Introduction
Prostate cancer (PCa) is the second most prevalent
cancer and the fifth leading cause of cancer-related deaths in the
male population worldwide (1). PCa
has a favorable prognosis when diagnosed in early stages, as
low-grade localized tumors progress slowly and are highly
treatable. Currently, PCa diagnosis is mostly based on serum
prostate-specific antigen (PSA) testing and digital rectal
examination (DRE) followed by confirmation using multi-core
prostatic biopsy (2).
Although PSA is the most widely used biomarker for
non-invasive PCa detection, its limitations are well known
(2). PSA is not cancer specific as
many nonmalignant conditions, such as benign prostatic hyperplasia
(BPH), prostatitis and urinary tract infections, may affect the PSA
serum levels (3). In fact, a
negative prostate biopsy was found in 70–80% of men with PSA levels
between 4–10 ng/ml (3). On the
other hand, up to 15% of men with a PSA level ≤4.0 ng/ml had
biopsy-detected PCa (4). In
addition, PSA is unable to distinguish between indolent and
aggressive PCa. PSA limited sensitivity and specificity results in
unnecessary biopsies, overdiagnosis and overtreatment of patients.
Thus, PCa is a major global health problem that imposes a
significant social and economic burden. Therefore, there is an
urgent need for novel non-invasive biomarkers that can accurately
detect PCa and improve disease risk stratification in order to
appropriately guide patient management.
Urine is an attractive source for PCa biomarker
discovery, it is readily available in large quantities and can be
sampled non-invasively (liquid biopsy) (5). Urine composition reflects the
physiological or pathological state of major urological tissues,
including the prostate (6,7). Therefore, prostate-derived molecules
found in urine, including DNA, RNA, proteins and peptides, may
represent potential biomarkers for PCa prognostic, diagnostic and
monitoring. Increasing evidences have shown that urinary biomarkers
are promising tools to improve PCa management (8).
Mass spectrometry (MS) is a powerful technique to
detect and monitor biomarkers in human biofluids. MS-based
approaches have been successfully used to profile urinary
proteins/peptides in the search for PCa biomarkers (8,9). In
this work, we used liquid chromatography-mass spectrometry (LC-MS)
to identify and quantify naturally occurring peptides in urine
samples from patients diagnosed with PCa, BPH, and healthy
individuals. We identified 5 urinary peptides derived from
uromodulin with potential for use as PCa biomarker, generating AUC
values between 0.788 and 0.951. In addition, the identified peptide
panel outperformed PSA in differentiating between PCa and BPH.
Thus, we hope to contribute to the identification of candidate
peptides as biomarkers for the early and accurate detection of
PCa.
Materials and methods
Patients and sample collection
The study cohort included men without prior PCa
diagnosis who underwent a transrectal ultrasound (TRUS)-guided
prostate biopsy from September 2018 to September 2019 in the
Urology Service of Hospital Ernesto Dornelles (Porto Alegre, Rio
Grande do Sul, Brazil) or in the Hospital Ana Nery (Santa Cruz do
Sul, Rio Grande do Sul, Brazil). Positive and negative prostate
biopsies were classification criteria for patients with PCa and
BPH, respectively. Tumor grading and staging of PCa patients were
based on histological analysis. The diagnosis of BPH was based on
lower urinary tract symptoms and evidence by palpation or
transrectal ultrasound of prostate enlargement. Urine samples were
collected just before prostate biopsy without prior DRE or
prostatic massage. Control urine samples were obtained from healthy
volunteers without any diagnosed prostate condition, no positive
DRE, no prostate alteration and PSA levels <4.0 ng/ml. A total
of 86 participants were included in the study, with age range from
42 to 88 years. Urine samples were centrifuged at 4,000 g for 10
min at 4°C and stored at −80°C until peptide isolation. This study
was approved by Institutional Review Boards of Universidade Federal
do Rio Grande do Sul (UFRGS), Universidade de Santa Cruz do Sul
(UNISC), Hospital Ernesto Dornelles (HED), and Hospital Ana Nery,
under the protocol CAAE number 69852617.1.1001.5347. The study was
conducted according to the guidelines of the Declaration of
Helsinki and written informed consent was obtained from all
participants.
Urinary peptide isolation
Urine endogenous peptides were isolated by
trichloroacetic acid (TCA) precipitation as described by Parker
et al (10). Briefly, 700 µl
of urine samples were concentrated by vacuum centrifugation using
SpeedVac. Samples were concentrated to more accurately reproduce
the methodology described by Parker et al (10), as the protocol was originally
standardized for human plasma samples, which contain larger amounts
of proteins/peptides than those normally found in urine. After
concentration, samples were mixed 1:1 with PBS and urinary proteins
were precipitated with 1 volume of 20% TCA for 1 h at 4°C. Samples
were centrifuged at 16,000 × g for 10 min at 4°C and the
peptide-containing supernatants were collected. Purified peptides
were desalted using HLB OASIS cartridges (Waters), following
manufacturer's instructions. Peptides were quantified using Pierce™
Quantitative Colorimetric Peptide Assay (Thermo Scientific, 23275)
and stored at −20°C until analysis.
