Introduction
Lymphoma is one of the most common cancers
worldwide, with an incidence approximately 6.68 per 100,000 people,
and develops in hematological and lymphoid compartments, including
lymph nodes, spleen and thymus, as well as extranodal lymphatic
tissues and organs (1,2). Diffuse large B-cell lymphoma (DLBCL)
accounts for ~30.0% of non-Hodgkin's lymphoma in western countries
and 45.8% in China (3). The
incidence of DLBCL continues to increase (4,5).
Notably, DLBCL is a clinically, pathologically and molecularly
heterogeneous entity, with a 5-year survival rate of 32–81%
(6). Despite improvements in its
response rate to rituximab, and combination with chemotherapy and
other novel treatment modalities, such as promising immunotherapy,
a significant proportion of patients with DLBCL exhibit strong
refractory and relapse trends, thus the long-term prognosis of this
disease remains unsatisfactory (7).
Thus, identification of novel prognostic biomarkers and therapeutic
targets for effective diagnosis and treatment of patients with
DLBCL is of great importance.
MicroRNAs (miRNAs/miRs) are small non-coding RNAs,
20–25 nucleotides in length (8).
Recently, several studies have demonstrated that miRNAs are
involved in an extensive range of biological processes, including
cell proliferation, migration, invasion, and apoptosis (9–11). In
addition, some miRNAs act as oncogenes or tumor suppressor genes in
different types of cancer (12–14).
miR-383-5p is located at chromosome 8p22 in humans (15), and its role as a tumor suppressor has
been investigated in several malignancies, including breast
(15), gastric (16,17) and
ovarian cancers (18), as well as
lung adenocarcinoma (19) and oral
squamous cell carcinoma (20). With
regards to DLBCL, several studies have reported that some miRNA
signatures are closely associated with characteristics of patients
with DLBCL, including cell proliferation, migration, invasion and
chemotherapy-resistance, as well as clinical practice (21–23).
However, the biological function and prognostic value of miR-383-5p
in DLBCL remains unclear.
The present study aimed to investigate the
biological functions and determine the prognostic value of
miR-383-5p in DLBCL progression. The expression pattern of
miR-383-5p, as well as its association with clinicopathological
characteristics of patients with DLBCL were assessed. The in
vitro effects of miR-383-5p on the proliferation and invasion
of DLBCL cells was also investigated.
Materials and methods
Patient samples and clinical
follow-up
A total of 80 patients with DLBCL at the Second
Affiliated Hospital of Harbin Medical University (Harbin, China)
were enrolled in the present study between January 2012 and
December 2016. The inclusion criteria were as follows: Patients
with DLBCL underwent routine R-CHOP and received no previous
chemotherapy or radiation. Patients with incomplete follow-up data,
including clinicopathological characteristics and survival time
were excluded from the present study.
DLBCL tissue samples were collected via biopsy and
stored at −80°C until subsequent experimentation. Control biopsy
tissue samples (n=80) were collected from patients with reactive
lymphoid hyperplasia (RLH, suspected of lymphoma). The pathological
status of all samples was independently confirmed by two
pathologists at the Second Affiliated Hospital of Harbin Medical
University. Patients with DLBCL were assessed once every 4–6
months, until death or dropout from the telephone follow-up
program. The follow-up program was between January 2012 and
December 2019. Clinical parameters and survival information were
recorded accordingly. The clinical parameters included age, sex, B
symptoms, clinical stage (24),
extranodal invasion, serum lactate dehydrogenase (LDH) levels and
International Prognostic Index (IPI) score (25). Patient follow-up was censored on
December 2019. The present study was approved by the Ethics
Committee of the Second Affiliated Hospital of Harbin Medical
University (Harbin, China; approval no. ASHHM000152), and performed
in accordance with the Declaration of Helsinki. Written informed
consent was provided by all patients prior to the study start.
Classification of patients
Patients were divided into two groups (miR-383-5p
low and high expression groups), based on median miR-383-5p
expression or the 25th percentile miR-383-5p expression. Overall
survival (OS) was defined from the time of treatment to death or
last follow-up, while disease-free survival (DFS) was defined from
the time of treatment to recurrence, death or last follow-up.
