Introduction
Hepatocellular carcinoma (HCC) is the third leading
cause of cancer-associated mortalities, with nearly 600,000
mortalities occurring worldwide each year (1). Although resection is considered a
potentially curative treatment for HCC patients, the 1-year
post-operative survival rate is only 30–40% (2). Thus, it is necessary to improve our
understanding of the disease-causing mechanisms and to identify
specific biomarkers for HCC progression to aid in the prediction
and improvement in clinical outcomes.
Non-coding RNAs (ncRNAs) are subdivided into two
groups: Small ncRNAs (<200 nt) and long ncRNAs (lncRNAs)
(3,4).
Small ncRNAs, including the well-documented microRNAs (miRNAs or
miRs), have received great attention, since they are important in
cancer (5). It has been proposed that
lncRNAs are involved in the epigenetic regulation of coding genes,
and thus, exert a powerful effect on a number of physiological and
pathological processes, including the pathogenesis of numerous
human cancers (6). It has been
reported that lncRNA HOX transcript antisense intergenic RNA
(HOTAIR) expression is significantly higher in HCC tissues than in
adjacent non-cancerous tissues (7).
Patients with HCC tumors that overexpress HOTAIR have an increased
risk of HCC recurrence following hepatectomy, and there is also a
correlation between HOTAIR overexpression and increased risk of
lymph node metastasis (8). The
overexpression of HOTAIR is an independent prognostic factor for
HCC recurrence in liver transplant patients (9). Furthermore, patients with high tumor
expression of HOTAIR have a significantly shorter recurrence-free
survival than patients with low expression of HOTAIR (10).
The mechanism by which HOTAIR exerts its oncogenic
activity remains largely unknown. A regulatory mechanism has been
proposed in which RNAs cross-talk via competing shared miRs
(11). In addition, lncRNAs directly
interact with RNA-binding proteins and localize to the gene
promoter region to regulate gene expression (12). The proposed competitive endogenous
RNAs (ceRNAs) mediate the bioavailability of miRs on their targets,
thus imposing another level of post-transcriptional regulation
(13). An example of this type of
regulation is the lncRNA colon cancer associated transcript 1,
(CCAT1), which binds miR-218-5p and forms a regulatory interaction
(14). Using jaspar bioinformatics
software (http://jaspar.genereg.net), forkhead
box C1 (FOXC1) shared binding sites with the upstream promoter
region of HOTAIR were predicted. Furthermore, miR-1 was identified
as the miR that shares binding sites with HOTAIR using starBase 2.0
(http://starbase.sysu.edu.cn). Thus, the
present authors hypothesized that transcription factors (TFs)
activate the transcription of HOTAIR. Furthermore, HOTAIR
overexpression inhibits the expression of miR-1. This hypothesis is
discussed to better elucidate the pathogenesis of HCC.
Materials and methods
Tissue specimens
Fresh-frozen and paraffin-embedded HCC tissues and
corresponding adjacent non-tumorous gastric samples were obtained
from Chinese patients at Shenzhen People's Hospital (Shenzhen,
China) between January 2010 and December 2014. All cases were
reviewed by a pathologist and histologically confirmed as HCC.
Informed consent was obtained from all patients and the study was
approved by the Institutional Ethics Committee of Shenzhen People's
Hospital.
Construction of recombinant HOTAIR
lentiviral expression vectors
The sequence of HOTAIR was obtained (https://www.ncbi.nlm.nih.gov/nuccore/383286742?report=fasta)
and synthesized. Upon double digestion with BamHI and EcoRI, HOTAIR
was directionally-connected to the pGLV3/H1/GFP/Puro vector and
then transformed into DH5α competent cells (Shanghai GenePharma
Co., Ltd., Shanghai, China). Recombinant, packaging and envelope
plasmids (Shanghai GenePharma Co., Ltd.) were co-transfected into
293T cells for 72 h. Viruses were then collected and the titers
were determined using the dilution hole measurement method
(15). The constructed recombinant
HOTAIR lentiviral expression vector was designated LV3-HOTAIR
(16).
Design and construction of a
eukaryotic expression vector for Homo sapiens (hsa)-miR-1 and
FOXC1
The mature hsa-miR-1 sequence
(5′-UGGAAUGUAAAGAAGUAUGUAU-3′) is available from the miR registry
(MIMAT000416; http://www.mirbase.org/cgi-bin/mirna_entry.pl?acc=MIMAT0000416).
To prevent the formation of a termination signal, TTGGCCACTGACT was
selected as the region in a miR expression vector template
(pcDNA3.1; Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA,
USA). The sequence TGCT was added to the 5′ sense strand template
of the miR expression vector, while GTCC was added to the 5′
antisense strand template (17). In
addition, a non-specific sequence was designed and synthesized by
Shanghai GenePharma Co., Ltd. The eukaryotic expression vector
plasmid targeting hsa-miR-1 was designated pmiR-1. The sequence of
FOXC1 was synthesized and sub-cloned into pcDNA3.1 (Invitrogen;
Thermo Fisher Scientific, Inc.) to generate pFOXC1. Empty pcDNA3.1
vector was used as a negative control (NC).
Cell culture
HCC HepG2 and LO2 cells were purchased from the
Shanghai Institute of Cell Biology (Shanghai, China). The cell
lines were cultured in Dulbecco's modified Eagle's medium (Gibco;
Thermo Fisher Scientific, Inc.) containing 10% fetal bovine serum
(HyClone; GE Healthcare Life Sciences, Logan, UT, USA), 100 U/ml
penicillin and 100 µg/ml streptomycin (Invitrogen; Thermo Fisher
Scientific, Inc.). Cells were maintained in a humidified incubator
at 37°C in the presence of 5% CO2. All cell lines were passaged for
<6 months.