LC-MS analysis
The isolated urinary peptides were analyzed by LC-MS
using a nanoACQUITY UPLC system coupled to a Xevo G2-XS Q-Tof mass
spectrometer (Waters) with a low-flow probe at the source. Peptides
were separated by analytical chromatography (Acquity UPLC BEH C18,
1.7 µm, 2.1×50 mm, Waters) at a flow rate of 8 µl/min, using a
7–85% water/ACN 0.1% formic acid linear gradient over 42 min. The
MS survey scan was set to 0.5 s and recorded from 50 to 2,000 m/z.
MS/MS scans were acquired from 50 to 2,000 m/z, and scan time was
set to 1 s. Data were collected in data-independent mode
(MSE). Two independent LC-MSE runs were
performed for each sample and each run contained 25 µg of
sample.
LC-MS data analysis
LC-MSE data were processed and searched
using ProteinLynx Global Server (PLGS 3.0.3, Waters Corporation).
Database searches were conducted against Homo sapiens
protein sequences retrieved from UniProtKB/Swiss-Prot database,
with the following parameters: oxidation of methionine (M), proline
(P) and lysine (K) as variable modifications, without any enzyme
specificity and maximal missed cleavage of 0. Peptides and protein
tolerances were set as automatic, allowing minimum fragment ion per
protein as 2, minimum fragment ion per peptide as 2, minimum
peptide matches per proteins as 1 and false discovery rate (FDR) as
4%. Only peptides detected in the two technical replicates were
considered for further analysis in order to improve confidence. The
mass spectrometry data have been deposited to the ProteomeXchange
Consortium via the PRIDE (11)
partner repository with the dataset identifier PXD037031.
Raw files containing MSE spectra and
peptide ID files (fragment.csv) generated by PLGS were imported
into Skyline software (12) to
create a comprehensive spectral library. MS1 precursor ion
chromatograms (M, M + 1, and M + 2) were extracted for each peptide
and the integrated areas of isotope peaks were used for label-free
peptide quantification. Log2-transformed values were submitted to
differential expression analysis using the NormalyzerDE tool
(13). Peptides with differential
abundance between study groups were submitted to receiver operating
characteristic (ROC) curve analyses using SPSS Statistics software,
version 18 (SPSS Inc., Chicago, III., USA).
In silico protease cleavage sites
prediction
The prediction of proteases potentially involved in
the generation of the identified urinary peptides was performed
using the Proteasix tool (14),
which uses the MEROPS peptidase database as a reference. Peptide
sequence data were prepared in the required input format and the
analysis was conducted using the default settings. In order to
retrieve high-confidence cleavage site predictions, we selected a
specificity threshold >80%. TCGA expression data of predicted
proteases in PCa and normal prostate tissue were retrieved from
UALCAN (15).
Statistical analysis
Demographic and clinical data of study groups were
analyzed using one-way ANOVA and Tukey's post hoc test or
Kruskal-Wallis and Dunn's post hoc test, depending on the normality
of the data. Peptide peak area values (log2-transformed) were used
for quantitative analysis using NormalyzerDE tool and differential
abundance between the study groups were analyzed by unpaired,
moderated t-test (empirical Bayes Limma approach) with
Benjamini-Hochberg correction. Comparisons between different age
groups were performed using one-way ANOVA or Kruskal-Wallis tests,
depending on the normality of the data. Peptide levels in low- and
high-grade tumors were compared using a Mann-Whitney non-parametric
test. Statistical significance was set at P<0.05. Peptides with
differential abundance between the study groups were submitted to
ROC curve analyses using SPSS Statistics software. Expression data
from predicted proteases in normal and PCa tissues were retrieved
from TCGA using UALCAN portal and analyzed by unpaired Welch's
t-test.
Results
Clinical data of the study cohort
A total of 86 participants were included in the
study: 33 patients with PCa, 25 patients with BPH and 28 healthy
controls (Table I). Men in the PCa
group were significantly older than men from BPH and control groups
(P=0.012 and P<0.0001, respectively). Median PSA levels were
significantly lower (P<0.0001) in control group compared to the
PCa and BPH groups. In addition, there was no statistically
significant difference in mean PSA levels between BPH and PCa
groups (P=0.368).
 | Table I.Demographic and clinical data of the
study cohort. |
Table I.