Reverse transcription-quantitative
(RT-q)PCR
Total miRNA was extracted from fresh tissues using
the RNA Extraction kit (Qiagen, Inc.), according to the
manufacturer's protocol, and reverse transcribed into cDNA using
the RT kit (Shanghai GenePharma Co., Ltd.). The temperature
protocol for RT was 51°C for 20 min, followed by 82°C for 10 min. A
miR-383-5p-specific primer and probe (TaqMan MicroRNA Assay kit,
Applied Biosystems; Thermo Fisher Scientific, Inc.) were used to
detect expression levels, using an ABI 7500 FAST Real-Time PCR
system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The
following thermocycling conditions were used for qPCR: Initial
denaturation at 94°C for 2 min, followed by 40 cycles of 94°C for
10 sec, 55°C for 30 sec and 72°C for 20 sec. The following primer
sequences were used for qPCR: miR-383-5p forward,
5′-GGGAGATCAGAAGGTGATTGTGGCT-3′ and reverse,
5′-CAGTGCGTGTCGTGGAGT-3′; and U6 forward, 5′-CTCGCTTCGGCAGCACA-3′
and reverse, 5′-AACGCTTCACGAATTTGCGT-3′. Relative miR-383-5p
expression levels were calculated using the 2−ΔΔCq
method (26) and normalized to the
internal reference gene U6.
Cell culture and transfection
Human DLBCL cell lines, OCI-LY7 and OCI-LY3, and
normal B lymphocytes, IM-9I, were purchased from the American Type
Culture Collection. OCI-LY7 and OCI-LY3 cells were maintained in
IMEM medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented
with 10% fetal bovine serum (FBS; Thermo Fisher Scientific, Inc.),
while IM-9I cells were maintained in RPMI-1640 culture medium
(Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS.
All cells were cultured at 37°C with 5% CO2.
miR-383-5p mimic, miR-383-5p inhibitor and
corresponding negative controls (NC) were purchased from Shanghai
GenePharma Co., Ltd.. Once they reached 70% confluence,
2×105 cells were transfected with 50 nM miR-383-5p
mimic, miR-383-5p inhibitor and/or respective controls, using
Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) at 37°C for 3 h, according to the manufacturer's
protocol. The following sequences were used: miR-383-5p mimic,
5′-AAAUCUCUGCAGGCAAAUGUGA-3′ and mimic-NC,
5′-UUCUCCGAACGUGUCACGUTT-3′; and miR-383-5p inhibitor,
5′-CAGUGGUUUUACCCUAUGGUAG-3′ and inhibitor-NC,
5′-UUCUCCGAACGUGUCACGUTT-3′. Cells were harvested for subsequent
experimentation 48 h post-transfection.
Cell counting kit-8 (CCK-8) assay
Transfected cells were seeded into 96-well plates at
a density of 1×104 cells/well. CCK-8 reagent (10 µl,
Dojindo Molecular Technologies, Inc.) was added to each well at 0,
24, 48, and 72 h, and incubated at 37°C for an additional 2 h. Cell
proliferation was subsequently analyzed at a wavelength of 450 nm,
using an automatic microplate reader (BioTek Instruments).
Crystal violet staining
Crystal violet staining was performed to assess cell
proliferation. Transfected cells were seeded into 6-well plates at
the density of 1,000 cells/well and cultured in IMEM medium (Gibco;
Thermo Fisher Scientific, Inc.) supplemented with 10% FBS at 37°C.
Culture medium was replaced every 3 days for a total of 2 weeks.
Cells were subsequently fixed with 100% methanol for 5 min at room
temperature and stained with 0.5% crystal violet solution for 10
min at room temperature. Cells were washed three times with PBS and
observed under a light microscope (magnification, ×4). Cell
proliferation was subsequently analyzed at a wavelength of 570 nm,
using an automatic microplate reader.
Transwell assay
The Transwell assay was performed to assess cell
invasion. A total of 100 µl cell suspension without FBS
(4×104 cells) was plated in the upper chambers of
24-well Transwell plates (Corning, Inc.), with 8 µm pore size
membranes. IMEM medium (Gibco; Thermo Fisher Scientific, Inc.) (500
µl) supplemented with 10% FBS was plated in the lower chambers.