Cell transfection
All plasmid vectors (pmiR-1, pFOXC1 and empty
vector) used for cell transfection were extracted using Hieff
Trans™ Liposomal Transfection Reagent (Yeasen Biotechnology Co.,
Ltd., Shanghai, China). HepG2 cells cultured on 6-well plates were
transfected with pmiR-1, pFOXC1 or empty vector using Lipofectamine
2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. Cells were harvested after 48 h for
quantitative polymerase chain reaction (qPCR). Methods for
transfecting cells with LV3-HOTAIR were adopted, as previously
described (17). Small interfering
RNAs (siRNAs) were designed and synthesized by Shanghai GenePharma
Co., Ltd. The siRNA sequence used in the present study was as
follows: Sense, 5′-GGGAGATGTTCGAGTCACAGA-3′ and antisense,
5′-GCGTAAAGCTCGGGTAAGTAG-3′. The siRNAs sequences targeting human
HOTAIR were as follows: Sense, 5′-GGAGAACACUUAAAUAAGUTT-3′ and
antisense, 5′-ACUUAUUUAAGUGUUCUCCTA-3′ (18).
Trypan blue exclusion assay
The density of the HepG2 cell line suspension was
determined by counting on a hemacytometer. A 0.4% solution of
trypan blue in phosphate-buffered saline (PBS) was prepared (pH
7.2–7.3). Trypan blue stock solution (0.1 ml) was added to 1 ml of
cells. A hemacytometer was loaded and examined immediately under a
microscope at low magnification. The number of blue-stained cells
and the number of total cells were counted. Cell viability was
considered to be ≥95% for healthy log-phase cultures. The
percentage of viable cells was calculated as follows: Viable cells
(%) = [1.00 − (number of blue-stained cells/number of total cells)]
× 100.
In vivo treatment
A total of 15 BALB/c (nu/nu) male mice (200±2.6 g;
age, 3 months) from the Animal Center of Guangzhou Province
(Guangdong, China) received subcutaneous injections of 2×106 HepG2
cells into the axillae bilaterally. The mice were housed in a
pathogen-free environment at a temperature of 20–26°C and were
exposed to 12 h light/dark cycles with free access to food and
water. When xenograft tumors became palpable (~0.1 mm3), mice were
randomly divided (n=5 mice/group) into a control group receiving a
PBS injection (100 µl), a transfection group receiving LV3-HOTAIR
(200 nM) and a NC group receiving LV3 + scramble sequence (200 nM).
There was no difference in the baseline tumor size between the
groups. Tumor volume was calculated every 3 days according to the
following formula: V=ab2π/6, where ‘a’ is the maximum tumor
diameter and ‘b’ is the minimum tumor diameter. After treatment for
20 days, the mice were euthanized, and the tumors were extirpated
and weighed.
Chromatin immunoprecipitation (ChIP)
assay
HepG2 cells were treated with formaldehyde and
incubated for 10 min to generate DNA-protein cross-links. Cell
lysates were then sonicated to generate chromatin fragments of
200–300 bp and immunoprecipitated with anti-FOXC1 (cat. no. 625905;
1:1,000; R&D Systems China Co., Ltd., Shanghai, China) for 1 h
at room temperature or Alexa Fluor® 488
conjugated-immunoglobulin G (IgG) antibody (cat. no. 4408; 1:1,000;
Cell Signaling Technology, Inc., Danvers, MA, USA) for 30 min at
room temperature as a control. Precipitated chromatin DNA was
recovered and analyzed by qPCR.
Reverse transcription (RT)-qPCR
Total RNA samples were extracted using TRIzol
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. RT-qPCR analysis was performed using the
Ultra SYBR Mixture with ROX (CWBio, Co., Ltd., Beijing, China) and
an Applied Biosystems 7500 Real-Time PCR System (Thermo Fisher
Scientific, Inc.). HOTAIR cDNA amplification was performed under
the following conditions: Initial denaturation at 95°C for 5 min
followed by 30 cycles of denaturation at 95°C for 30 sec and primer
annealing at 55°C for 30 sec, with a final extension step at 72°C
for 60 sec. FOXC1 cDNA amplification was performed under the
following conditions: Initial denaturation at 95°C for 5 min
followed by 30 cycles of denaturation at 95°C for 30 sec and primer
annealing at 55°C for 30 sec, with a final extension step at 72°C
for 60 sec. miR-1 cDNA was amplified under the following
conditions: Initial denaturation at 95°C for 10 min followed by 40
cycles of denaturation at 95°C for 10 sec and primer annealing at
57°C for 20 sec, with a final extension step at 72°C for 10 sec.
The level of 18S expression was used as an internal control for
messenger RNAs, while the U6 level was used as an internal control
for miRs. The primers used in RT-qPCR are indicated in Table I. The expression levels were
calculated using the 2-∆∆Cq method (19).
 | Table I.Primers for quantitative polymerase
chain reaction. |
Table I.