Demographic and clinical data of the
study cohort.
|
| Groups | Comparisons
(P-value) |
|---|
|
|
|
|
|---|
| Parameters | PCa (n=33) | BPH (n=25) | Control (n=28) | PCa vs. BPH | PCa vs.
Control | BPH vs.
Control |
|---|
| Median age, years
(95% CI) | 69 (64–73) | 61
(58–66)a | 57
(53–60)b | 0.012c |
1.98×10−6c | 0.076c |
| PSA median, ng/ml
(95% CI) | 7.93
(7.00–9.00)d | 6.65
(5.41–9.31)e | 0.93
(0.58–1.52)e | 0.368f |
2.56×10−10f |
1.02×10−6f |
| Gleason score,
n |
|
|
|
|
|
|
| Gleason
6 | 13 | n.a. | n.a. | n.a. | n.a | n.a |
| Gleason
3+4/4+3 | 17 | n.a. | n.a. | n.a. | n.a. | n.a. |
| Gleason
8 | 3 | n.a. | n.a. | n.a. | n.a. | n.a. |
Endogenous urinary peptide
profiles
Endogenous peptides were isolated from urine samples
by protein precipitation using TCA. Varying amounts of peptides
were recovered from each sample, from 105 to 1,260 µg. Twenty-five
micrograms of each sample were analyzed by LC-MSE. The
analysis resulted in the identification of 10 peptides derived from
uromodulin and 9 peptides derived from alpha-1-antitrypsin
(Table SI). Peptides detected in
>95% of the analyzed samples were subjected to label-free
quantitative analysis based on precursor (MS1) peak area using
NormalyzerDE tool. These peptides, highlighted in Table SI, were named UMOD-P1 to UMOD-P7.
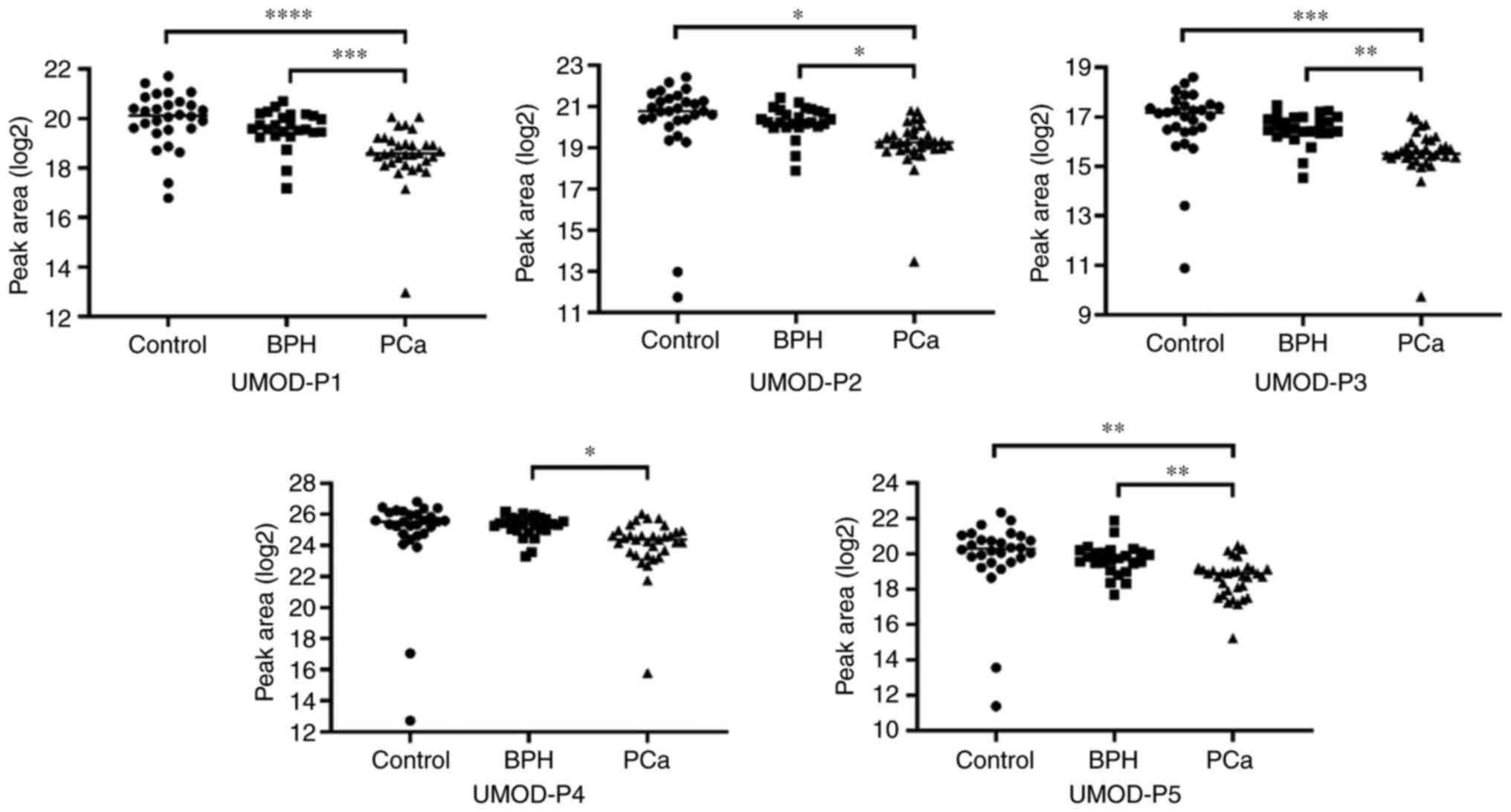
Comparative analyzes revealed 5 peptides (UMOD-P1 to UMOD-P5) with
differential abundance between the study groups, all displaying
lower abundance in the PCa group (Table II; Fig.
