcriptional status (60–62).
Upon 3C, it was demonstrated that the estrogen
responsive element located ~1.5 kb downstream from the
transcription start site of the estrogen responsive gene
bcl-2 bridged through a complex containing the estrogen
receptor with the transcription complex located onto the promoter
of the same gene. Moreover, looping between the promoter at the
5′-end, and the 3′-end where the polyadenylation site is located,
has been widely assessed, supporting the concept that 3′
end-processing factors bridge with the transcriptional machinery
(63,64).
DNA-picked chromatin (DPC)
3C certainly represents a classical, strong tool to
identify gene looping; nevertheless, it requires the existence of
suitable restriction sites strategically located along the genes
under investigation and setting up the right balance between the
concentration of DNA and the restriction enzyme to enhance
intramolecular ligations. To try to overcome these bottlenecks, a
novel experimental strategy conceived by introducing several
changes into the method designed to perform proteomic analysis of
multiprotein complexes assembled on chromatin (65) was proposed. This strategy, called
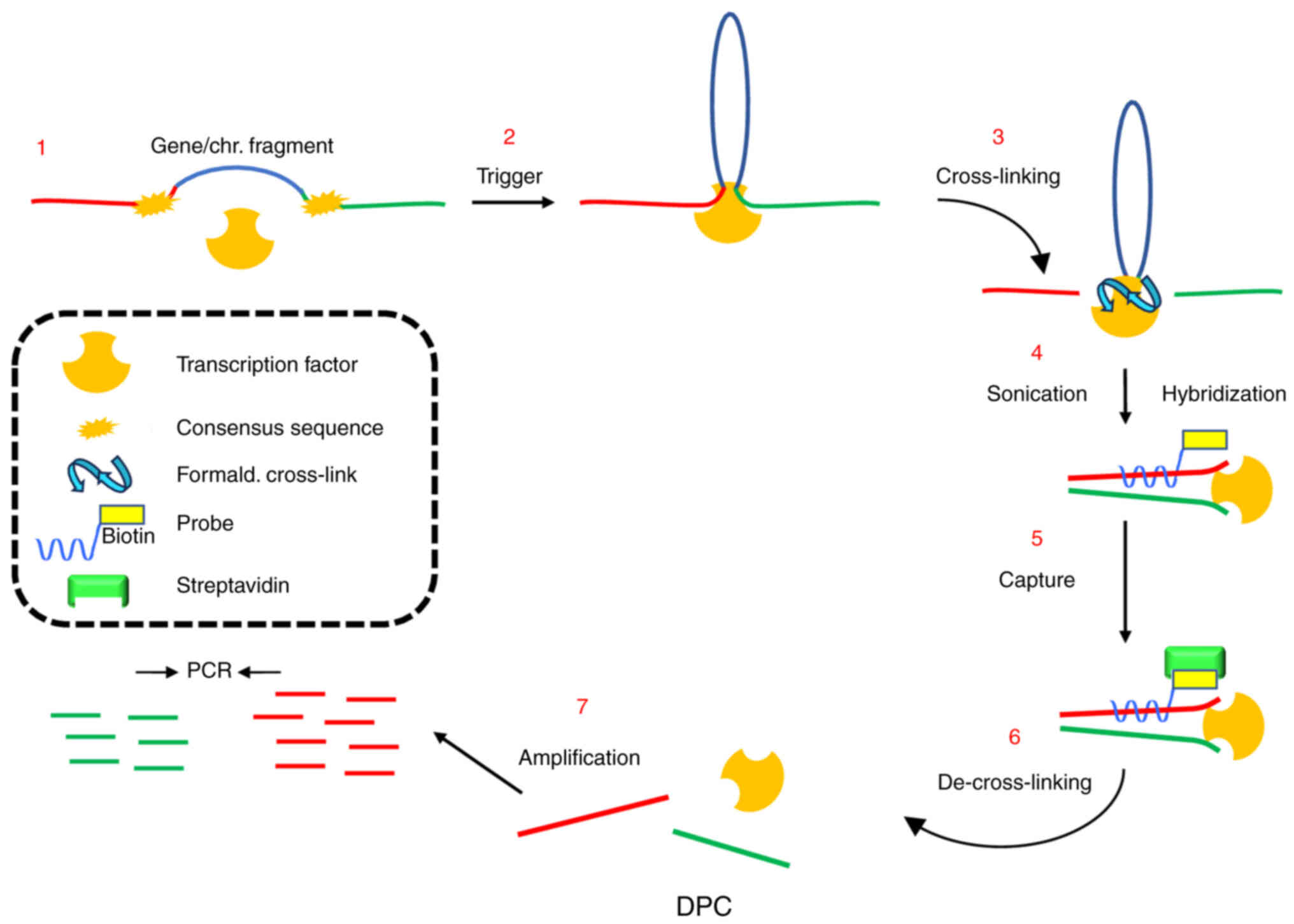
DPC (DNA-picked chromatin) by the authors, consisted in
cross-linking chromatin from cells challenged or not with
estrogens, DNA cleavage to 500–600 base-pair fragments by
sonication, and hybridization with a biotinylated oligonucleotide
probe complementary to the site under investigation, followed by
capture on magnetic streptavidin beads (Fig. 3). Presence of DNA loci bordering
the site hybridized by the probe was, then, revealed by qPCR
amplifications of the DNA extracted and purified from the eluted
fragments after de-crosslinking. As a control, PCR reactions were
carried out using chromatin before hybridization or naked DNA as