Primers for quantitative polymerase
chain reaction.
| Gene | Primers
(5′-3′) |
|---|
| U6 | F:
CTCGCTTCGGCAGCACA |
| U6 | R:
AACGCTTCACGAATTTGCGT |
| 18S | F:
CCTGGATACCGCAGCTAGGA |
| 18S | R:
AACGCTTCACGAATTTGCGT |
| HOTAIR | F:
CAGTGGGGAACTCTGACTCG |
| HOTAIR | R:
GTGCCTGGTGCTCTCTTACC |
| FOXC1 | F:
CTCAACGAGTGCTTCGTCAA |
| FOXC1 | R:
ACATGTTGTAGGAGTCCGGG |
| miR-1 | F:
CACTCCAGCTGGGTGGAATGTAAAGAAGTAT |
Luciferase reporter assay
The full sequence HOTAIR gene was obtained by qPCR
amplification using the Ultra SYBR Mixture with ROX and an Applied
Biosystems 7500 Real-Time PCR System. HOTAIR cDNA was amplified
under the following conditions: Initial denaturation at 95°C for 5
min followed by 30 cycles of denaturation at 95°C for 30 sec and
primer annealing at 55°C for 30 sec, with a final extension step at
72°C for 60 sec. The gene was cloned separately into the multiple
cloning site of the psi-CHECK™-2 luciferase miR expression reporter
vector. HepG2 cells were transfected with miR mimic, miR inhibitor,
control miR, negative control (NC), negative control inhibitor (all
purchased from Guangzhou Leader Bio-Technology Co., Ltd.,
Guangzhou, China) or empty plasmid using Lipofectamine 2000,
according to the manufacturer's protocol. Nucleotide-substitution
mutation analysis was conducted using direct oligomer synthesis of
full sequences. All constructs were verified by sequencing.
Luciferase activity was measured using the Dual Luciferase Reporter
Assay System Kit (Promega Corporation, Madison, WI, USA) on an
Infinite M200 luminescence reader (Tecan Group Ltd., Männedorf,
Switzerland), according to the manufacturer's protocol.
Data analysis
All results are the average of ≥3 independent
experiments from separately treated and transfected cultures. Data
are expressed as the mean ± standard deviation. Statistical
comparisons were performed by one-way analysis of variance.
P<0.05 was considered to indicate a statistically significant
difference.
Results
High expression of HOTAIR in HCC
tissues and cells
HCC HepG2 and LO2 cells were cultured
conventionally. Total RNA was extracted from tissues and cells
using TRIzol. The relative expression levels of FOXC1, HOTAIR and
miR-1 were detected by RT-qPCR. The relative level of FOXC1 and
HOTAIR expression in HCC tissues and HepG2 cells was significantly
higher than that in normal liver LO2 cells and adjacent carcinoma
tissues (Fig. 1A and B). The relative
expression of miR-1 exhibited the opposite pattern (Fig. 1C). These results indicate that HOTAIR
has a tumor-promoting role in HCC, while miR-1 has a
tumor-suppressor role, and there may be negative regulation at the
level of expression between HOTAIR and miR-1. The high expression
of FOXC1 may activate the transcription of HOTAIR. These results
provided a foundation to further explore the mechanism of action of
the HOTAIR transcript.
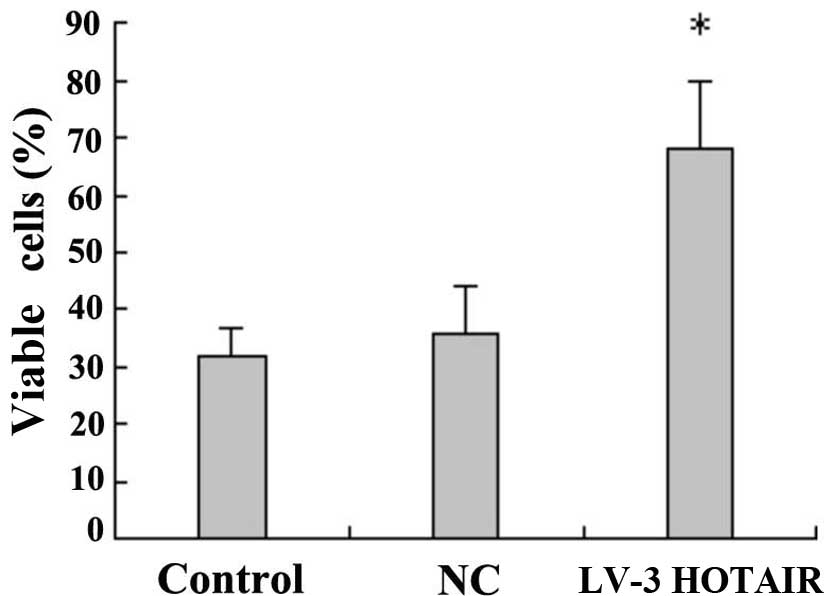
LV3-HOTAIR promotes HepG2 cell
proliferation
Following Lipofectamine 2000 transfection of HepG2
cells with the lentiviral expression vector LV3-HOTAIR, cells were
stained 72 h later with 0.04% trypan blue, and the cell survival
rate was calculated. In the experimental group (LV3-HOTAIR), the
effects on cell proliferation activity decreased significantly
compared with those in the blank control and NC groups (P=0.01),
indicating that LV3-HOTAIR can promote the proliferation of HepG2
cells (Fig. 2).