1). These five peptides showed statistically significant
differences between the BPH and PCa groups, with four of them also
significantly altered between the control and PCa groups. No
peptide showed significant differences between control and BPH
groups. Therefore, our data indicate that urine endogenous UMOD
peptides have the potential to discriminate between benign and
malignant prostate conditions. We subdivided the study groups into
distinct age groups to evaluate possible differences in UMOD
peptide levels between younger and older individuals. No
statistically significant differences were found between age groups
in any of the study groups (Table
SII). In addition, no significant differences were observed in
UMOD peptide levels between low-(Gleason score 6) and high-grade
(Gleason score >6) tumors (Table
SIII).
 | Table II.Comparison of peak area values of
UMOD peptides between study groups. |
Table II.
Comparison of peak area values of
UMOD peptides between study groups.
|
|
| Peptide peak area
(log2) | Comparisons |
|---|
|
|
|
|
|
|---|
|
|
| Control | BPH | PCa | Control vs.
PCa | BPH vs. PCa |
|---|
|
|
|
|
|
|
|
|
|---|
| Peptide | Peptide
sequence | Min-Max | Mean | SD | Min-Max | Mean | SD | Min-Max | Mean | SD |
P-valuea | Adjusted
P-valueb |
P-valuea | Adjusted
P-valueb |
|---|
| UMOD-P1 | DQSRVLNLGPITR | 16.79-21.71 | 19.96 | 1.11 | 17.18-20.70 | 19.58 | 0.76 | 12.98-20.07 | 18.53 | 1.20 |
1.89×10−6 |
1.32×10−5 | 0.00044 | 0.00311 |