template (input), and recovery of the probing region in retrieved
DNA from each experimental condition was assumed as the baseline to
be compared with the relative amount of retrieved associated sites
in the same experimental point. Moreover, to solve troubles emerged
by possible changes of spatial chromatin organization throughout
transcriptional stimulation, the use of multiple probes was
recommended. In summary, the strategy assumes that two sites can be
co-captured by the probe only if they are bridged by proteins
recruited upon the transcriptional trigger, independently from
their distance on linear DNA, and can be imagined as a sort of
chromatin immunoprecipitation (ChIP) where the antibody is
substituted with the DNA probe (Fig.
3) (66).
Owing to DPC strategy, the spatial relationship
between the enhancer and polyadenylation sites of bcl-2 gene
upon estrogen addition was first analyzed and it was observed that
the last was increased in the DNA purified after hybridization with
the enhancer probe (65). More
important, this technique allowed to assess the connection within
the nuclear space between this gene, located on chromosome 18, and
another estrogen-sensitive gene, RIZ, located on chromosome
1, and the authors were able to detect an increase of bcl-2
enhancer and polyadenylation sites when the rescued DNA hybridized
with RIZ promoter was used as bait (65). Noteworthy, two different
chromosomes, 1 and 18, have been demonstrated to occupy contiguous
territories within the nuclear space and previous data by the
authors suggested that this is dependent on assembly of the cognate
transcription complex after hormone stimulation (67). It was also assessed that the
demethylation of H3K9me2 is essential to the formation of
transcription-induced looping of hormone-dependent genes (66).