Membranes were pre-coated with 50 µl Matrigel (BD Biosciences) for
4 h at 37°C. Following incubation for 24 h at 37°C with 5%
CO2, the invasive cells were fixed with 100% methanol
for 5 min at room temperature and stained with 0.1% crystal violet
for 10 min at room temperature. Stained cells were counted in five
randomly selected fields using a light microscope (magnification,
×200; Olympus Corporation).
Statistical analysis
Statistical analysis was performed using GraphPad
Prism 5.0 software (GraphPad Software Inc.) and SPSS 20.0 software
(IBM Corp.). All experiments were performed in triplicate and data
are presented as the mean ± standard deviation. The χ2
test was used to assess the association between miR-383-5p
expression and the clinicopathological characteristics of patients
with DLBCL. Unpaired Student's t-test was used to compare
differences between two groups, while one-way ANOVA followed by LSD
test were used to compare differences between multiple groups.
Kaplan-Meier curves were generated for both OS and DFS. Cox
regression analysis was performed to determine the prognostic
factors. P<0.05 was considered to indicate a statistically
significant difference.
Results
miR-383-5p expression is downregulated
in DLBCL tissues and cell lines
RT-qPCR analysis was performed to detect miR-383-5p
expression in 80 paired DLBCL tissues and normal tissues from
patients with RLH. The results demonstrated that miR-383-5p
expression was significantly downregulated in DLBCL tissues
compared with RLH tissues (P<0.001; Fig. 1A). miR-383-5p expression was also
detected in the DLBCL cell lines (OCI-LY7 and OCI-LY3 cells) and
normal B lymphocytes (IM-9I cells). As presented in Fig. 1B, miR-383-5p expression was
significantly lower in DLBCL cells compared with IM-9I cells
(P<0.001).
Patients with DLBCL were classified into two groups,
miR-383-5p low expression (n=40) and miR-383-5p high expression
(n=40) groups, based on median miR-383-5p expression.
Association between miR-383-5p
expression and the clinicopathological characteristics of patients
with DLBCL
The association between miR-383-5p expression and
the clinicopathological characteristics of patients with DLBCL was
assessed. As presented in Table I,
patients with low miR-383-5p expression were significantly
associated with advanced clinical stages of the disease (P=0.004).
In addition, low miR-383-5p expression was significantly associated
with a higher rate of extranodal invasion compared with the high
miR-383-5p expression group (P=0.001). Notably, no significant
differences in age, sex, B symptoms, IPI score and serum LDH levels
were observed between the two groups.
 | Table I.Association between miR-383-5p
expression and the clinicopathological characteristics of patients
with diffuse large B-cell lymphoma (n=80). |
Table I.
Association between miR-383-5p
expression and the clinicopathological characteristics of patients
with diffuse large B-cell lymphoma (n=80).