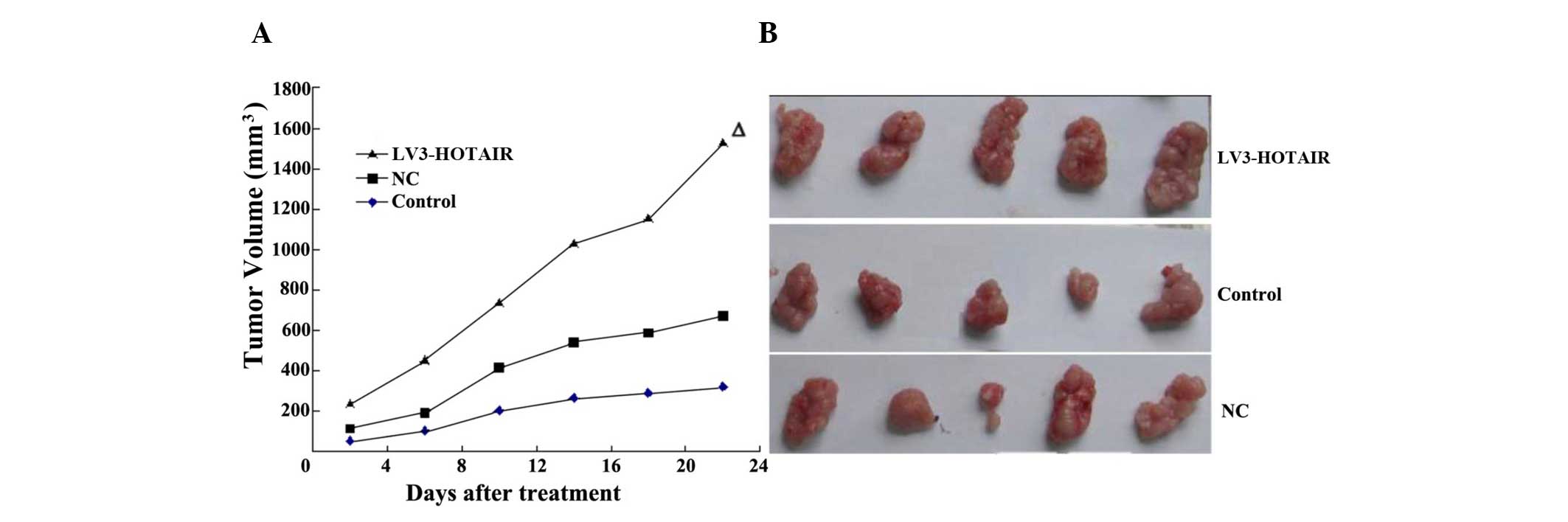
LV3-HOTAIR promotes the progression of
tumor xenografts
HepG2 tumor xenografts were established in athymic
nude mice to evaluate the effects of LV3-HOTAIR on HCC growth in
vivo. Compared with the untreated animals, application of
LV3-HOTAIR significantly diminished the tumor volume, whereas no
effect was observed in the NC group (Fig.
3). No body weight loss or diarrhea was observed, and all
animals (treated and non-treated) survived. The results
demonstrated that the overexpression of HOTAIR can effectively
inhibit HCC cancer growth in vivo.
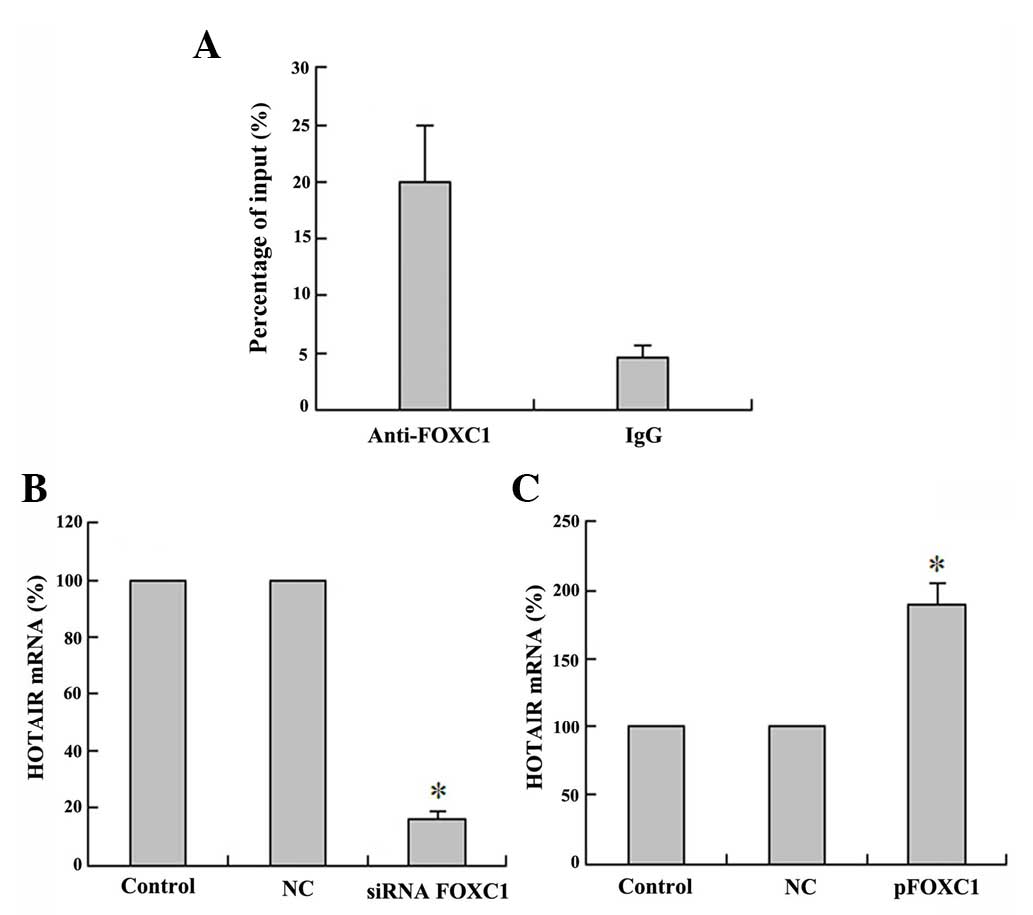
FOXC1 binds to the promoter region of
HOTAIR and activates lncRNA HOTAIR expression
Recently, numerous important TFs have been
demonstrated to be involved in regulating lncRNA transcription
(20). To determine which TFs
activate DD3 expression, the potential TF binding sites in the
promoter region of HOTAIR were analyzed using the jaspar database
(http://jaspar.genereg.net). The results
revealed that there is one E-box element recognized by FOXC1. To
further demonstrate that FOXC1 can directly bind to the HOTAIR
promoter regions and activate the expression of HOTAIR, FOXC1
immunoprecipitates were observed to be highly enriched in DNA
fragments compared with the NC IgG immunoprecipitates in a ChIP
assay. The ChIP assays indicated that FOXC1 directly bound to
HOTAIR promoter regions (Fig. 4A).