Circular chromosome conformation
capture (4C)
Having demonstrated the intimate correlation between
changes induced on chromatin flexibility by transcription (through
induction of single-strand breaks by an increase of ROS production
due to the demethylase activity) and establishment of loops that
allow allocation of concurrently transcribed genes within the same
transcription factories, the further step is represented by the
identification of the genes involved in the assembly of such
factories, starting by a gene whose responsiveness to the
investigated trigger is universally recognized (in the present
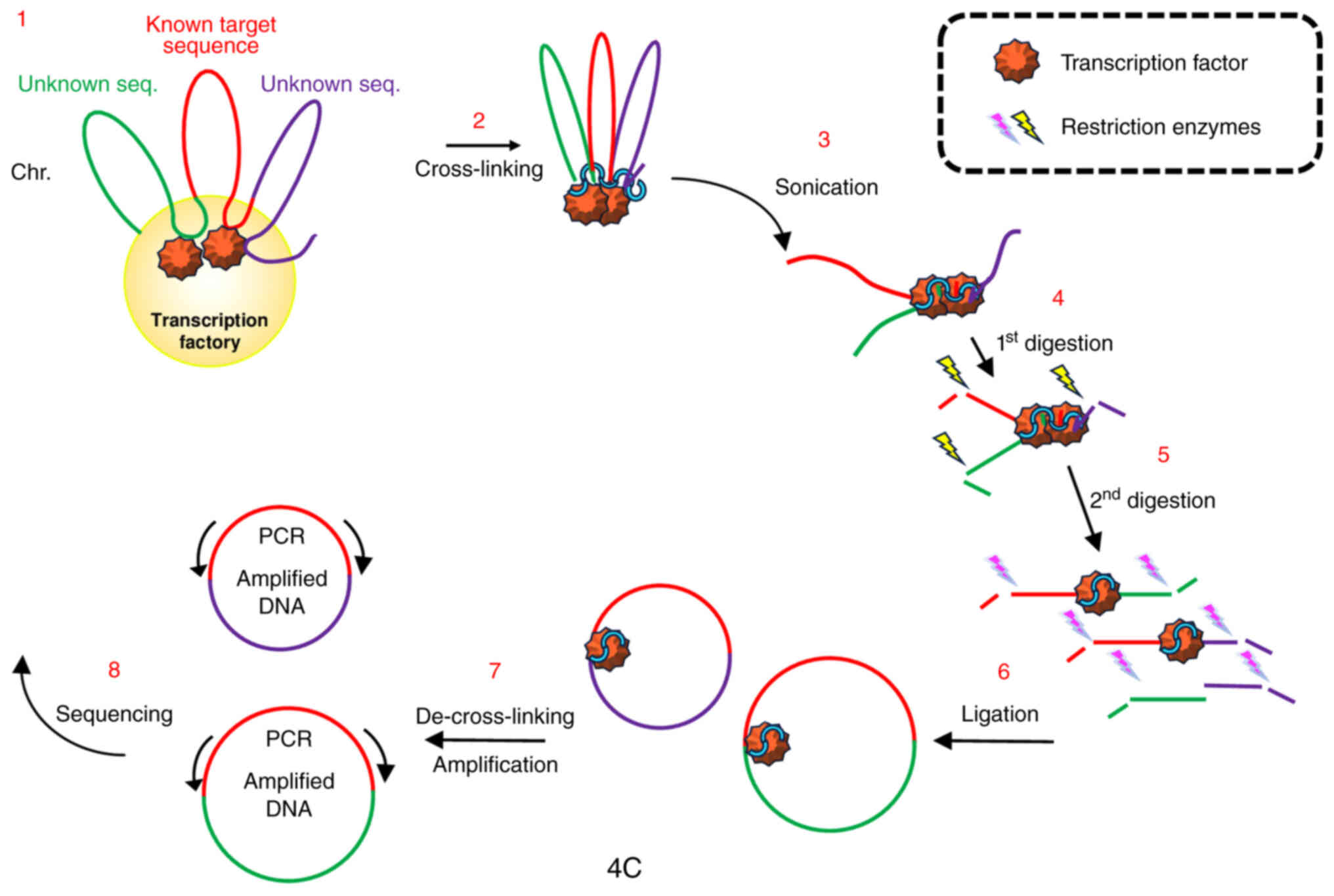
case, the bait is represented by an estrogen-sensitive gene). In
fact, as has been graphically represented in Fig. 1, if the estrogen responsive factory
is imagined as a flower with the transcribed genes being the
pistils and the intervening DNA the petals, then the goal should be
the elucidation of the genes that form the factory/flower
establishing contacts to the bait (Fig. 3).