|
|
| miR-383-5p
expression, n (%) |
|
|---|
|
|
|
|
|
|---|
| Characteristic | Patients, n | High (n=40) | Low (n=40) | P-value |
|---|
| Age, years |
|
|
| 0.654 |
|
<60 | 38 | 18 (45.0) | 20 (50.0) |
|
| ≥60 | 42 | 22 (55.0) | 20 (50.0) |
|
| Sex |
|
|
| 0.072 |
|
Male | 36 | 14 (35.0) | 22 (55.0) |
|
|
Female | 44 | 26 (65.0) | 18 (45.0) |
|
| B symptoms |
|
|
| 0.653 |
|
Absent | 36 | 19 (47.5) | 17 (42.5) |
|
|
Present | 44 | 21 (52.5) | 23 (57.5) |
|
| Clinical stage |
|
|
| 0.004a |
| I/II | 43 | 28 (70.0) | 15 (37.5) |
|
|
III/IV | 37 | 12 (30.0) | 25 (62.5) |
|
| Extranodal
invasion |
|
|
| 0.001a |
|
<2 | 39 | 27 (57.5) | 12 (20.0) |
|
| ≥2 | 41 | 13 (42.5) | 28 (80.0) |
|
| IPI score |
|
|
| 0.651 |
|
0-2 | 34 | 18 (45.0) | 16 (40.0) |
|
|
3-5 | 46 | 22 (55.0) | 24 (60.0) |
|
| Serum LDH level,
IU/l |
|
|
| 0.073 |
|
<300 | 38 | 23 (57.5) | 15 (37.5) |
|
|
≥300 | 42 | 17 (42.5) | 25 (62.5) |
|
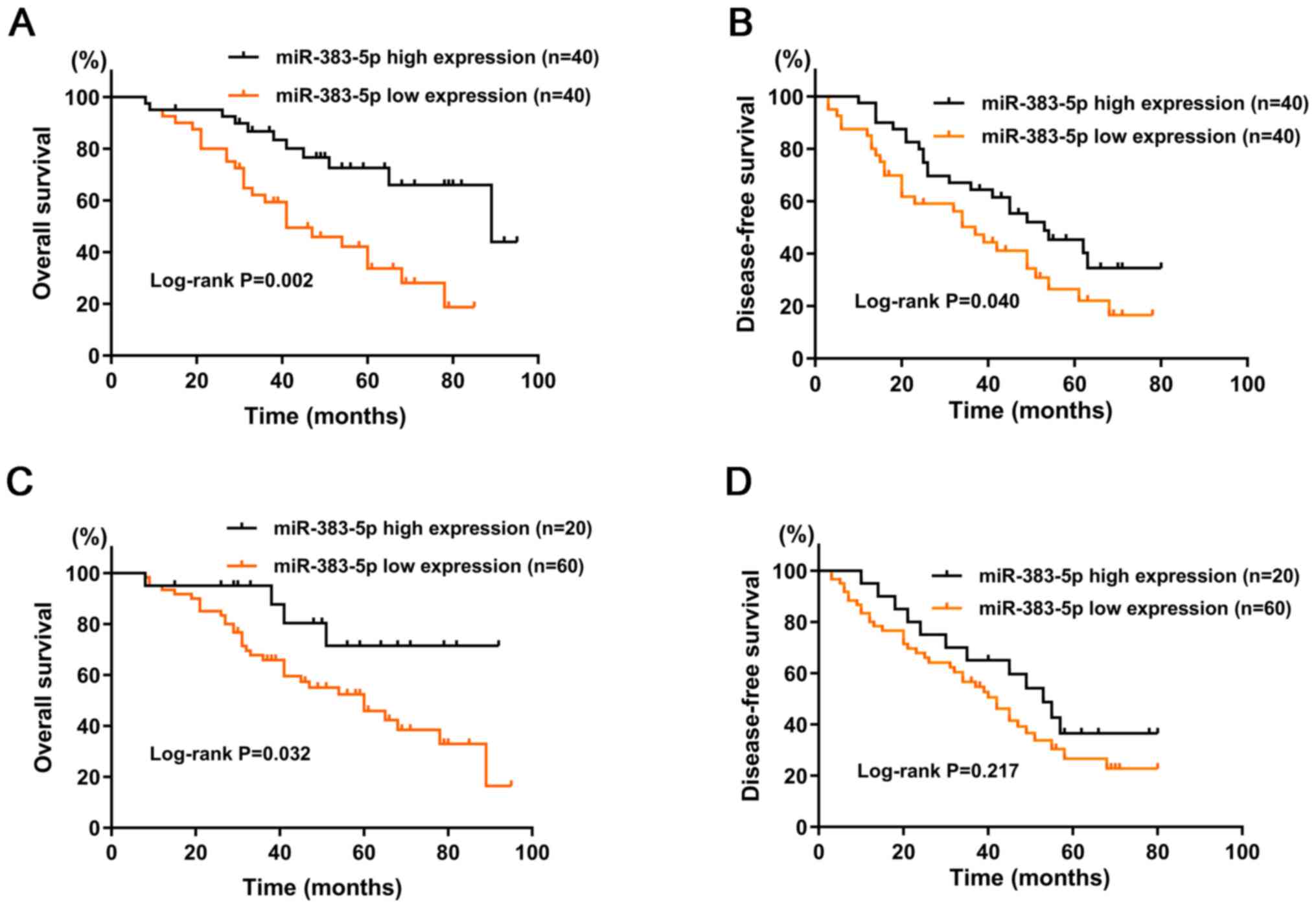
Kaplan-Meier survival analysis demonstrated that
patients with high miR-383-5p expression had significantly higher
OS (P=0.002; Fig. 2A) and DFS
(P=0.04; Fig. 2B) rates than those
with low miR-383-5p expression. Univariate and multivariate Cox
regression analyses were performed to determine the prognostic
factors associated with OS and DFS.