| UMOD-P2 | IDQSRVLNLGPITR | 11.74-22.43 | 20.26 | 2.37 | 17.89-21.40 | 20.26 | 0.76 | 13.47-20.79 | 19.19 | 1.22 | 0.01102 | 0.01928 | 0.01410 | 0.02467 |
| UMOD-P3 | QSRVLNLGPITR | 10.89-18.61 | 16.77 | 1.53 | 14.54-17.47 | 16.52 | 0.65 | 9.75-17.02 | 15.52 | 1.19 | 0.00012 | 0.00042 | 0.00248 | 0.00867 |
| UMOD-P4 |
SGSVIDQSRVLNLGPITR | - | - | - | 23.30-26.17 | 25.23 | 0.70 | 15.80-26.04 | 24.06 | 1.76 | - | - | 0.03157 | 0.04420 |
| UMOD-P5 |
SGSVIDQSRVLNLGPITRK | 11.38-22.34 | 19.84 | 2.26 | 17.69-21.88 | 19.69 | 0.88 | 15.24-20.47 | 18.61 | 1.08 | 0.00242 | 0.00565 | 0.00929 | 0.02168 |
Diagnostic performance of urine
endogenous peptides
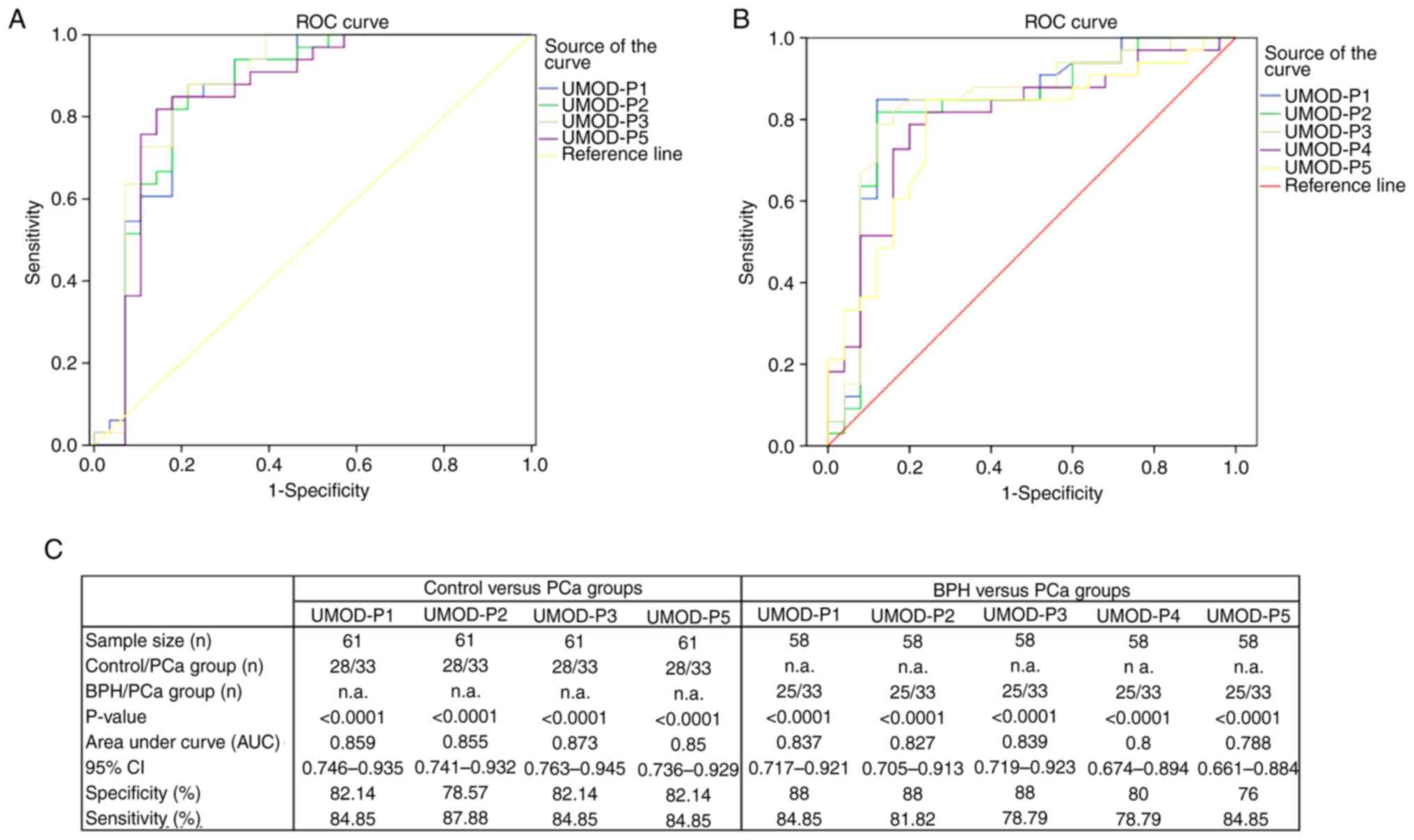
ROC analyses were performed for urinary peptides
that showed differential abundance between the study groups. ROC
curves were constructed using the log2-transformed precursor peak
area values of UMOD-P1, UMOD-P2, UMOD-P3, UMOD-P4 and UMOD-P5 in
samples from PCa, BPH and control groups (Fig. 2). UMOD peptides showed similar
potential to discriminate between control and PCa groups,
displaying AUC values between 0.850-0.873, with sensitivity and
specificity ranging from 84.85 to 87.88% and 78.57 to 82.14%,
respectively (Fig. 2A and C). In
BPH × PCa analysis, UMOD peptides showed AUC values between 0.788
and 0.839, with sensitivity and specificity ranging from 78.79 to
84.85% and 76 to 88%, respectively (Fig. 2B and C). UMOD-P1 showed the best
diagnostic performance for differentiating BPH and PCa groups, with
sensitivity and specificity levels of 84.85 and 88%, respectively.