To this aim, since either 3C as well as DPC focus on
interactions between two already known loci (for this reason called
‘one versus one’) and can, then, be used exclusively to detect
spatial interactions between already known regions (68), an-omics approach is needed, using
the products generated by the 3C or DPC procedures as a library
that needs further treatment. On this regard, a growing family of
related strategies has been reported, among which the 3C-on-chip
(69), and the open-ended 3C
(70), mostly based on
circularization of the generated chromatin fragments using a second
restriction enzyme. Thus, the authors' attention will be now
pointed to the 4C experimental approach that represents a paramount
protocol to assess the involvement of co-responsive genes within
the same transcription factory (a ‘one versus all’ approach)
(71). In synthesis, the procedure
may be summarized as follows: Cross-linked chromatin is treated to
generate circular DNA molecules from restricted hybrid fragments by
use of a second restriction enzyme under high ligase concentration
and prolonged incubation times. After reversal of cross-linking,
nested PCR primers spanning the opposite ends of the DNA site
chosen as probe are used to amplify any sequence fallen in strict
proximity with the bait (also called the ‘viewpoint’) and then,
ligated. The amplified ligation products are sequenced to assess
all the spatial partners of the gene (or a specific locus of it)
under investigation, providing the identity of all the pistils of
the flower (Fig. 4) (72).
Conclusions and future perspectives
4C has been demonstrated to represent a useful
device to highlight either short-range interactions as well as
long-range spatial cross-talks between very distant sites (73); therefore, it appears as the
strategy of choice to obtain a complete detection of genes that
co-localize with the estrogen-responsive gene PS2 after
hormone challenge (74). The same
strategy will presumably allow assessment of components of any of
the transcription factories established by any transcriptional
trigger throughout cellular life-span, either in physiological as
well as pathological conditions.
In conclusion, the scope of this brief review was to
emphasize the importance of studying the three-dimensional
structure of chromatin with the hope of providing a succinct
summary, based on the authors' experience of the evolution of the
principal methods used to deepen knowledge about this issue; the
respective advantages and limits have been highlighted in Table I.
 | Table I.Although the 4C appears undoubtedly
as the most performing method to discover and analyze new
transcription factories, it should be reiterated that 3C and the
less widespread DPC techniques still remain very effective tools
when a specific target is under investigation. |
Table I.
Although the 4C appears undoubtedly
as the most performing method to discover and analyze new
transcription factories, it should be reiterated that 3C and the
less widespread DPC techniques still remain very effective tools
when a specific target is under investigation.
| Technique | Advantages | Limits |
|---|
| 3C | Easy to use, high
sensitivity, inexpensive | Possible
unavailability of restriction sites, undesired structure alteration
due to cross-linking, informative only for two known sites |
| DPC | Independent on
availability of restriction sites, free of possible experimental
artifacts, very fast, high sensitivity, negligible costs | Informative only
for two known sites, limited by the interference of some
three-dimensional chromatin structure |
| 4C | Allows a
genome-wide analysis of the transcription factories | Possible
unavailability of restriction sites, rather expensive, requires
preliminary ‘in silico’ analysis |
Acknowledgements
Not applicable.
Funding
The present study was supported by The Italian Ministry of
Health-Project of finalized research RF-2019-12368937 ‘Targeting
novel insulin/IGF-driven signaling networks in breast cancer’
(grant no. CUP D69J21001840001).
Availability of data and materials
Data sharing is not applicable to this article, as
no data sets were generated or analyzed during the current
study.
