Introduction
Cholangiocarcinoma (CCA) are epithelial cell
malignancies arising from various locations within the biliary
tree, showing markers of cholangiocyte differentiation. Most CCA
are adenocarcinomas, which are anatomically classified as either of
intrahepatic (ICCA) or extrahepatic (ECCA) origin (1). In general this heterogeneous group of
CCA are aggressive malignancies with poor survival rates
(5-year-survival of <10%) (2).
Resectability is a critical factor associated with a better
outcome. However, the majority of cases is diagnosed at the time of
progressive disease, without the opportunity of surgical resection
in curative intent (3,4). These data emphasize the need of new
therapeutic options besides surgery, chemo- (CTX) and radiotherapy
(RTX). So far, there are only very limited options of available
targeted therapies, which are based on specific biomarkers of
individual tumors like e.g. FGFR2-inhibitors (5). For this reason new biomarkers are
urgently required, that might contribute to a better understanding
of tumor biology in specific subgroups of the different CCA types.
ICCAs are subdivided into large duct type and small duct type,
according to their occurrence and cell of origin (6–8). Large
duct ICCAs arise in the large intrahepatic bile ducts near the
hepatic hilus and resemble ECCAs, whereas small duct ICCAs
typically develop in the hepatic periphery (7,9). The
ICCA subtypes, which were included into the WHO classification of
2019, go along with different clinical, histomorphological and
molecular features, diverse risk factors and prognosis (10,11).
Recently increasing interest has been paid to
nucleosome remodeling complexes in many different cancers, e.g.
esophageal-, pancreatic-, ovarian-, renal cell and hepatocellular
carcinoma and they appear to be promising new opportunities for
novel prognostic markers and therapeutic targets (12–14). The
mammalian SWI/SNF-complex (SWI/SNF) functions as tumor suppressor
in many human malignancies and plays an ATP-dependent
chromatin-remodeling role, contributing to transcriptional
regulation by altering chromatin structure and controlling the
accessibility of DNA (15,16).
SWI/SNF are heterogeneous complexes of 12–15 protein
subunits with diverse and variable functions required in different
cellular and developmental contexts. Mammalian SWI/SNF complexes
always contain two mutually exclusive ATPases: BRM (SMARCA2) or
BRG1 (SMARCA4). Three additional subunits [INI-1 (SMARCB1), SMARCC1
and SMARCC2] form the core complex (17–19). The
mutation frequency of the complexes is ~20% in cancer and AT-rich
interactive domain-containing protein 1A (ARID1A) is the most
frequently mutated gene subunit (12). Moreover, mutations of SWI/SNF might
provide a potential target for therapy (13).
To date, inactivating mutations of ARID1a and PBRM-1
have been detected in 17–19% of ICCA by exome sequencing and have
been correlated with worse survival (20). ARID1a and PBRM-1 gene mutations have
even been described as predictors for poor prognosis in ICCA,
however histomorphological subtypes were not considered (21). Simbolo et al demonstrated that
specific molecular SWI/SNF alterations, including ARID1a, PBRM-1
and INI-1, were associated with different CCA subtypes and that
potentially actionable pathways for small molecule inhibitors are
evident in 68% of cases (16).
Despite advances in the analysis of SWI/SNF, so far
little is known about its relevance in CCA. In our study we
concentrated on the analysis of the ATPase-dependent core subunits
of SWI/SNF (ARID1a, INI-1, BRG1, PBRM-1 and BRM) which, if mutated,
all separately result in loss-of-function of SWI/SNF (22). These mutations can be detected by
immunohistochemistry (IHC), demonstrating the loss of the nuclear
protein.
Cases of large- and small duct ICCA as well as cases
of ECCA were included in order to analyze the frequency of
mutations of SWI/SNF in correlation to the subtype of CCA and
survival. Simultaneously we assessed FGFR2-translocations,
HER2-amplification status and P53 mutations to exclude other
underlying alterations, which may bias our survival data.
Due to the abundance of actionable mutations and the
absence of effective systemic therapy options against CCA, the aim
of our study was, to reveal the influence of SWI/SNF core subunits
protein-loss on overall survival of CCA patients corresponding to
its subtype and further to identify possible new promising
therapeutic targets.
Materials and methods
Case selection
Between 2000 and 2019, a cohort of 52 patients with
the diagnosis of primary CCA was collected. All these patients
underwent surgical treatment with curative intent within the
surgical department of the University Hospital of Cologne (Cologne,
Germany). All patients gave their written informed consent for the
procedure. The current retrospective study was conducted with the
approval of the Ethics Committee of the University of Cologne,
utilizing clinical follow-up data that were collected
retrospectively according to a standardized follow-up within the
oncological outpatient clinic (application 18–269). The following
exclusion criteria were defined: i) Administration of systematic
therapy prior to surgery to avoid any bias affecting survival
analysis. ii) Survival <14 day after surgery, to exclude
short-term deaths due to surgical complications. iii) Age <42 or
>89 years. Detailed patient cohort information is displayed in
(Table I).
 | Table I.Specifications of the patient
cohort. |
Table I.