The prognostic factors found to be significant in
the univariate analysis (P<0.05) were subjected to multivariate
analysis with the Cox proportional hazards regression model.
Univariate and multivariate analyses indicated that OS was
significantly associated with clinical stage [hazard ratio (HR),
1.570; 95% confidence interval (CI), 1.402–1.759; P=0.001],
extranodal invasion (HR, 1.444; 95% CI, 1.265–1.649; P=0.001) and
miR-383-5p expression (HR, 0.844; 95% CI, 0.754–0.945; P=0.003)
(Table II). Similarly, multivariate
analysis indicated that DFS was significantly associated with
clinical stage (HR, 1.605; 95% CI, 1.433–1.797; P<0.001),
extranodal invasion (HR, 1.734; 95% CI, 1.520–1.981; P<0.001)
and miR-383-5p expression (HR, 0.753; 95% CI, 0.672–0.842;
P<0.001) (Table III).
 | Table II.Univariate and multivariate analyses
of prognostic factors associated with overall survival. |
Table II.
Univariate and multivariate analyses
of prognostic factors associated with overall survival.
|
| Univariate | Multivariate |
|---|
|
|
|
|
|---|
| Variable | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Age, years (≥60 vs.
<60) | 1.005
(0.841–1.200) | 0.931 | – | – |
| Sex (Male vs.
Female) | 1.036
(0.830–1.292) | 0.755 | – | – |
| B symptoms (Absent
vs. Present) | 0.923
(0.761–1.121) | 0.419 | – | – |
| Clinical stage
(I/II vs. III/IV) | 1.592
(1.422–1.782) |
<0.001b | 1.570
(1.402–1.759) | 0.001a |
| Extranodal invasion
(≥2 vs. <2) | 1.448
(1.268–1.656) |
<0.001b | 1.444
(1.265–1.649) | 0.001a |
| IPI score (3–5 vs.
0–2) | 1.231
(0.686–2.423) | 0.376 | – | – |
| Serum LDH level,
IU/l (≥300 vs. <300) | 1.983
(0.881–3.123) | 0.556 | – | – |
| miR-383-5p
expression (High vs. Low) | 0.783
(0.700–0.876) | 0.001a | 0.844
(0.754–0.945) | 0.003a |
 | Table III.Univariate and multivariate analyses
of prognostic factors associated with disease-free survival. |
Table III.
Univariate and multivariate analyses
of prognostic factors associated with disease-free survival.
|
| Univariate | Multivariate |
|---|
|
|
|
|
|---|
| Variable | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Age, years (≥60 vs.
<60) | 1.163
(0.960–1.411) | 0.112 | – | – |
| Sex (Male vs.
Female) | 1.009
(0.845–1.206) | 0.920 | – | – |
| B symptoms (Absent
vs. Present) | 1.006
(0.878–1.152) | 0.931 | – | – |
| Clinical stage
(I/II vs. III/IV) | 1.570
(1.402–1.757) |
<0.001a | 1.605
(1.433–1.797) |
<0.001a |
| Extranodal invasion
(≥2 vs. <2) | 1.701
(1.490–1.944) |
<0.001a | 1.734
(1.520–1.981) |
<0.001a |
| IPI score (0–2 vs.