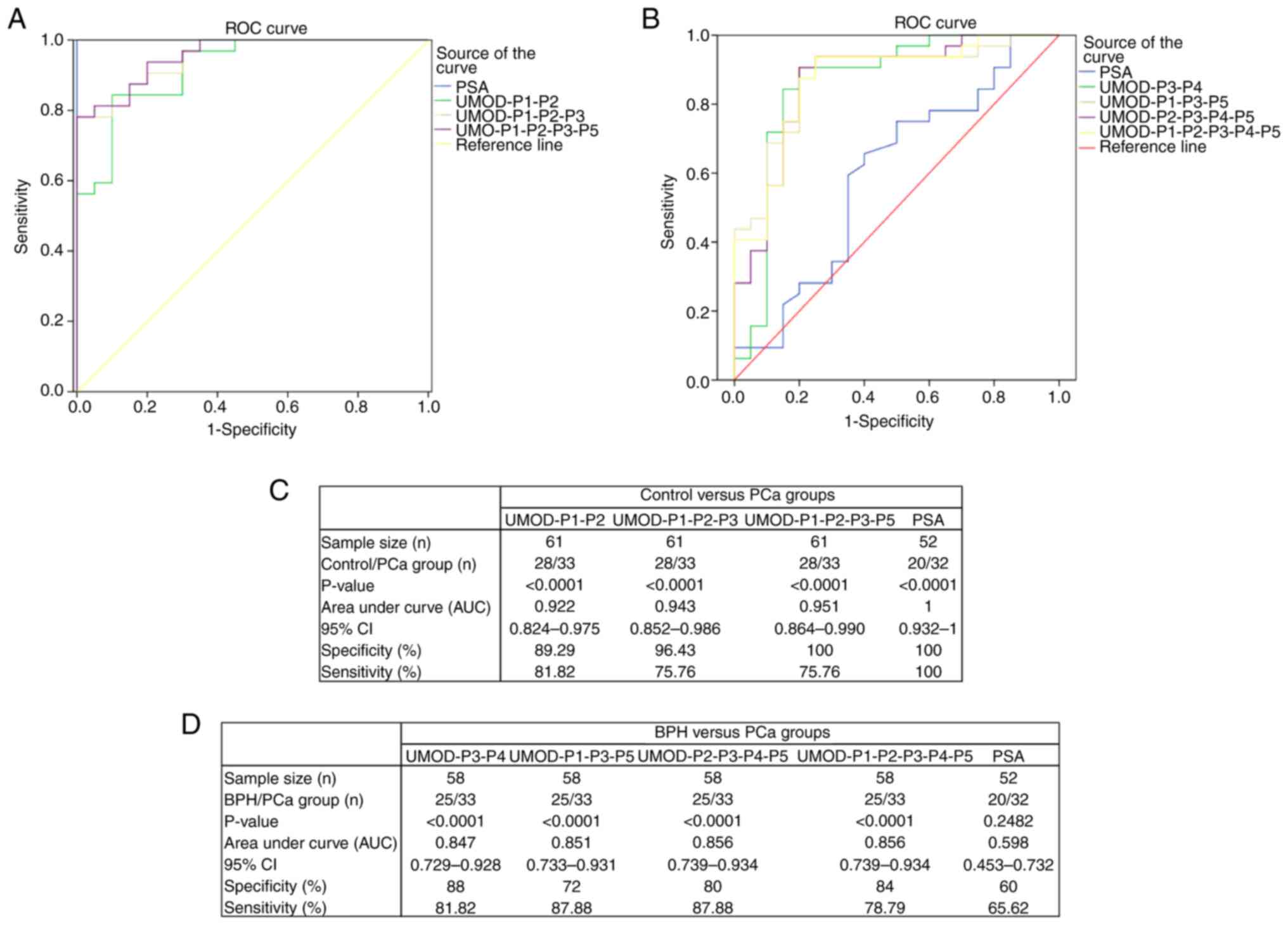
We then evaluated the ability of combinations of two or more
peptides to discriminate between study groups (Fig. 3). Peptide combinations resulted in
AUC values between 0.922 and 0.951 for control × PCa analysis, with
sensitivity and specificity levels ranging from 75.76 to 81.82% and
89.29 to 100%, respectively (Fig. 3A
and C). The combination of 4 peptides (UMOD-P1, UMOD-P2,
UMOD-P3 and UMOD-P5) showed the best performance for discriminating
control and PCa groups, with sensitivity and specificity levels of
75.76 and 100%, respectively. AUC values between 0.847 and 0.856
were obtained for BPH × PCa analysis using peptide combinations,
with sensitivity and specificity levels ranging from 78.79 to
87.88% and 72 to 88%, respectively (Fig. 3B and D). The combination of UMOD-P3
and UMOD-P4 showed high sensitivity and specificity (81.82 and 88%,
respectively) in discriminating between benign and malignant
prostate conditions.
We also performed a direct comparison of our
biomarker panel with PSA levels (Fig.
3). Peptide panel resulted in specificity levels similar to
those seen for PSA (100%) in a control vs. PCa analysis (Fig. 3A and C), but with lower sensitivity.