Specifications of the patient
cohort.
|
Characteristics | Total | ECCA | ICCA | ICCA-large
duct | ICCA-small
duct |
|---|
| Total patients | 52 | 17 | 35 | 8 | 27 |
| Sex |
|
Male | 31 | 10 | 21 | 4 | 17 |
|
Female | 21 | 6 | 15 | 3 | 12 |
| Portal vein
embolization | 8 | 3 | 5 | 1 | 4 |
| Postoperative
chemotherapy | 22 | 2 | 20 | 5 | 15 |
| Postoperative
radiotherapy | 2 | 1 | 1 | 1 | 0 |
| Alive at time of
investigation | 15 | 3 | 12 | 2 | 10 |
|
pT-statius |
|
pT1 | 12 | 0 | 12 | 2 | 10 |
|
pT2 | 25 | 12 | 13 | 1 | 12 |
|
pT3 | 9 | 2 | 7 | 1 | 6 |
|
pT4 | 6 | 2 | 4 | 3 | 1 |
|
pN1 | 23 | 10 | 13 | 2 | 11 |
| M1 | 3 | 1 | 2 | 0 | 2 |
| L1 | 44 | 14 | 30 | 5 | 25 |
| V1 | 22 | 4 | 18 | 3 | 15 |
|
Pn1 | 34 | 15 | 19 | 3 | 16 |
| R-status |
| R0 | 38 | 10 | 28 | 3 | 25 |
| R1 | 13 | 6 | 7 | 4 | 3 |
| R2 | 1 | 0 | 1 | 0 | 1 |
| Grading |
| G1 | 1 | 1 | 0 | 0 | 0 |
| G2 | 30 | 11 | 19 | 6 | 13 |
| G3 | 20 | 4 | 16 | 1 | 15 |
| G4 | 1 | 0 | 1 | 0 | 1 |
| Operation |
|
Hemihepatectomy right | 8 | 0 | 8 | 1 | 7 |
|
Extended hemihepatectomy
right | 8 | 1 | 7 | 2 | 5 |
|
Hemihepatectomy left | 9 | 1 | 8 | 1 | 7 |
|
Extended hemihepatectomy
left | 4 | 1 | 3 | 0 | 3 |
| Liver
segment resection | 5 | 0 | 5 | 1 | 4 |
|
Atypical liver resection | 2 | 0 | 2 | 1 | 1 |
|
Pancreas operation (Whipple or
Traverso-modification) | 2 | 2 | 0 | 0 | 0 |
|
Trisegmentectomy | 4 | 3 | 1 | 0 | 1 |
|
Extrahepatic bile duct
resection | 5 | 5 | 0 | 0 | 0 |
|
Hemihepatectomy right +
extrahepatic bile duct resection | 4 | 2 | 2 | 1 | 1 |
|
Hemihepatectomy left +
pancreas operation | 1 | 1 | 0 | 0 | 0 |
Classification and pathological
features
CCAs were divided in ECCA (n=17) and ICCA (n=35)
corresponding to their location in the biliary tree. Moreover, ICCA
were subdivided into small duct (n=27) and large duct type (n=8)
corresponding to the WHO classification of 2019 (10). This subdivision was based on standard
histomorphological and cellular criteria (HE and PAS staining), as
well as immunohistochemical staining (see below).
Immunohistochemical study (IHC)
Tissue microarray analysis (TMA) construction was
performed as previously described (23,24). In
brief, four tissue cylinders from each tumor center with a diameter
of 1.2 mm were punched out from the tumor center of selected tumor
tissue blocks using a semi-automated precision instrument and
embedded in empty recipient paraffin blocks. Four-micrometer
sections of the resulting TMA blocks were transferred to an
adhesive-coated slide system (Instrumedics Inc.) for further
staining. IHC was performed on TMA slides using primary antibodies
specific for ARID1A (Abcam, clone EPR13501, 1:1,000 citrate
buffer), BRG1 (Abcam, clone EPNCIR111A, 1:300 citrate buffer), BRM
(Cell Signaling, Inc., clone D9E8B, 1:50 citrate buffer), INI1
(Dako, clone DO-7, 1:1200, citrate buffer), PBRM-1 (Abcam, BAF180,
EPR15860, 1:1000, EDTA buffer) and P53 (Dako, clone DO-7, 1:800,
citrate buffer) with a Bond Max automated system (Leica). All these
markers showed a nuclear staining pattern. For subdivision of ICCA
into small duct- and large duct type the primary antibodies S100P
(Cell Marque, 16/f5, 1:1600, enzyme buffer), N-cadherin
(Novocastra, IAR06, 1:100, EDTA buffer), CD56 (Thermo Fisher
Scientific, Inc., 123C3, 1:500, EDTA buffer) and MUC5ac (Abcam,
MUC5AC/917, 1:500, EDTA buffer) were used in the same manner. S100P
and MUC5ac showed a cytoplasmatic -, N-cadherin and CD56 a
membranous staining pattern. The expression frequency of these
markers corresponding to the CC subtypes is depicted in Fig. 1. Lymphoid tissue served as an
internal control. Two pathologists (either U.D. or A.Q. and B.J.W.)
manually performed IHC analysis. ARID1A-, BRG1-, BRM-, PBRM-1and
INI-1 staining was assessed according to a three-tier scoring
system (score 0, 1 and 2). A score of 0 associated with the loss of
protein and defined as unequivocal clean absent staining in the
nuclei of viable tumor cells for the SWI/SNF-complex subunits was
interpreted as an underlying mutation, deletion or promotor
alteration. Score 1 was determined as nuclear staining of tumor
cells and interpreted as an intact, unmuted ARID1-, BRG1-, BRM-,
PBRM-1 or INI1 gene with regular protein expression. Score 2 was
used in case of heterogeneous expression, defined as loss and
intact nuclear staining within different tumor areas of the same
tumor sample and interpreted as partial underlying mutation,
deletion or promotor alteration. Strong nuclear stainability of the
surrounding non-tumor cells served as an internal control.