Authors' contributions
BP conceived, wrote and edited the manuscript. AM
discussed and revised the manuscript and prepared figures. GC
discussed and revised the manuscript. All authors read and approved
the final manuscript. Data authentication is not applicable.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Fritz AJ, Sehgal N, Piss A, Xu J and
Berezney R: Chromosome territories and the global regulation of the
genome. Genes Chromosomes Cancer. 58:407–426. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rajapakse I, Perlman MD, Scalzo D,
Kooperberg C, Groudine M and Kosak ST: The emergence of
lineage-specific chromosomal topologies from coordinate gene
regulation. Proc Natl Acad Sci USA. 106:6679–6684. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cremer T, Cremer M, Dietzel S, Müller S,
Solovei I and Fakan S: Chromosome territories-a functional nuclear
landscape. Curr Opin Cell Biol. 18:307–316. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chuang CH, Carpenter AE, Fuchsova B,
Johnson T, de Lanerolle P and Belmont AS: Long-range directional
movement of an interphase chromosome site. Curr Biol. 18:825–831.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Xu M and Cook PR: Similar active genes
cluster in specialized transcription factories. J Cell Biol.
181:615–623. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sutherland H and Bickmore WA:
Transcription factories: Gene expression in unions? Nat Rev Genet.
10:457–466. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Pombo A, Hollinshead M and Cook PR:
Bridging the resolution gap: Imaging the same transcription
factories in cryosections by light and electron microscopy. J
Histochem Cytochem. 47:471–480. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Faro-Trindade I and Cook PR: A conserved
organization of transcription during embryonic stem cell
differentiation and in cells with high C value. Mol Biol Cell.
17:2910–2920. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
de Laat W and Grosveld F: Spatial
organization of gene expression: The active chromatin hub.
Chromosome Res. 11:447–459. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Papantonis A and Cook PR: Transcription
factories: Genome organization and gene regulation. Chem Rev.
13:8683–8705. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Osborne CS, Chakalova L, Brown KE, Carter
D, Horton A, Debrand E, Goyenechea B, Mitchell JA, Lopes S, Reik W
and Fraser P: Active genes dynamically colocalize to shared sites
of ongoing transcription. Nat Genet. 36:1065–1071. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Faro-Trindade I and Cook PR: Transcription
factories: Structures conserved during differentiation and
evolution. Biochem Soc Trans. 34:1133–1137. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Larkin JD, Cook PR and Papantonis A:
Dynamic reconfiguration of long human genes during one
transcription cycle. Mol Cell Biol. 32:2738–2747. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mahy NL, Perry PE and Bickmore WA: Gene
density and transcription influence the localization of chromatin
outside of chromosome territories detectable by FISH. J Cell Biol.
159:753–763. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chambeyron S and Bickmore WA: Chromatin
decondensation and nuclear reorganization of the HoxB locus upon
induction of transcription. Genes Dev. 18:1119–1130. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Krivega I and Dean A: Enhancer and
promoter interactions-long distance calls. Curr Opin Genet Dev.
22:1–7. 2012. View Article : Google Scholar
|
|
17
|
Stadhouders R, Thongjuea S, Andrieu-Soler
C, Palstra RJ, Bryne JC, van den Heuvel A, Stevens M, de Boer E,
Kockx C, van der Sloot A, et al: Dynamic long-range chromatin
interactions control Myb proto-oncogene transcription during
erythroid development. EMBO J. 31:986–999. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kagey MH, Newman JJ, Bilodeau S, Zhan Y,
Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine
SS, et al: Mediator and cohesion connect gene expression and
chromatin architecture. Nature. 467:430–435. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhou Q, Yu M, Tirado-Magallanes M, Li B,
Kong L, Guo M, Tan ZH, Lee S, Chai L, Numata A, et al: ZNF143
mediates CTCF-bound promoter-enhancer loops required for murine
hematopoietic stem and progenitor cell function. Nat Commun.
12:432021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Weintraub AS, Li CH, Zamudio AV, Sigova
AA, Hannett NM, Day DS, Abraham BJ, Cohen MA, Nabet B, Buckley DL,
et al: YY1 is a structural regulator of enhancer-promoter loops.