These results suggest that HOTAIR interacts with the FOXC1
responsive element in the HOTAIR promoter to induce transcription.
Furthermore, the qPCR results indicated that HOTAIR expression was
increased upon pFOXC1 transfection (Fig.
4B), while HOTAIR expression was decreased following
siRNA-FOXC1 transfection (Fig. 4C).
These results indicated that there is a positive regulation between
the TF FOXC1 and the lncRNA HOTAIR. Taken together, these data
suggest that the TF FOXC1 activates the expression of the lncRNA
HOTAIR in HCC HepG2 cells.
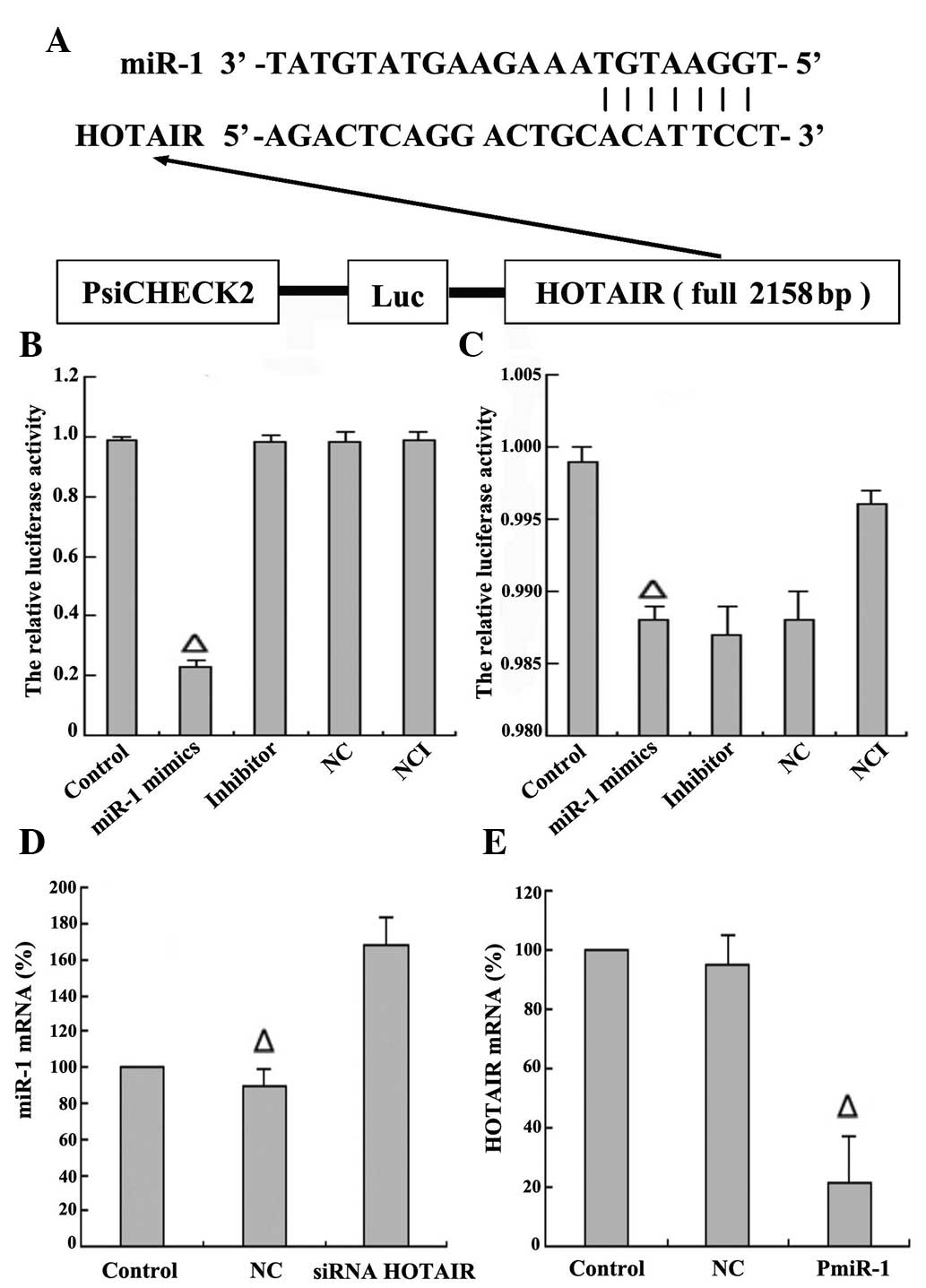
HOTAIR negatively regulates the
expression of miRNA-1
The function of lncRNAs in human diseases may
reflect their ability to regulate gene expression. Increasing
evidence has suggested that ncRNAs may participate in the ceRNA
regulatory network (21). For
example, there is a negative correlation between CCAT1 and let-7
(22). To confirm the direct binding
between HOTAIR and miR-1, luciferase reporter constructs were
generated (Fig. 5A). As predicted,
there was one putative binding site in HOTAIR (1,615–1,638 bp). It
was observed that co-transfection of HepG2 cells with miR-1 mimics
and wild type psi-CHECK™-2-HOTAIR significantly inhibited
luciferase activity (P=0.01; Fig.
5B); however, co-transfection of HepG2 cells with miR-1 mimics
and mutant psi-CHECK™-2-HOTAIR had little effect on the activity of
luciferase (P=0.09; Fig. 5C). These
data confirmed that miR-1 regulates the expression of HOTAIR by
directly binding to target sites within the HOTAIR sequence. To
further explore the regulatory association between HOTAIR and
miR-1, the siRNA sequencing for HOTAIR was designed and transfected
into HepG2 cells for 48 h to knock down the expression of HOTAIR.