Discrepant results were resolved by consensus between the
reviewers. In case of PBRM-1 to proof absent nuclear staining
certain tumor samples were reanalyzed on large tumor slides. S100P,
N-cadherin, CD56 and MUC5ac staining was classed as positive if
≥10% of tumor cells were positive. P53 was considered as altered in
case of nuclear loss or homogeneous nuclear overexpression.
 | Figure 1.Subtyping of CCA. (A) IHC staining
results. Positive IHC staining results for either N-cadherin, CD56,
S100P or MUC5ac are depicted in percent (y-axes) compared to the
CCA subtype (x-axes). The CCA sample was assessed as positive if
staining was present in ≥10%. Small duct ICCA (n=27), large duct
ICCA (n=8), ECCA (n=17). (B) Representative images of IHC staining,
corresponding to the CCA subtype. CCA, cholangiocarcinoma; IHC,
immunohistochemistry; CD56, cluster of differentiation 56; S100P,
S100 calcium-binding protein P; MUC5ac, mucin 5AC; ICCA,
intrahepatic cholangiocarcinoma; ECCA, extrahepatic
cholangiocarcinoma. |
FISH-analysis
Fluorescence in situ hybridization (FISH) for
FGFR2-translocation and HER2-amplification was
performed on formalin-fixed, paraffin-embedded tissue specimens
(TMA). Sections of 1.5 µm tumor material were cut and hybridized
overnight. FGFR2-translocation was assessed by using a
ZytoLight SPEC, FGFR2 Dual Color Break Apart probe
(Z-2169-200), HER2-amplification by using a ZytoLight SPEC
ERBB2/CEN 17 Dual Color probe (Z-2015-200). Review of fluorescence
signals was performed at ×630 magnification with a Leica CTR 5500
fluorescence microscope. FGFR2 was scored as positive, if
separate Spectrum red and/or Spectrum green signals were present in
>20% of nuclei throughout the tumor. HER2 analysis was
performed according to defined guidelines (25).
Data analysis and statistics
For statistical analysis and graphic presentation of
the results, IBM SPSS v26.0 was used. Descriptive analysis included
the frequency of nominal parameters, the median with lower (LQ) and
upper (UQ) quartiles for numeric variables (ordinal or asymmetric
distribution) and the mean for numeric variables with a normal
distribution. Prognosis was calculated including all types of
mortality beginning 14 days after the date of surgery. In this way,
mortality associated to the surgical procedure itself was excluded.
Kaplan-Meier univariate analysis was used to describe survival
distribution, and log-rank tests were performed to evaluate
survival differences. Significant differences between patient
cohorts were defined as P<0.05 for all analyses.
Results
IHC analysis of SWI/SNF complex
subunits
ARID1a-, BRG1-, BRM-, PBRM-1and INI-1 IHC staining
shows a clear nuclear staining pattern, if proficiently expressed
(Fig. 2). In case of protein-loss
the nuclear staining turned negative. For ARID1a and PBRM-1 only
complete protein-loss was detected whereas BRG1-, BRM and INI1 also
showed a heterogeneous staining pattern. Assuming that a
heterogeneous protein expression at least results in a reduced
function of SWI/SNF, we summarized both immunophenotypes as
protein-loss for further analysis. This approach was confirmed by
the finding that heterogeneous protein expression and protein-loss,
separately analyzed, both resulted in reduced overall survival
(data not shown).
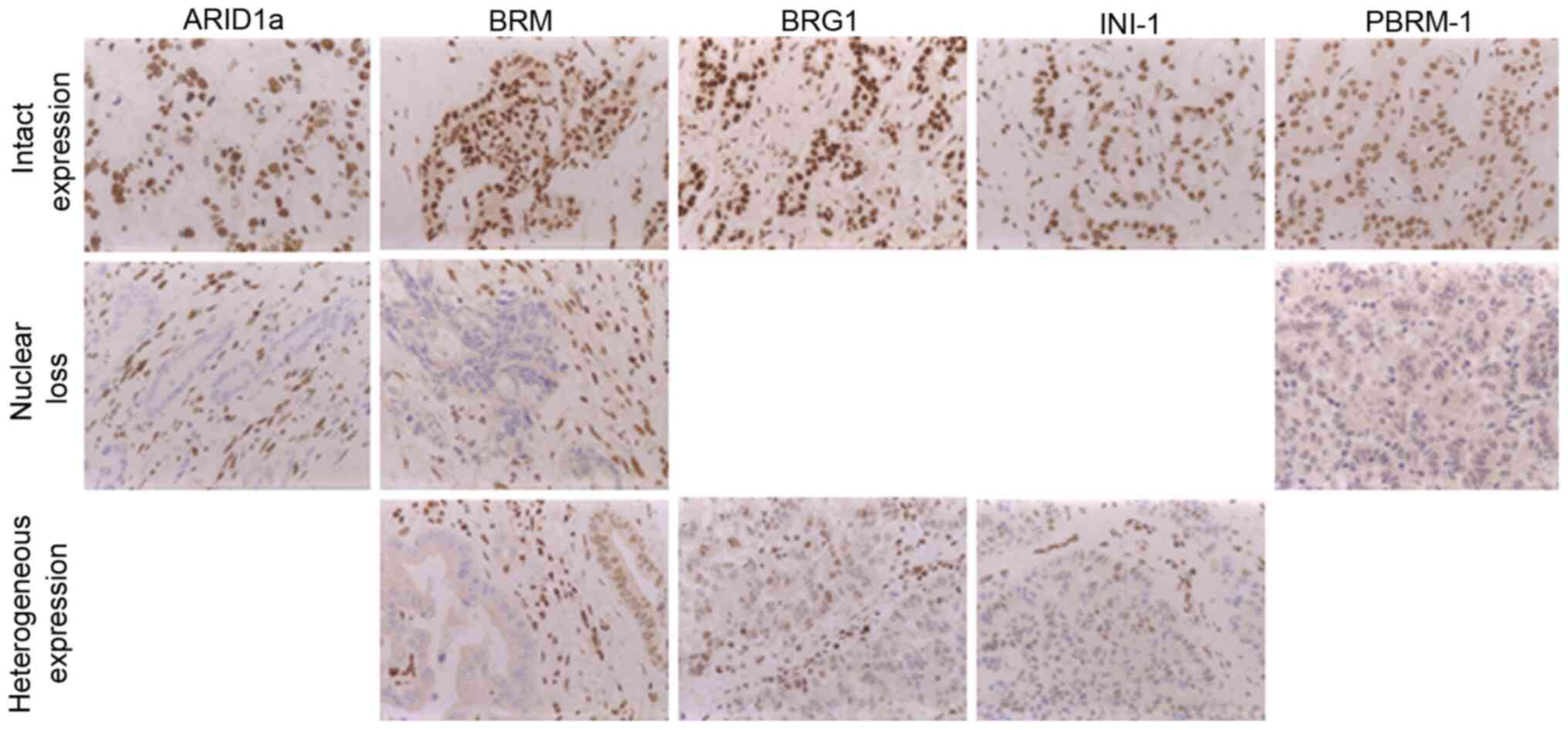 | Figure 2.IHC analysis of SWI/SNF-complex
subunits. All tested SWI/SNF subunits (ARID1A, BRG1-, BRM-, PBRM-1
and INI1) show a nuclear positive staining pattern, in case of IHC
protein-loss nuclear staining was absent. A mosaic-like nuclear
staining pattern was detected for BRG1, BRM and INI1 and was
defined as heterogeneous expression (magnification, ×40). IHC,
immunohistochemistry; SWI/SNF, SWI/SNF-complex; ARID1A, AT-rich