Cell. 171:1573–1588. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jiao W, Chen Y, Song H, Li D, Mei H, Yang
F, Fang E, Wang X, Huang K, Zheng L and Tong Q: HPSE enhancer RNA
promotes cancer progression through driving chromatin looping and
regulating hnRNPU/p300/EGR1/HPSE axis. Oncogene. 36:2728–2745.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hsieh CL, Fei T, Chen Y, Li T, Gao Y, Wang
X, Sun T, Sweeney CJ, Lee GS, Chen S, et al: Enhancer RNAs
participate in androgen receptor-driven looping that selectively
enhances gene activation. Proc Natl Acad Sci USA. 111:7319–7324.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tan SH, Leong WZ, Ngoc PCT, Tan TK,
Bertulfo FC, Lim MC, An O, Li Z, Yeoh AEJ, Fullwood MJ, et al: The
enhancer RNA ARIEL activates the oncogenic transcriptional program
in T-cell acute lymphoblastic leukemia. Blood. 134:239–251. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
He X, Khan AU, Cheng H, Pappas DL Jr,
Hampsey M and Moore CL: Functional interactions between the
transcription and the mRNA 3′ end processing machineries mediated
by Ssu72 and Sub1. Genes Dev. 17:1030–1042. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Deng W, Lee J, Wang H, Miller J, Reik A,
Gregory PD, Dean A and Blobel GA: Controlling long-range genomic
interactions at a native locus by targeted tethering of a looping
factor. Cell. 149:1233–1244. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chubb JR and Bickmore WA: Considering
nuclear compartmentalization in the light of nuclear dynamics.
Cell. 112:403–406. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chubb JR, Boyle S, Perry P and Bickmore
WA: Chromatin motion is constrained by association with nuclear
compartments in human cells. Curr Biol. 12:439–445. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Meshorer E, Yellajoshula D, George E,
Scambler PJ, Brown DT and Misteli T: Hyper-dynamic plasticity of
chromatin proteins in pluripotent embryonic stem cells. Dev Cell.
10:105–116. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Turner BM: Open chromatin and
hypertranscription in embryonic stem cells. Cell Stem Cell.
2:408–410. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chang HH, Hemberg M, Barahona M, Ingber DE
and Huang S: Transcriptome-wide noise controls lineage choice in
mammalian progenitor cells. Nature. 453:544–547. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Guelen L, Pagie L, Brasset E, Meuleman W,
Faza MB, Talhout W, Eussen BH, de Klein A, Wesels L, de Laat W and
van Steensel B: Domain organization of human chromosomes revealed
by mapping of nuclear lamina interactions. Nature. 453:948–951.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Solovei I, Kreysing M, Lanctôt C, Kösem S,
Peichl L, Cremer T, Guck J and Joffe B: Nuclear architecture of rod
photoreceptor cells adapts to vision in mammalian evolution. Cell.
137:356–368. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Nitzsche A, Paszkowski-Rogacz M, Matarese
F, Janssen-Megens EM, Hubner NC, Schulz H, de Vries I, Ding L,
Huebner N, Mann M, et al: RAD21 cooperates with pluripotency
transcription factors in the maintenance of embryonic stem cell
identity. PLoS One. 6:e194702011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bertolini JA, Favaro R, Zhu Y, Pagin M,
Ngan CY, Wong CH, Tjong H, Vermunt MW, Martynoga B, Barone C, et
al: Mapping the global chromatin connectivity network for Sox2
function in neural stem cell maintenance. Cell Stem Cell.