3–5) | 1.056
(0.931–1.197) | 0.398 | – | – |
| Serum LDH level
(≥300 vs. <300) | 1.068
(0.927–1.230) | 0.365 | – | – |
| miR-383-5p
expression (High vs. Low) | 0.726
(0.649–0.812) |
<0.001a | 0.753
(0.672–0.842) |
<0.001a |
Patients were classified into two groups, miR-383-5p
high expression (n=20) and miR-383-5p low expression (n=60) groups,
based on the 25th percentile. Kaplan-Meier survival analysis
demonstrated that patients with high miR-383-5p expression had a
significantly higher OS rate (P=0.032; Fig. 2C). However, no significant difference
in DFS rate was observed between the two groups (P=0.217; Fig. 2D). Taken together, these results
suggest that miR-383-5p expression is downregulated in patients
with DLBCL, and high miR-383-5p expression is associated with a
favorable clinical prognosis.
Overexpression of miR-383-5p inhibits
the proliferation and invasion of DLBCL cells
The biological function of miR-383-5p during DLBCL
progression was investigated. OCI-LY7 and OCI-LY3 cells were
transfected with miR-383-5p mimic, and the effect of overexpressing
miR-383-5p on the proliferation and invasion of lymphoma cells was
assessed in vitro. The results confirmed that miR-383-5p
mimic significantly increased miR-383-5p expression in OCI-LY7 and
OCI-LY3 cells (P<0.001; Fig. 3A and
B).
The results of the CCK-8 assay demonstrated that the
number of OCI-LY7 and OCI-LY3 cells significantly decreased at days
3 and 4 following transfection with miR-383-5p mimic compared with
the untreated cells (P<0.01 and P<0.001; Fig. 3C and D). In addition, crystal violet
staining demonstrated that overexpression of miR-383-5p
significantly inhibited the proliferation of both OCI-LY7 and
OCI-LY3 cells (P<0.001; Fig. 3E).
The results of the Transwell assay demonstrated that overexpression
of miR-383-5p significantly inhibited the invasive ability of both
OCI-LY7 and OCI-LY3 cells (P<0.001; Fig. 3F).
miR-383-5p knockdown promotes the
proliferation and invasion of DLBCL cells
OCI-LY7 and OCI-LY3 cells were transfected with
miR-383-5p inhibitor to decrease miR-383-5p expression, and the
results indicated that miR-383-5p inhibitor markedly suppressed
miR-383-5p expression in OCI-LY7 and OCI-LY3 cells (P<0.001;
Fig. 4A and B). The results of the
CCK-8 assay and crystal violet staining demonstrated that
miR-383-5p knockdown significantly promoted the proliferative
ability of both OCI-LY7 and OCI-LY3 cells (P<0.05 Fig. 4C-E). Similarly, the results of the
Transwell assay demonstrated that miR-383-5p knockdown
significantly promoted the invasive ability of OCI-LY7 and OCI-LY3
(P<0.001; Fig. 4F).
Discussion
Several miRNAs have been demonstrated to play
fundamental roles in the initiation and progression of DLBCL. For
example, miR-155 knockdown inhibits cell proliferation and
facilitates apoptosis of DLBCL in vitro by upregulating
SOCS3 expression to suppress JAK-STAT3 signaling (27). In addition, low miR-27b expression is
associated with poor overall survival of patients with DLBCL, while
high miR-27b expression suppresses the proliferation of DLBCL cells
(28).
The results of the present study demonstrated that
miR-383-5p expression was downregulated in human DLBCL tissues and
related cell lines. In addition, miR-383-5p expression was closely
associated with advanced clinical stage and extranodal invasion.
Notably, high miR-383-5p expression predicted favorable clinical
prognosis for patients with DLBCL. Taken together, these results
suggest that miR-383-5p may serve as an independent prognostic
biomarker, as well as a tumor suppressor in patients with DLBCL.
Overexpression and knockdown of miR-383-5p were performed in two
independent DLBCL cell lines using miR-383-5p mimic and inhibitor,
respectively. The effect of altering miR-383-5p expression on the
proliferation and invasion of DLBCL cells was assessed. The results
demonstrated that overexpression of miR-383-5p significantly
inhibited the proliferation and invasion of DLBCL cells, the
effects of which were reversed following miR-383-5p knockdown.