interactive domain 1A; BRG1, Brahma-related gene 1; BRM, Brahma,
PBRM-1, polybromo-1; INI1, integrase interactor1. |
In total, 14 of 52 patients (~35%) showed a
protein-loss of any tested SWI/SNF core subunit, 2 of them with
heterogeneous expression, and 4 of them with protein-loss of more
than one SWI/SNF subunits. The highest frequency showed ARID1a with
17%. The CCA subtypes showed different frequencies of the lost
SWI/SNF subunits (Fig. 3). In small
duct ICCA, all tested subunits showed protein loss. The most
frequent alterations were detected for ARID1a and PBRM-1 with 11%
each. In opposite, in large duct type ICCA only protein-loss of
ARID1a (25%) and BRM (12.5%) were revealed. With 6 of 17 ECCA
patients this CCA subtype showed the highest frequency (35%) of any
lost SWI/SNF subunit. ARID1a, BRM and PBRM-1 alterations were found
in decreasing frequency. Besides these findings, alterations of
HER2, P53 and FGFR2 were observed as follows:
HER2 amplification in 13.46%, P53 alterations in 25% and
FGFR2 translocation in 5.77% (Fig. 3). Whereas HER2- and P53 alterations
were found in each CCA subtype, FGFR2 translocations were
only observed in ICCA. Further, 7 of the 14 patients showed
protein-loss of SWI/SNF subunits parallel to HER2, FGFR2- or
P53 alterations.
 | Figure 3.Frequency of SWI/SNF-complex-,
HER2-, P53- and FGFR-2-alterations. (A) IHC proven
protein-loss (heterogeneous expression included) of SWI/SNF
subunits are depicted in percent (y-axes) compared to the CCA
subtype (x-axes); (B) in the same manner HER2-amplification,
FGFR2 translocation and P53 alterations are illustrated.
Small duct ICCA n=27, large duct ICCA n=8, ECCA n=17. (C)
Fluorescence microscopy photographs of HER2-amplificated
(left) and FGFR2-translocated (right) CCAs compared to
wild-type each (magnification, ×63). HER2, human epidermal
growth factor receptor 2; P53, protein 53; FGFR-2, fibroblast
growth factor receptor 2; SWI/SNF, SWI/SNF-complex; CCA,
cholangiocarcinoma; ICCA, intrahepatic cholangiocarcinoma; n,
number of patients; ECCA, extrahepatic cholangiocarcinoma. |
Overall survival analysis
Kaplan-Meier analysis (log-rank test) was used for
overall survival. We combined IHC proven protein-loss of ARID1A,
BRG1, BRM, PBRM-1 or INI-1 as SWI/SNF altered cohort (n=14) and
compared it to patients with intact expression pattern (n=38)
(Fig. 4A). The median survival of
the SWI/SNF subunit protein-loss cohort was 13±4.677 months versus
34±7.200 months in the wild-type CCA patients, which was
statistically significant (P=0.013). Further, we analyzed the
effect of molecular alterations (HER2-amplification,
FGFR2-translocation or P53 alteration) on the overall
survival and excluded CCA patients with these alterations from the
cohorts (Fig. 4B). Again, the
difference in overall survival (P=0.002) was statistically
significant: The median survival of SWI/SNF subunit protein loss
cohort (n=25) was 10±7.071 months compared to 43±13.835 months for
the cohort with intact expression pattern (n=7).
 | Figure 4.Kaplan-Meier survival analysis of the
SWI/SNF subunit protein-loss cohort versus patients with proficient
expression. (A) Overall survival of the SWI/SNF altered patients
(n=14, blue line) compared to the cohort with intact expression
pattern (n=38, red line). (B) Overall survival of SWI/SNF altered
patients (n=7, blue line) compared to the cohort intact expression
pattern (n=25, red line) after exclusion of HER2-,
FGFR2- or P53 alterations. With or without exclusion of
HER2-, FGFR2- or P53 alterations, the SWI/SNF-subunit
protein loss cohort showed a highly significant worse survival than
the SWI/SNF wild-type subgroup [log-rank test: (A) P=0.013 and for
(B) P=0.002]. In each section the x-axes shows cumulative survival,
postsurgical follow-up in months is depicted on the y-axes.