24:462–476. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Drissen R, Palstra RJ, Gillemans N,
Splinter E, Grosveld F, Philipsen S and de Laat W: The active
spatial organization of the beta-globin locus requires the
transcription factor EKLF. Genes Dev. 18:2485–2490. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Magli A, Baik J, Pota P, Cordero CO, Kwak
IY, Garry DJ, Love PE, Dynlacht BD and Perlingeiro RCR: Pax3
cooperates with Ldb1 to direct local chromosome architecture during
myogenic lineage specification. Nat Commun. 10:23162019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Dall'Agnese A, Caputo L, Nicoletti C, di
Iulio J, Schmitt A, Gatto S, Diao Y, Ye Z, Forcato M, Perera R, et
al: Transcription factor-directed re-wiring of chromatin
architecture for somatic cell nuclear reprogramming toward
trans-differentiation. Mol Cell. 76:453–472. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Johanson TM, Lun ATL, Coughlan HD, Tan T,
Smyth GK, Nutt SL and Allan RS: Transcription-factor-mediated
supervision of global genome architecture maintains B cell
identity. Nat Immunol. 19:1257–1264. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sołtys K and Ozyhar A: Transcription
regulators and membrane less organelles challenges to investigate
them. Int J Mol Sci. 22:127582021. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Shrinivas K, Sabari BR, Coffey EL, Klein
IA, Boija A, Zamudio AV, Schuijers J, Hannett NM, Sharp PA, Young
RA and Chakraborty AK: Enhancer features that drive formation of
transcriptional condensates. Mol Cell. 75:549–561.e7. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Pederson T: The nucleolus. Cold Spring
Harbor Perspect Biol. 3:a0003682011. View Article : Google Scholar
|
|
42
|
Gondor A and Ohlsson R: Chromosome
crosstalk in three dimensions. Nature. 461:212–217. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Pujadas E and Feinberg AP: Regulated noise
in the epigenetic landscape of development and disease. Cell.
148:1123–1131. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Osborne CS, Chakalova L, Mitchell JA,
Horton A, Wood AL, Bolland DJ, Corcoran AE and Fraser P: Myc
dynamically and preferentially relocates to a transcription factory
occupied by Igh. PLoS Biol. 5:e1922007. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Misteli T: Higher-order genome
organization in human disease. Cold Spring Harb Perspect Biol.
2:a0007942010. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kohwi-Shigematsu T, Poterlowicz K,
Ordinario E, Han HJ, Botchkarev V and Kohwi Y: Genome organizing
function of SATB1 in tumor progression. Semin Cancer Biol.
23:72–79. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wijchers PJ and de Laat W: Genome
organization influences partner selection for chromosomal
rearrangements. Trends Genet. 27:63–71. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
De S and Michor F: DNA replication timing
and long-range DNA interactions predict mutational landscapes of
cancer genomes. Nat Biotechnol. 29:1103–1108. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Fudenberg G, Getz G, Meyerson M and Mirny
LA: High order chromatin architecture shapes the landscape of
chromosomal alterations in cancer. Nat Biotechnol. 29:1109–1113.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Strahl BD and Allis CD: The language of
covalent histone modifications. Nature. 403:41–45. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Berger SL: The complex language of
chromatin regulation during transcription. Nature. 447:407–412.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Kouzarides T: Chromatin modifications and
their functions. Cell. 128:693–705. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Peters AH, Kubicek S, Mechtler K,
O'Sullivan RJ, Derijck AAH, Perez-Burgos L, Kohlmaier A, Opravil S,
Tachibana M, Shinkai Y, et al: Partitioning and plasticity of
repressive histone methylation states in mammalian chromatin. Mol
Cell. 12:1577–1589. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Rice JC, Briggs SD, Ueberheide B, Barber
CM, Shabanowitz J, Hunt DF, Shinkai Y and Allis CD: Histone
methyltransferases direct different degrees of methylation to
define distinct chromatin domains. Mol Cell. 12:1591–1598. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Closs PAC, Christensen J, Agger K and
Helin K: Erasing the methyl mark: Histone demethylases at the
center of cellular differentiation and disease. Genes Dev.
22:1115–1140. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Perillo B, Ombra MN, Bertoni A, Cuozzo C,
Sacchetti S, Sasso A, Chiariotti L, Malorni A, Abbondanza C and
Avvedimento EV: DNA oxidation as triggered by H3K9me2 demethylation
drives estrogen-induced gene expression. Science. 319:202–206.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Papantonis A and Cook PR: Genome
architecture and the role of transcription. Curr Opin Cell Biol.