It is important to consider that PSA levels <4.0 ng/ml was an
inclusion criterion for the control group, which may be influencing
the high levels of sensitivity and specificity observed for PSA. On
the other hand, our urinary peptide panel significantly
outperformed the PSA in BPH × PCa comparison (Fig. 3B and D), showing significantly
higher AUC values and sensitivity/specificity levels
(P<0.0001).
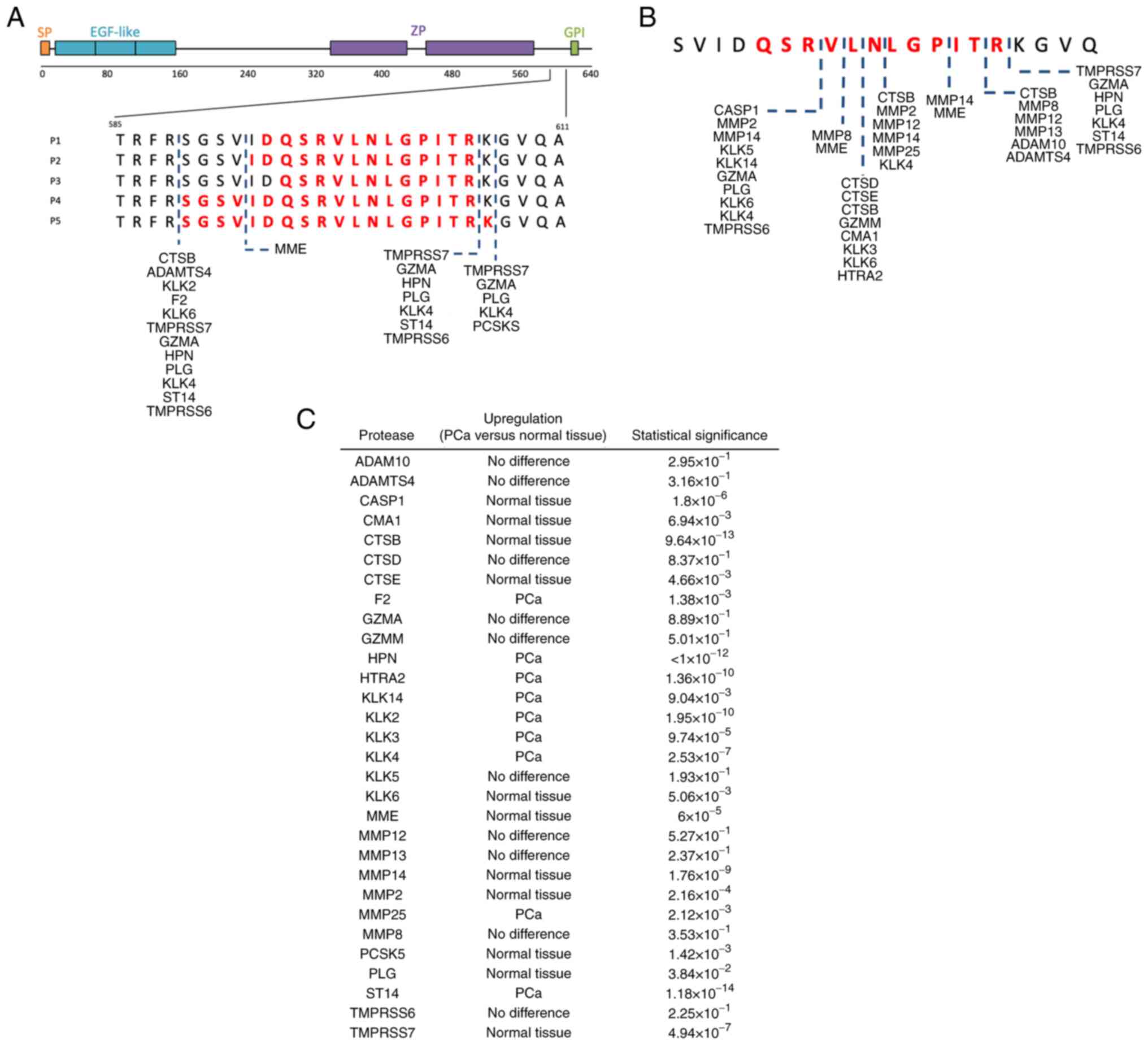
Prediction of proteases potentially
involved in the generation of UMOD urinary peptides
In silico analyses using Proteasix indicated
that different proteolytic events could be involved in generating
UMOD-P1 to UMOD-P5. A total of 104 combinations of predicted
proteases and cleavage sites were identified using a >80%
specificity threshold (Table SIV).