SWI/SNF, SWI/SNF-complex; n, number of patients; HER2, human
epidermal growth factor receptor 2; FGFR-2, fibroblast growth
factor receptor 2; P53, protein 53. |
As we did not see an effect of HER2, P53 and FGFR2
alterations on overall survival in ratio SWI/SNF altered patients
and patients with intact expression pattern, we analyzed the
overall survival corresponding to the histological CCA subtypes in
the total patient cohort (Fig. 5).
Only for small duct (Fig. 5A) and
large duct ICCA (Fig. 5B) a
significant worse survival for the SWI/SNF subunit protein-loss
cohort was detected (small duct ICCA P=0.031, large duct ICCA
P=0.010), but not for ECCA (Fig.
5C). For small duct ICCA the median overall survival of the
cohort with intact expression pattern was 43.911±7.356 months,
whereas SWI/SNF altered patients only survived 13±7.088 months in
average. Similar results were detected for large duct ICC: overall
survival of the cohort with intact expression pattern was
46.5±6.759 months compared with 16±8.667 months in SWI/SNF altered
patients. General overall survival of small duct ICCA was the
longest (39.709±6.849 months), followed by large duct ICCA
(33.571±7.480 months). The shortest overall survival was detected
for ECCA patients (21.486±6.381 months). Each SWI/SNF subtype
protein-loss in relation to overall survival and CCA subtype was
analyzed separately (data not shown). For this analysis other
underlying SWI/SNF alterations were excluded. ARID1a protein loss
correlated with lower overall survival significantly (P=0.025).
Median overall survival of the CCA patient cohort with intact
expression pattern was 39.4±5.626 months versus 16.125±4.725 months
of ARID1a altered patients. Corresponding to the histological
subtypes, median survival of ARID1a altered patients was also
shorter, whereas only significant for large duct ICCA (P=0.010).
For PBRM-1 survival of the cohort with intact expression pattern
was longer compared to the altered PRBM-1 patients. However, this
finding was non-significant. BRM protein-loss also decreased
overall survival in all CCA subtypes, whereas significant results
were only detected for the large duct ICCA (P=0.046).
 | Figure 5.Kaplan-Meier survival analysis
(log-rank test) for the patient cohort with SWI/SNF-complex-
subunit protein loss versus proficient expression corresponding to
CCA subtypes. (A) Small duct ICCA: Overall survival of the SWI/SNF
protein-loss cohort (n=6, blue line) compared to patients with
intact expression pattern (n=21, red line). (B) Large duct ICCA:
Overall survival of the SWI/SNF protein-loss cohort (n=2, blue
line) compared to patients with intact expression pattern (n=6, red
line). (C) ECCA: Overall survival the SWI/SNF protein-loss cohort
(n=6, blue line) compared to patients' intact expression pattern
(n=11, red line). A significant worse overall survival of the
SWI/SNF protein-loss cohort was detected for small duct ICCA
(P=0.031) and large duct ICCA (P=0.010), but not in ECCA. In each
section the x-axes shows cumulative survival, postsurgical
follow-up in months is depicted on the y-axes. CCA,
cholangiocarcinoma; ICCA, intrahepatic cholangiocarcinoma; SWI/SNF,
SWI/SNF-complex; n, number of patients; ECCA, extrahepatic
cholangiocarcinoma. |
Discussion
In our study we established a cohort of 52 cases of
resected CCAs. Consistent with prognostic data given in the
literature (26), our patients had a
poor median overall survival of 35 months, despite all of them
underwent surgery in curative intent. Overall incidence of CCA has
increased progressively worldwide over the past four decades
(27,28). CCA are heterogeneous tumors with
different risk factors and precursor lesions, clinical features and
prognosis. Previous studies have documented that their origins form
different parts of the biliary tree result in different
histopathological and molecular pathological features (6). Within the last few years, new molecular
alterations have been discovered in CCA, which are improving the
pathological characterization and which might be transformed into
personalized targeted therapy algorithms, which are imperatively
needed.
We divided our cohort of tumors into the subtypes
ECCA, large duct and small duct type ICCA according to a
comprehensive analysis of macroscopic and histopathological aspects
together with an immunohistochemical and molecular pathological
characterization (ECCA, 17; small duct ICCA, 27; large duct ICCA,
8) (10).
In line with literature, our analysis of molecular
pathological changes revealed in all CCA subtypes mutations of the
tumor suppressor gene P53, with a maximum of 38% in ICCA, large
duct type. FISH-analysis for HER2 revealed amplification in
all three tumor types with a maximum of 15% in small duct type
ICCA. HER2 amplification is a relevant marker, serving as
putative therapeutical target with the availability of drugs.
According to other studies, FGFR2 fusions are
typically found in ICCA and are present in ~10–16% of patients
(29,30). The efficiency of FGFR2
inhibitors is tested to date in several clinical trials in patients
with advanced ICCA. Targeted therapies e.g. the pan-selective FGFR
kinase inhibitor BGJ398 lately showed promising antitumor activity
in a multicenter, open label phase II trial in CCA patients with
FGFR2-fusions (26). In our
cohort FGFR2-fusions were detected in ICCA with a frequency
of 8% in small duct type and 15% in large duct type. As described
earlier, FGFR2 translocation were absent in ECCA (29,30).
Recent studies demonstrated somatic mutations of
chromatin remodelers in a number of human cancers. Notably, Jiao
et al documented mutations in at least one
chromatin-remodeling gene in 47% of CCA (20). Mutations in the genes coding for
ARID1a-, BRG1-, BRM-, PBRM-1and INI-1 are inactivating and result
in a loss of protein expression. Previous studies demonstrated that
loss of protein expression correlates strongly with a mutational
status in these genes (31).