22:271–276. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Dekker J, Rippe K, Dekker M and Kleckner
N: Capturing chromosome conformation. Science. 295:1306–1311. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Simonis M, Kooren J and de Laat W: An
evaluation of 3C-based methods to capture DNA interactions. Nat
Methods. 4:895–901. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Murrell A, Heeson S and Reik W:
Interaction between differentially methylated regions partitions
the imprinted genes Igf2 and H19 into parent-specific chromatin
loops. Nat Genet. 36:889–893. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
61
|
Gheldof N, Smith EM, Tabuchi TM, Koch CM,
Dunham I, Stamatoyannopoulos JA and Dekker J: Cell-type-specific
long-range looping interactions identify distant regulatory
elements of the CFTR gene. Nucleic Acids Res. 38:4325–4336. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Dekker J, Marti-Renom MA and Mirny LA:
Exploring the three-dimensional organization of genomes:
Interpreting chromatin interaction data. Nat Rev Genet. 14:390–403.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
O'Sullivan JM, Tan-Wong SM, Morillon A,
Lee B, Coles J, Mellor J and Proudfoot NJ: Gene loops juxtapose
promoters and terminators in yeast. Nat Genet. 36:1014–1018. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Bentley DL: Rules of engagement:
Co-transcriptional recruitment of pre-mRNA processing factors. Curr
Opin Cell Biol. 17:251–256. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Déjardin J and Kingston RE: Purification
of proteins associated with specific genomic loci. Cell.
136:175–186. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Abbondanza C, De Rosa C, Ombra MN, Aceto
F, Medici N, Altucci L, Moncharmont B, Puca GA, Porcellini A,
Avvedimento EV and Perillo B: Highlighting chromosome loops in
DNA-picked chromatin (DPC). Epigenetics. 6:979–986. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Bolzer A, Kreth G, Solovei I, Koehler D,
Sarakoglu K, Fauth C, Muller S, Eils R, Cremer C, Speicher MR and
Cremer T: Three-dimensional maps of all chromosomes in human male
fibroblast nuclei and prometaphase rosettes. PLoS Biol. 3:e1572005.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Han J, Zhang Z and Wang K: 3C and 3C-based
techniques: The powerful tools for spatial genome organization
deciphering. Mol Cytogen. 9:11–21. 2018.
|
|
69
|
Simonis M, Klous P, Splinter E, Moshkin Y,
Willemsen R, de Wit E, van Steensel B and de Laat W: Nuclear
organization of active and inactive chromatin domains uncovered by
chromosome conformation capture-on-chip (4C). Nat Genet.
38:1348–1354. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
70
|
Wurtele H and Chartrand P: Genome-wide
scanning of HoxB1-associated loci in mouse ES cells using an
open-ended chromosome conformation capture methodology. Chromosome
Res. 14:477–495. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Zhao Z, Tavoosidana G, Sjölinder M, Göndör
A, Mariano P, Wang S, Kanduri C, Lezcano M, Sandhu KS, Singh U, et
al: Circular chromosome conformation capture (4C) uncovers
extensive networks of epigenetically regulated intra- and
interchromosomal interactions. Nat Genet. 38:1341–1347. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
van de Werken HJ, Landan G, Holwerda SJ,
Hoichman M, Klous P, Chachik R, Splinter E, Valdes-Quezada C, Oz Y,
Bouwman BAM, et al: Robust 4C-seq data analysis to screen for
regulatory DNA interactions. Nat Methods. 9:969–972. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Symmons O, Pan L, Remeseiro S, Aktas T,
Klein F, Huber W and Spitz F: The Shh topological domain
facilitates the action of remote enhancers reducing the effects of
genomic distances. Dev Cell. 5:529–543. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Tomasetto C, Rockel N, Mattei MG, Fujita R
and Rio MC: The gene encoding the human spasmolytic protein
(SML1/hSP) is in 21q physically linked to the homologous breast
cancer marker gene BCEI/pS2. Genomics. 13:1328–1330. 1992.
View Article : Google Scholar : PubMed/NCBI
|