It was observed that the levels of miR-1 expression were
significantly upregulated in contrast to those in the blank control
(non-transfected siRNA HOTAIR) and NC (transfected with random
sequences) groups (Fig. 5D). The
eukaryotic expression vector plasmid of miR-1 was further
constructed and transfected into HepG2 cells for 48 h to induce
overexpression of miR-1. Upon overexpression of miR-1, the levels
of HOTAIR expression were significantly downregulated in contrast
to those in the blank control (non-transfected miR-1) and NC
(transfected with plasmid and random sequences) groups (Fig. 5E). Thus, there may be a regulatory
effect between HOTAIR and miR-1. To summarize, HOTAIR negatively
regulated the expression of miR-1 in HepG2 cells.
Discussion
Recent evidence has suggested that ncRNAs serve an
important role in cancer pathogenesis and can provide a novel
insight into the biology of cancer (23). Over the past decade, research
involving miRs has dominated the field of ncRNA regulation
(24); however, the role of lncRNAs
in the tumorigenesis of HCC remains largely unknown. Understanding
the precise molecular mechanism by which lncRNAs function would
facilitate the development of lncRNA-directed diagnostics and
therapeutics against cancer. The present study provided evidence
that FOXC1, a TF, activates the expression of HOTAIR, and that
HOTAIR exhibits oncogenic activities in part through the modulation
of miR-1. Therefore, HOTAIR promotes the tumorigenesis of HCC.
HOTAIR lncRNA was introduced by Kornienko et
al (24) as a spliced and
polyadenylated RNA with 2,158 nt and six exons (25). This RNA arises from the transcription
of the antisense strand of the HOXC gene, which is specifically
situated between HOXC11 and HOXC12 on chromosome 12q13.13 (25). HOTAIR is an oncogenic factor and can
be used as a prognostic biomarker in different cancer types, since
HOTAIR plays a key role in the initiation and progression of
different types of cancer, including cervical cancer and
nasopharyngeal carcinoma (26).
HOTAIR, a lncRNA initially identified in breast cancer, was
demonstrated to be upregulated in a variety of carcinomas (27,28). A
large number of studies have focused on the biological role and
association of HOTAIR with clinical prognosis (9,29,30), yet the precise factors regulating its
expression remain largely unknown. HOTAIR is transcriptionally
regulated by estradiol in breast cancer, which is tumor-specific
(31). In the current study, a
putative binding site of FOXC1 in the promoter region of HOTAIR was
predicted. In addition, miR-1 was observed to have a binding site
on HOTAIR. RT-qPCR results revealed that the relative level of
FOXC1 and HOTAIR expression in HCC tissues and HepG2 cells was
significantly higher than that in normal liver LO2 cells and
adjacent carcinoma tissues. The expression of miR-1 exhibited the
opposite pattern (Fig. 1). FOXC1 is a
well-known TF. Knockdown of FOXC1 expression leads to cytoskeleton
modification accompanied by a decreased ability of HCC cell
proliferation, migration and invasion (32). miR-1 has been reported to be a
tumor-suppressor gene that represses cancer cell proliferation and
metastasis and promotes apoptosis by ectopic expression (33). The present results support the
findings reported in the literature (33). Using technology in which LV3-HOTAIR
can promote the expression of the HepG2 gene and tumor growth in
animals (Figs. 2 and 3), HOTAIR was observed to promote the
migration and invasion of HCC cells by inhibiting RNA binding motif
protein 38 (RBM38), which indicates critical roles of HOTAIR and
RBM38 in HCC progression (27). In
contrast, knockdown of HOTAIR by siRNAs resulted in the reduction
of motility and invasion of the human melanoma cell line A375
(34).
lncRNAs regulate gene expression through a variety
of mechanisms, including transcription, post-transcriptional
processing, chromatin modification, genomic imprinting and
regulation of protein function (35,36).
Recently, it has been reported that lncRNA transcription can be
regulated by key TFs and epigenetic modification (23). For example, p53 can promote lncRNA-p21
transcription, and E2F1 regulates lncRNA E2F1-regulated inhibitor
of cell death expression, while the core catalytic subunit of the
polycomb repressive complex 2, enhancer of zeste homolog 2,
represses lncRNA sprouty RTK signaling antagonist 4-intronic
transcript 1 transcription via epigenetic maintenance of
tri-methylation of lysine 27 on histone H3 (37). c-Myc directly binds to the CCAT1
promoter region and activates lncRNA CCAT1 expression in colon
cancer cells (38). In the present
study, ChIP assays determined that FOXC1 directly binds to the
HOTAIR promoter region. Furthermore, overexpression of FOXC1
increased HOTAIR expression in HCC cells, and knockdown of FOXC1
expression decreased HOTAIR expression in HepG2 cells. FOXC1 also
activated lncRNA HOTAIR expression in HCC cells. Taken together,
these findings provide clues for exploring the mechanism underlying
HOTAIR transcription.
Previous studies have established that lncRNAs can
also regulate other non-coding RNAs, in particular miRs, and miRNAs
may have an effect on the regulation of lncRNAs (39,40). In
support of this notion, it was demonstrated in the current study
that HOTAIR-mediated oncogenic activity occurs, at least in part,
through the suppression of miR-1. Knockdown of HOTAIR induced the
upregulation of miR-1. By contrast, overexpression of miR-1 could
downregulate the HOTAIR level, while miR-1 overexpression repressed
the HOTAIR level. Thus, HOTAIR and miR-1 may form a reciprocal
repression feedback loop. In addition, the mechanism of such a
feedback loop was explored in the present study, and it was
observed that miR-1 regulates the expression of HOTAIR by directly
binding to target sites within the HOTAIR sequence. lncRNA H19 has
been demonstrated to inhibit muscle differentiation by antagonizing
let-7 (41). It has been reported
that long intergenic non-coding RNA muscle differentiation 1
‘sponges’ miR-133, and that miR-133 regulates muscle
differentiation (42). A recently
identified lncRNA, cardiac hypertrophy related factor, has been
reported to regulate cardiac hypertrophy by targeting miR-489
(43).