In our analysis, 35% of the cases showed protein
loss in at least one of the complex-proteins, suggesting
inactivating mutations in the respective gene. ARID1a was the most
frequent lost SWI/SNF protein, followed by BRM. ARID1a- and
BRM-negative cases were found in a small percentage of all tumor
subtypes. In literature, alterations in the ARID1a gene were
detected in 7.2–36% of ICCAs and 5%-12.3% of ECCAs (32). In this aspect, our results are in
line with previous studies. The absence of ARID1a in the different
subtypes of biliary carcinoma appears to reflect similar mechanisms
of carcinogenesis in the different subtypes of CCA.
In our study, BRG1 and INI-1 protein loss was only
seen in the group of small duct ICCA, while PRMB-1 protein loss was
found in small duct type ICCA and ECCA. Luchini et al
analyzed PBRM-1 loss in large duct and small duct ICCA and found a
prevalence of 20–30% in both subtypes (33). In our study we did not find cases
with PRBM-1 loss in the group of large duct type ICCA, which might
be explained by the low number of cases of this subtype (n=8).
Within the whole group 7.6% of cases showed protein loss of
PBRM-1.
There is still controversy about the loss of
proteins of the SWI/SNF complex subunits and its correlation to
prognosis of CCA. In the study of Jiao et al, there was no
correlation between overall survival and ARID1a alteration
(20). This was in line with other
studies (32,34). However, they subdivided ECCA and ICCA
and did not look at small duct and large duct type ICCA separately,
which might be an explanation for the varying results, compared to
our data. We identified that median survival of ARID1a altered
patients was reduced, but it was only significant for large duct
ICCA. Sarcognato et al saw in their cohort of ICCA a
correlation with retained protein expression of PBRM-1 and longer
overall survival and disease-free survival (35). Other results presented by Misumi
et al, demonstrated PBRM1 protein loss in both, small-duct
type and large-duct type ICCA. However, it was not associated
significantly with any specific characteristics, including
prognosis (36).
Our current data give strong evidence, that IHC
proven protein-loss of SWI/SNF core subunits (namely ARID1A-,
BRG1-, BRM-, PBRM-1and INI1) is associated with a highly
significant worse survival of small duct and large duct ICCA,
whereas no significant change in survival for ECCA was detected.
Furthermore, significant worse overall survival could also be shown
for the SWI/SNF-altered cohort after exclusion of
HER2-amplificated, FGFR2-translocated or P53 altered
patients, which was partly observed in coincidence. These data
underline the importance of this chromatin remodeler complex in
tumorigenesis of CCA. Our results strengthen preceding data, that
the SWI/SNF complex and its core subunits is worth further
investigation for targeted therapy options as well as predictive
marker for ICCA in future (16,20,21).
Furthermore, several studies suggest a possible
increased therapeutic vulnerability of ARID1a-deficient carcinoma.
Shen et al were able to show in cell culture analyses and in
mouse models that therapeutic inhibition of the enzyme
poly-ADP-ribose polymerase (PARP) is effective in ARID1a-deficient
ovarian and colon carcinoma (37).
ARID1a-deficient carcinoma cells also show activation of the PIk3
pathway. Therapeutic inhibition of this pathway could also be
promising in this tumor subgroup. Lee et al successfully
inhibited ARID1a-deficient gastric cancer cells with AKT inhibitors
(38). Several studies indicate an
increased expression of the programmed cell death-ligand 1 (PD-L1)
in ARID1a deficient tumor cells. First results show a significantly
better response to PD1-/PD-L1 blockade in this setting than in
ARID1a intact tumors (39). The vast
majority of studies focus on the loss of function of ARID1a. There
are no reliable findings as to whether the loss of function of the
other SWI/SNF subunits investigated offers comparable therapeutic
intervention options. This should be the subject of future clinical
studies.
However, our study has some limitations. As a
monocentric study, the case number is limited. The statistical
analysis gives interesting results and should be confirmed by the
analysis of a larger, separate and prospective cohort.
In conclusion, our data prove that CCAs are
heterogeneous tumors with different immunohistochemical and
molecular marker profiles. The proteins of the SWI/SNF complex are
lost in 35% of cases with an impact on prognosis. The core subunits
of SWI/SNF, with ARID1A and PBRM-1 for small duct ICCA and ARID1a
and BRM for large duct ICCA leading the way, may be possible
promising predictive and therapeutic targets and worth further
investigation in CCA patients.
Acknowledgements
Mrs. Wiebke Jeske, medical technical assistant, and
Ms. Elke Binot, biological- technical assistant (Institute of
Pathology, University of Cologne, Faculty of Medicine and
University Hospital Cologne, Cologne, Germany) produced the TMA
samples and performed the hybridization steps for FISH analysis,
respectively.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
BJW, RB, AQ and UD designed the study, planned the
study and were the primary writers. PSP, KA, MS, DS, DB and SB
contributed to the study design. BJW, AQ and UD conducted and
evaluated histopathological and immunohistochemical analyses. BJW
conducted the FISH analysis. PSP performed statistical analysis.
PSP, MS, DS, DB, and SB chose, collected and assembled the clinical
data, including patient follow-up from 2000 to 2019. BJW, UD and KA
chose and prepared the samples, and collected and assembled
pathological data. BJW, AQ and UD performed clinical-pathological
data analysis and interpretated the results related to the CCA
subtype and clinical implications. BJW and UD confirm the
authenticity of all raw data. All authors participated in writing,
and read and approved the final manuscript.