The tumor suppressive role of miR-383-5p has been
reported in several malignancies, including breast (15), gastric (16) and ovarian cancers (18), as well as lung adenocarcinoma
(19). Zhang et al (15) demonstrated that miR-383-5p expression
is downregulated in breast cancer tissues and respective cell
lines. In addition, miR-383-5p can inhibit the proliferation,
migration and invasion of breast cancer cells, the findings of
which are consistent with the results of the present study. Xu
et al (17) reported that
overexpression of miR-383-5p suppresses the proliferation and
increases the apoptosis of gastric cancer cells, suggesting its
prominent role in gastric carcinogenesis. In addition,
overexpression of miR-383-5p inhibits cell proliferation, tumor
growth and enhances chemosensitivity of ovarian cancer cells by
downregulating TRIM27 expression (18). miR-383-5p expression is downregulated
in lung adenocarcinoma tissues, and also exerts antiproliferative
effects on lung adenocarcinoma cells (19). Further studies are required to
determine the underlying mechanism of miR-383-5p in regulating the
progression of DLBCL cells.
In conclusion, the results of the present study
demonstrated that miR-383-5p expression is downregulated in DLBCL
tissues, and thus may serve as a prognostic biomarker for patients
with DLBCL. In addition, miR-383-5p appears to play a critical role
in inhibiting the proliferation and invasion of DLBCL cells, and
thus may act as a putative therapeutic target for effective
treatment of DLBCL.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Science and
Technology Project of Heilongjiang Provincial Department of
Education (grant no. 12541372) and the Science and Technology
Project of Heilongjiang Provincial Health and Family Planning
Commission (grant no. 2013055).
Availability of data and materials
All data generated or analyzed during the present
study are included in this published article.
Authors' contributions
LYC, BQH and XMZ designed the present study,
performed the experiments and analyzed the data. XBY, DDY and LQY
collected the clinical samples and analyzed the data. LYC and LQY
confirm the authenticity of all the raw data. All authors have read
and approved the final manuscript.
Ethics approval and consent for
participation
The present study was approved by the Ethics
Committee of the Second Affiliated Hospital of Harbin Medical
University (Harbin, China; approval no. ASHHM000152), and performed
in accordance with the Declaration of Helsinki. Written informed
consent was provided by all patients prior to the study start.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Mugnaini EN and Ghosh N: Lymphoma. Prim
Care. 43:661–675. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Monfardini S, Zagonel V and Noordijk EM:
Lymphomas. Crit Rev Oncol Hematol. 27:157–160. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Xu-Monette ZY, Wu L, Visco C, Tai YC,
Tzankov A, Liu WM, Montes-Moreno S, Dybkaer K, Chiu A, Orazi A, et
al: Mutational profile and prognostic significance of TP53 in
diffuse large B-cell lymphoma patients treated with R-CHOP: Report
from an international DLBCL rituximab-CHOP consortium program
study. Blood. 120:3986–3996. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Martelli M, Ferreri AJ, Agostinelli C, Di
Rocco A, Pfreundschuh M and Pileri SA: Diffuse large B-cell
lymphoma. Crit Rev Oncol Hematol. 87:146–171. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jiang W and Huang H: The evolution and
curative effect of diffuse large B cell lymphoma treatment in
China. Zhonghua Xue Ye Xue Za Zhi. 35:357–360. 2014.(In Chinese).
PubMed/NCBI
|
|
6
|
Schürch CM, Federmann B,
Quintanilla-Martinez L and Fend F: Tumor heterogeneity in
lymphomas: A different breed. Pathobiology. 85:130–145. 2018.
View Article : Google Scholar
|
|
7
|
Matasar MJ and Zelenetz AD: Overview of
lymphoma diagnosis and management. Radiol Clin North Am. 46175–198.
(vii)2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Larrabeiti-Etxebarria A, Lopez-Santillan
M, Santos-Zorrozua B, Lopez-Lopez E and Garcia-Orad A: Systematic
review of the potential of MicroRNAs in diffuse large B cell
lymphoma. Cancers (Basel). 11:1442019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lee YS and Dutta A: MicroRNAs in cancer.
Annu Rev Pathol. 4:199–227. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wu M, Wang G, Tian W, Deng Y and Xu Y:
MiRNA-based therapeutics for lung cancer. Curr Pharm Des.