It has been recently reported that HOTAIR is a
c-Myc-activated driver of malignancy, which acts in part through
repressing miR-130a (44). The
current study demonstrated that LV3-HOTAIR effectively promotes
HepG2 cell formation of clones, as well as growth of animal tumor
xenografts. The underlying molecular mechanism may involve the TF
FOXC1, which activates the transcription of the lncRNA HOTAIR. The
current study suggested another layer of regulation involving
ncRNAs (molecular and biological). A better understanding of the
ncRNA interaction regulatory network will clearly advance the
research in the tumorigenesis of HCC.
Acknowledgements
The present study was supported by the Natural
Science Foundation of Guangdong Province (Guangzhou, China; grant
no. 04006966).
Glossary
Abbreviations
Abbreviations:
|
lncRNA
|
long non-coding RNA
|
|
miR
|
microRNA
|
|
siRNA
|
small interfering RNA
|
|
TF
|
transcription factor
|
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction
|
|
HCC
|
hepatocellular carcinoma
|
|
HOTAIR
|
HOX transcript antisense intergenic
RNA
|
|
FOXC1
|
forkhead bo
|
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wang P, Ouyang L, Zheng L and Wang Z:
Identifying hepatocellular carcinoma-related genes and pathways by
system biology analysis. Ir J Med Sci. 184:357–364. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Geisler S and Coller J: RNA in unexpected
places: Long non-coding RNA functions in diverse cellular contexts.
Nat Rev Mol Cell Biol. 14:699–712. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ellis BC, Graham LD and Molloy PL: CRNDE,
a long non-coding RNA responsive to insulin/IGF signaling,
regulates genes involved in central metabolism. Biochim Biophys
Acta. 1843:372–386. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ma L, Bajic VB and Zhang Z: On the
classification of long non-coding RNAs. RNA Biol. 10:925–934. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang Z, Zhu Z, Watabe K, Zhang X, Bai C,
Xu M, Wu F and Mo YY: Negative regulation of lncRNA GAS5 by miR-21.
Cell Death Differ. 20:1558–1568. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Geng YJ, Xie SL, Li Q, Ma J and Wang GY:
Large intervening non-coding RNA HOTAIR is associated with
hepatocellular carcinoma progression. J Int Med Res. 39:2119–2128.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gao JZ, Li J, Du JL and Li XL: Long
non-coding RNA HOTAIR is a marker for hepatocellular carcinoma
progression and tumor recurrence. Oncol Lett. 11:1791–1798.
2016.PubMed/NCBI
|
|
9
|
Yang Z, Zhou L, Wu LM, Lai MC, Xie HY,
Zhang F and Zheng SS: Overexpression of long non-coding RNA HOTAIR
predicts tumor recur-rence in hepatocellular carcinoma patients
following liver transplantation. Ann Surg Oncol. 18:1243–1250.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xu ZY, Yu QM, Du YA, Yang LT, Dong RZ,
Huang L, Yu PF and Cheng XD: Knockdown of long non-coding RNA
HOTAIR suppresses tumor invasion and reverses
epithelial-mesenchymal transition in gastric cancer. Int J Biol
Sci. 9:587–597. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kugel JF and Goodrich JA: Non-coding RNAs:
Key regulators of mammalian transcription. Trends Biochem Sci.
37:144–151. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Beermann J, Piccoli MT, Viereck J and Thum
T: Non-coding RNAs in development and disease: Background,
mechanisms, and therapeutic approaches. Physiol Rev. 96:1297–1325.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ma MZ, Chu BF, Zhang Y, Weng MZ, Qin YY,
Gong W and Quan ZW: Long non-coding RNA CCAT1 promotes gallbladder
cancer development via negative modulation of miRNA-218-5p. Cell
death Dis. 6:e15832015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Meng YX, Niu X and Deng ZH: Inhibitory
effect of lentivirus-mediated hTERTp-TK combined with
hTERTp-tumstatin on human hepatocarcinoma HepG2 cells. Zhonghua Gan
Zang Bing Za Zhi. 23:837–843. 2015.(In Chinese). PubMed/NCBI
|
|
16
|
Braconi C, Kogure T, Valeri N, Huang N,
Nuovo G, Costinean S, Negrini M, Miotto E, Croce CM and Patel T:
microRNA-29 can regulate expression of the long non-coding RNA gene
MEG3 in hepatocellular cancer. Oncogene. 30:4750–4756. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
He JH, Li YM, Li YG, Xie XY, Wang L, Chun
SY and Cheng WJ: hsa-miR-203 enhances the sensitivity of leukemia
cells to arsenic trioxide. Exp Ther Med. 5:1315–1321.
2013.PubMed/NCBI
|
|
18
|
Li D, Feng J, Wu T, Wang Y, Sun Y, Ren J
and Liu M: Long intergenic noncoding RNA HOTAIR is overexpressed
and regulates PTEN methylation in laryngeal squamous cell
carcinoma. Am J Pathol. 182:64–70. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2-ΔΔCT method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Karreth FA and Pandolfi PP: ceRNA
cross-talk in cancer: When ce-bling rivalries goawry. Cancer
Discov. 3:1113–1121. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lipovich L, Johnson R and Lin CY: MacroRNA
underdogs in a microRNA world: Evolutionary, regulatory and
biomedical significance of mammalian long non-protein-coding RNA.