Ethics approval and consent to
participate
The present retrospective study was conducted with
the approval of the Ethics Committee of the University of Cologne,
utilizing clinical follow-up data that was collected
retrospectively according to a standardized protocol (application
18–269). All patients gave their appropriate informed consent to
the procedure.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CCA
|
cholangiocarcinoma
|
|
CTX
|
chemotherapy
|
|
ECCA
|
extrahepatic cholangiocarcinoma
|
|
ICCA
|
intrahepatic cholangiocarcinoma
|
|
IHC
|
immunohistochemistry/immunohistochemical
|
|
n
|
number of patients
|
|
RTX
|
radiotherapy
|
|
SWI/SNF
|
SWI/SNF-complex
|
References
|
1
|
Blechacz B, Komuta M, Roskams T and Gores
GJ: Clinical diagnosis and staging of cholangiocarcinoma. Nat Rev
Gastroenterol Hepatol. 8:512–522. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Everhart JE and Ruhl CE: Burden of
digestive diseases in the United States Part III: Liver, biliary
tract, and pancreas. Gastroenterology. 136:1134–1144. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bridgewater J, Galle PR, Khan SA, Llovet
JM, Park JW, Patel T, Pawlik TM and Gores GJ: Guidelines for the
diagnosis and management of intrahepatic cholangiocarcinoma. J
Hepatol. 60:1268–1289. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
DeOliveira ML, Cunningham SC, Cameron JL,
Kamangar F, Winter JM, Lillemoe KD, Choti MA, Yeo CJ and Schulick
RD: Cholangiocarcinoma: Thirty-one-year experience with 564
patients at a single institution. Ann Surg. 245:755–762. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rizvi S, Khan SA, Hallemeier CL, Kelley RK
and Gores GJ: Cholangiocarcinoma - evolving concepts and
therapeutic strategies. Nat Rev Clin Oncol. 15:95–111. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Komuta M, Govaere O, Vandecaveye V, Akiba
J, Van Steenbergen W, Verslype C, Laleman W, Pirenne J, Aerts R,
Yano H, et al: Histological diversity in cholangiocellular
carcinoma reflects the different cholangiocyte phenotypes.
Hepatology. 55:1876–1888. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Akita M, Fujikura K, Ajiki T, Fukumoto T,
Otani K, Azuma T, Itoh T, Ku Y and Zen Y: Dichotomy in intrahepatic
cholangiocarcinomas based on histologic similarities to hilar
cholangiocarcinomas. Mod Pathol. 30:986–997. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liau JY, Tsai JH, Yuan RH, Chang CN, Lee
HJ and Jeng YM: Morphological subclassification of intrahepatic
cholangiocarcinoma: Etiological, clinicopathological, and molecular
features. Mod Pathol. 27:1163–1173. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Aishima S and Oda Y: Pathogenesis and
classification of intrahepatic cholangiocarcinoma: Different
characters of perihilar large duct type versus peripheral small
duct type. J Hepatobiliary Pancreat Sci. 22:94–100. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nagtegaal ID, Odze RD, Klimstra D, Paradis
V, Rugge M, Schirmacher P, Washington KM, Carneiro F and Cree IA;
WHO Classification of Tumours Editorial Board, : The 2019 WHO
classification of tumours of the digestive system. Histopathology.
76:182–188. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gupta A and Dixon E: Epidemiology and risk
factors: Intrahepatic cholangiocarcinoma. Hepatobiliary Surg Nutr.
6:101–104. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shain AH and Pollack JR: The spectrum of
SWI/SNF mutations, ubiquitous in human cancers. PLoS One.
8:e551192013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Schallenberg S, Bork J, Essakly A, Alakus
H, Buettner R, Hillmer AM, Bruns C, Schroeder W, Zander T, Loeser
H, et al: Loss of the SWI/SNF-ATPase subunit members SMARCF1
(ARID1A), SMARCA2 (BRM), SMARCA4 (BRG1) and SMARCB1 (INI1) in
oesophageal adenocarcinoma. BMC Cancer. 20:122020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Numata M, Morinaga S, Watanabe T, Tamagawa
H, Yamamoto N, Shiozawa M, Nakamura Y, Kameda Y, Okawa S, Rino Y,
et al: The clinical significance of SWI/SNF complex in pancreatic
cancer. Int J Oncol. 42:403–410. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hodges C, Kirkland JG and Crabtree GR: The
Many Roles of BAF (mSWI/SNF) and PBAF Complexes in Cancer. Cold
Spring Harb Perspect Med. 6:62016. View Article : Google Scholar
|
|
16
|
Simbolo M, Fassan M, Ruzzenente A,
Mafficini A, Wood LD, Corbo V, Melisi D, Malleo G, Vicentini C,
Malpeli G, et al: Multigene mutational profiling of
cholangiocarcinomas identifies actionable molecular subgroups.
Oncotarget. 5:2839–2852. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Reisman D, Glaros S and Thompson EA: The
SWI/SNF complex and cancer. Oncogene. 28:1653–1668. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Phelan ML, Sif S, Narlikar GJ and Kingston
RE: Reconstitution of a core chromatin remodeling complex from
SWI/SNF subunits. Mol Cell. 3:247–253. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Pulice JL and Kadoch C: Composition and
Function of Mammalian SWI/SNF Chromatin Remodeling Complexes in
Human Disease. Cold Spring Harb Symp Quant Biol. 81:53–60. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jiao Y, Pawlik TM, Anders RA, Selaru FM,
Streppel MM, Lucas DJ, Niknafs N, Guthrie VB, Maitra A, Argani P,
et al: Exome sequencing identifies frequent inactivating mutations
in BAP1, ARID1A and PBRM1 in intrahepatic cholangiocarcinomas. Nat
Genet. 45:1470–1473. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Simbolo M, Vicentini C, Ruzzenente A,
Brunelli M, Conci S, Fassan M, Mafficini A, Rusev B, Corbo V,
Capelli P, et al: Genetic alterations analysis in prognostic
stratified groups identified TP53 and ARID1A as poor clinical
performance markers in intrahepatic cholangiocarcinoma. Sci Rep.