23:5989–5996. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Du X, Zhang J, Wang J, Lin X and Ding F:
Role of miRNA in lung cancer-potential biomarkers and therapies.
Curr Pharm Des. 23:5997–6010. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kabekkodu SP, Shukla V, Varghese VK,
D'Souza J, Chakrabarty S and Satyamoorthy K: Clustered miRNAs and
their role in biological functions and diseases. Biol Rev Camb
Philos Soc. 93:1955–1986. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lu TX and Rothenberg ME: MicroRNA. J
Allergy Clin Immunol. 141:1202–1207. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Vishnoi A and Rani S: MiRNA biogenesis and
regulation of diseases: An overview. Methods Mol Biol. 1509:1–10.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang J, Kong X, Shi Q and Zhao B:
MicroRNA-383-5p acts as a potential prognostic biomarker and an
inhibitor of tumor cell proliferation, migration, and invasion in
breast cancer. Cancer Biomark. 27:423–432. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wei C and Gao JJ: Downregulated miR-383-5p
contributes to the proliferation and migration of gastric cancer
cells and is associated with poor prognosis. PeerJ. 7:e78822019.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xu G, Li N, Zhang Y, Zhang J, Xu R and Wu
Y: MicroRNA-383-5p inhibits the progression of gastric carcinoma
via targeting HDAC9 expression. Braz J Med Biol Res. 52:e83412019.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jiang J, Xie C, Liu Y, Shi Q and Chen Y:
Up-regulation of miR-383-5p suppresses proliferation and enhances
chemosensitivity in ovarian cancer cells by targeting TRIM27.
Biomed Pharmacother. 109:595–601. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhao S, Gao X, Zang S, Li Y, Feng X and
Yuan X: MicroRNA-383-5p acts as a prognostic marker and inhibitor
of cell proliferation in lung adenocarcinoma by cancerous inhibitor
of protein phosphatase 2A. Oncol Lett. 14:3573–3579. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shao B, Fu X, Li X, Li Y and Gan N:
RP11-284F21.9 promotes oral squamous cell carcinoma development via
the miR-383-5p/MAL2 axis. J Oral Pathol Med. 49:21–29. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zheng B, Xi Z, Liu R, Yin W, Sui Z, Ren B,
Miller H, Gong Q and Liu C: The function of MicroRNAs in B-cell
development, lymphoma, and their potential in clinical practice.
Front Immunol. 9:9362018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fernando TR, Rodriguez-Malave NI and Rao
DS: MicroRNAs in B cell development and malignancy. J Hematol
Oncol. 5:72012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
de Yébenes VG, Bartolomé-Izquierdo N and
Ramiro AR: Regulation of B-cell development and function by
microRNAs. Immunol Rev. 253:25–39. 2013. View Article : Google Scholar
|
|
24
|
Cheson BD, Fisher RI, Barrington SF,
Cavalli F, Schwartz LH, Zucca E, Lister TA; Alliance, Australasian
Leukaemia; Lymphoma Group and Eastern Cooperative Oncology Group, ;
et al: Recommendations for initial evaluation, staging, and
response assessment of Hodgkin and non-Hodgkin lymphoma: The Lugano
classification. J Clin Oncol. 32:3059–3068. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yang Y, Wang L, Ma Y, Han T and Huang M:
The enhanced international prognostic index for diffuse large
B-cell lymphoma. Am J Med Sci. 353:459–465. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li XD, Li XM, Gu JW and Sun XC: MiR-155
regulates lymphoma cell proliferation and apoptosis through
targeting SOCS3/JAK-STAT3 signaling pathway. Eur Rev Med Pharmacol
Sci. 21:5153–5159. 2017.PubMed/NCBI
|
|
28
|
Jia YJ, Liu ZB, Wang WG, Sun CB, Wei P,
Yang YL, You MJ, Yu BH, Li XQ and Zhou XY: HDAC6 regulates
microRNA-27b that suppresses proliferation, promotes apoptosis and
target MET in diffuse large B-cell lymphoma. Leukemia. 32:703–711.
2018. View Article : Google Scholar : PubMed/NCBI
|