Biochim Biophys Acta. 1799:597–615. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Deng L, Yang SB, Xu FF and Zhang JH: Long
noncoding RNA CCAT1 promotes hepatocellular carcinoma progression
by functioning as let-7 sponge. J Exp Clin Cancer Res. 34:182015.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lang JH, Li JH, Jiang S, Zhou H and Qu LH:
ChIPBase: A database for decoding the transcriptional regulation of
long non-coding RNA and microRNA genes fromChIP-Seq data. Nucleic
Acids Res. 41:D177–D187. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kornienko AE, Guenzl PM, Barlow DP and
Pauler FM: Gene regulation by the act of long non-coding RNA
transcription. BMC biology. 11:592013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Rinn JL, Kertesz M, Wang JK, Squazzo SL,
Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E and
Chang HY: Functional demarcation of active and silent chromatin
domains in human HOX loci by noncoding RNAs. Cell. 129:1311–1323.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Huang L, Liao LM, Liu AW, Wu JB, Cheng XL,
Lin JX and Zheng M: Overexpression of long noncoding RNA HOTAIR
predicts a poor prognosis in patients with cervical cancer. Arch
Gynecol Obstet. 290:717–723. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ding C, Cheng S, Yang Z, Lv Z, Xiao H, Du
C, Peng C, Xie H, Zhou L, Wu J and Zheng S: Long non-coding RNA
HOTAIR promotes cell migration and invasion via down-regulation of
RNA binding motif protein 38 in hepatocellular carcinoma cells. Int
J Mol Sci. 15:4060–4076. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kim K, Jutooru I, Chadalapaka G, Johnson
G, Frank J, Burghardt R, Kim S and Safe S: HOTAIR is a negative
prognostic factor and exhibits pro-oncogenic activity in pancreatic
cancer. Oncogene. 32:1616–1625. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Fu WM, Zhu X, Wang WM, Lu YF, Hu BG, Wang
H, Liang WC, Wang SS, Ko CH, Waye MM, et al: Hotair mediates
hepatocarcinogenesis through suppressing miRNA-218 expression and
activating P14 and P16 signaling. J Hepatol. 63:886–895. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li C, Chen J, Zhang K, Feng B, Wang R and
Chen L: Progress and prospects of long noncoding RNAs (lncRNAs) in
hepatocellular carcinoma. Cell Physiol Biochem. 36:423–434. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bhan A, Hussain I, Ansari KI, Kasiri S,
Bashyal A and Mandal SS: Antisense transcript long noncoding RNA
(lncRNA) HOTAIR is transcriptionally induced by estradiol. J Mol
Biol. 425:3707–3722. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xu ZY, Ding SM, Zhou L, Xie HY, Chen KJ,
Zhang W, Xing CY, Guo HJ and Zheng SS: FOXC1 contributes to
microvascular invasion in primary hepatocellular carcinoma via
regulating epithelial-mesenchymal transition. Int J Biol Sci.
8:1130–1141. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Han C, Yu Z, Duan Z and Kan Q: Role of
MicroRNA-1 in human cancer and its therapeutic potentials. Biomed
Res Int. 2014:4283712014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tang L, Zhang W, Su B and Yu B: Long
noncoding RNA HOTAIR is associated with motility, invasion and
metastatic potential of metastatic melanoma. Biomed Res Int.
2013:2510982013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Gibb EA, Brown CJ and Lam WL: The
functional role of long non-coding RNA in human carcinomas. Mol
Cancer. 10:382011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Bernard D, Prasanth KV, Tripathi V,
Colasse S, Nakamura T, Xuan Z, Zhang MQ, Sedel F, Jourdren L,
Coulpier F, et al: A long nuclear-retained non-coding RNA regulates
synaptogenesis by modulating gene expression. EMBO J. 29:3082–3093.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liu Q, Huang J, Zhou N, Zhang Z, Zhang A,
Lu Z, Wu F and Mo YY: LncRNA loc285194 is a p53-regulated tumor
suppressor. Nuclei Acids Res. 41:4976–4987. 2013. View Article : Google Scholar
|
|
38
|
Yang F, Xue X, Bi J, Zheng L, Zhi K, Gu Y
and Fang G: Long noncoding RNA CCAT1, which could be activated by
c-Myc, promotes the progression of gastric carcinoma. J Cancer Res
Clin Oncol. 139:437–445. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Mattick JS and Makunin IV: Non-coding RNA.
Hum Mol Genet. 15:R17–R29. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Gibb EA, Brown CJ and Lam WL: The
functional role of long non-coding RNA in human carcinomas. Mol
Cancer. 10:38–55. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kallen AN, Zhou XB, Xu J, Qiao C, Ma J,
Yan L, Lu L, Liu C, Yi JS, Zhang H, et al: The imprinted H19 lncRNA
antagonizes let-7 microRNAs. Molecular cell. 52:101–112. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Legnini I, Morlando M, Mangiavacchi A,
Fatica A and Bozzoni I: A feedforward regulatory loop between HuR
and the long noncoding RNA linc-MD1 controls early phases of
myogenesis. Mol Cell. 53:506–514. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wang K, Liu F, Zhou LY, Long B, Yuan SM,
Wang Y, Liu CY, Sun T, Zhang XJ and Li PF: The long noncoding RNA
CHRF regulates cardiac hypertrophy by targeting miR-489. Circ Res.
114:1377–1388. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ma MZ, Li CX, Zhang Y, Weng MZ, Zhang MD,
Qin YY, Gong W and Quan ZW: Long non-coding RNA HOTAIR, a c-Myc
activated driver of malignancy, negatively regulates miRNA-130a in
gallbladder cancer. Mol Cancer. 13:1562014. View Article : Google Scholar : PubMed/NCBI
|