8:71192018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wilson BG and Roberts CW: SWI/SNF
nucleosome remodellers and cancer. Nat Rev Cancer. 11:481–492.
2011. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Helbig D, Ihle MA, Pütz K, Tantcheva-Poor
I, Mauch C, Büttner R and Quaas A: Oncogene and therapeutic target
analyses in atypical fibroxanthomas and pleomorphic dermal
sarcomas. Oncotarget. 7:21763–21774. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Simon R, Mirlacher M and Sauter G: Tissue
microarrays. Methods Mol Med. 114:257–268. 2005.PubMed/NCBI
|
|
25
|
Wolff AC, Hammond ME, Hicks DG, Dowsett M,
McShane LM, Allison KH, Allred DC, Bartlett JM, Bilous M,
Fitzgibbons P, et al American Society of Clinical Oncology; College
of American Pathologists, : Recommendations for human epidermal
growth factor receptor 2 testing in breast cancer: American Society
of Clinical Oncology/College of American Pathologists clinical
practice guideline update. J Clin Oncol. 31:3997–4013. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Javle M, Lowery M, Shroff RT, Weiss KH,
Springfeld C, Borad MJ, Ramanathan RK, Goyal L, Sadeghi S,
Macarulla T, et al: Phase II Study of BGJ398 in Patients With
FGFR-Altered Advanced Cholangiocarcinoma. J Clin Oncol. 36:276–282.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Khan SA, Taylor-Robinson SD, Toledano MB,
Beck A, Elliott P and Thomas HC: Changing international trends in
mortality rates for liver, biliary and pancreatic tumours. J
Hepatol. 37:806–813. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Saha SK, Zhu AX, Fuchs CS and Brooks GA:
Forty-Year Trends in Cholangiocarcinoma Incidence in the U.S.:
Intrahepatic Disease on the Rise. Oncologist. 21:594–599. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Graham RP, Barr Fritcher EG, Pestova E,
Schulz J, Sitailo LA, Vasmatzis G, Murphy SJ, McWilliams RR, Hart
SN, Halling KC, et al: Fibroblast growth factor receptor 2
translocations in intrahepatic cholangiocarcinoma. Hum Pathol.
45:1630–1638. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wu YM, Su F, Kalyana-Sundaram S, Khazanov
N, Ateeq B, Cao X, Lonigro RJ, Vats P, Wang R, Lin SF, et al:
Identification of targetable FGFR gene fusions in diverse cancers.
Cancer Discov. 3:636–647. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Rao Q, Xia QY, Wang ZY, Li L, Shen Q, Shi
SS, Wang X, Liu B, Wang YF, Shi QL, et al: Frequent co-inactivation
of the SWI/SNF subunits SMARCB1, SMARCA2 and PBRM1 in malignant
rhabdoid tumours. Histopathology. 67:121–129. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sasaki M, Nitta T, Sato Y and Nakanuma Y:
Loss of ARID1A Expression Presents a Novel Pathway of
Carcinogenesis in Biliary Carcinomas. Am J Clin Pathol.
145:815–825. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Luchini C, Robertson SA, Hong SM,
Felsenstein M, Anders RA, Pea A, Nottegar A, Veronese N, He J,
Weiss MJ, et al: PBRM1 loss is a late event during the development
of cholangiocarcinoma. Histopathology. 71:375–382. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Churi CR, Shroff R, Wang Y, Rashid A, Kang
HC, Weatherly J, Zuo M, Zinner R, Hong D, Meric-Bernstam F, et al:
Mutation profiling in cholangiocarcinoma: Prognostic and
therapeutic implications. PLoS One. 9:e1153832014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sarcognato S, Gringeri E, Fassan M, Di
Giunta M, Maffeis V, Guzzardo V, Cillo U and Guido M: Prognostic
role of BAP-1 and PBRM-1 expression in intrahepatic
cholangiocarcinoma. Virchows Arch. 474:29–37. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Misumi K, Hayashi A, Shibahara J, Arita J,
Sakamoto Y, Hasegawa K, Kokudo N and Fukayama M: Intrahepatic
cholangiocarcinoma frequently shows loss of BAP1 and PBRM1
expression, and demonstrates specific clinicopathological and
genetic characteristics with BAP1 loss. Histopathology. 70:766–774.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Shen J, Peng Y, Wei L, Zhang W, Yang L,
Lan L, Kapoor P, Ju Z, Mo Q, Shih IeM, et al: ARID1A Deficiency
Impairs the DNA Damage Checkpoint and Sensitizes Cells to PARP
Inhibitors. Cancer Discov. 5:752–767. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lee D, Yu EJ, Ham IH, Hur H and Kim YS:
AKT inhibition is an effective treatment strategy in
ARID1A-deficient gastric cancer cells. OncoTargets Ther.
10:4153–4159. 2017. View Article : Google Scholar
|
|
39
|
Shen J, Ju Z, Zhao W, Wang L, Peng Y, Ge
Z, Nagel ZD, Zou J, Wang C, Kapoor P, et al: ARID1A deficiency
promotes mutability and potentiates therapeutic antitumor immunity
unleashed by immune checkpoint blockade. Nat Med. 24:556–562. 2018.
View Article : Google Scholar : PubMed/NCBI
|



