The predicted proteases are summarized in Fig. 4A. Among predicted proteases, 13
(~93%) predominantly cleave at the carboxyl side of arginine or
lysine residues. No N-terminal cleavage site was identified for
UMOD-P1 and UMOD-P3 peptides with the filtering criteria used.
Since UMOD-P1 to UMOD-P5 showed significantly
reduced abundance in PCa samples, we speculate that these peptides
are generated under normal physiological conditions and undergo
additional proteolytic events under malignant conditions of the
prostate. Therefore, we searched in silico for proteases
able to hydrolyze the smallest identified UMOD peptide (UMOD-P3).
Proteins potentially involved in the proteolysis of UMOD-P3 were
summarized in Fig. 4B and Table SV. The expression profile of the
identified proteases in PCa and normal tissue were retrieved from
UALCAN portal (Fig. 4C). Proteases
KLK3, KLK4, KLK14, HTRA2 and MMP25 were reported to be
overexpressed in PCa tumor and could be potentially involved in the
degradation of UMOD peptides in individuals with PCa. These
additional cleavage events may explain the reduced abundance of
UMOD peptides in the urine of individuals with PCa compared to BPH
and control groups.
Discussion
Most prostate tumors grow slowly and are confined to
the prostate gland, with a good prognosis if diagnosed early.
However, the PSA-based tests currently used for PCa diagnosis,
including age-adjusted PSA ranges, PSA velocity, PSA density and
percentage of free PSA, have important limitations (4,16). PSA
has low specificity for discriminating between malignant and benign
prostatic conditions, as well as to differentiate between indolent
and aggressive disease, leading to overdiagnosis and overtreatment
(4). Thus, new biomarkers have been
proposed to improve PCa diagnosis, including the urinary RNAs PCA3
and TMPRSS2-ERG (17). The clinical
utility of these biomarkers for PCa diagnosis is still conflicting
across different studies, which describe AUC values ranging from
0.660 to 0.770 (8,9,17).
Assays based on these molecules, such as Progensa and Mi-Prostate
Score (MiPS), have been used mainly to assist in the decision to
perform a new biopsy in the case of an inconclusive first biopsy
(8,9).
Reliable biomarkers able to provide an early and
accurate diagnosis may improve patient management, reducing
biopsies and overtreatment. In this work, we applied simple and
low-cost protocols for peptide biomarker discovery in urine for PCa
diagnosis. Using this approach, we identified a panel of urinary
peptides with high specificity in differentiating between PCa and
control groups (AUC=0.951). Most importantly, these urinary
peptides outperformed PSA in discriminating between malignant and
benign prostate conditions (AUC=0.847), showing high sensitivity
(81.82%) and specificity (88%). Overall, urinary peptides resulted
in AUC values comparable to or greater than those observed for
other urinary PCa biomarkers based on RNA, DNA or proteins
(8,17).
Our sample preparation protocol is simplest,
cheapest and fastest when compared to experimental approaches
traditionally used in the isolation of urinary protein/peptide
biomarkers for LC-MS analysis (6,18–23).
We analyzed naturally occurring urinary peptides and, therefore,
our protocol did not include an enzymatic protein digestion step
(e.g., trypsin digestion). Furthermore, we used TCA protein
precipitation to isolate endogenous urinary peptides, eliminating
expensive devices and time-consuming centrifugation steps typical
of ultrafiltration-based protocols (10). Overall, the standardized methodology
is compatible with LC-MS-based assays for routine clinical
applications, which could facilitate its future translation for
medicine and patient care.
Numerous studies have revealed the high intertumoral
and intratumoral heterogeneity in PCa (24). Therefore, a single biomarker is
unlikely to provide the sensitivity and specificity needed to
accurately diagnose and stratify PCa, as every single biomarker has
its own performance limits. Thus, more recent studies have focused
on the identification and assessment of multiple biomarkers to
improve diagnostic accuracy and disease risk stratification
(25,26). Here, we found that combining
peptides in a biomarker panel improved diagnostic performance
compared to individual biomarkers, showing higher AUC values in
discriminating PCa from control and BPH groups.
The invasive techniques for cancer diagnosis and
monitoring are slowly being replaced by liquid biopsies. Liquid
biopsies allow for easy and minimally invasive sample collection
for biomarker detection and quantification (7,27). In
this scenario, urine has been recognized as a good source of
biomarkers for urological tumors, including PCa (28). However, many studies use prostate
massage prior sample collection for biomarker discovery with the
aim of increasing the amount of prostate-derived molecules released
in the urine (29,30). Furthermore, it is important to
consider that a significant portion of men still refuse DRE on
account of discomfort or embarrassment. Thus, our study was based
on the analysis of biomarkers present in urine without previous
prostate massage in order to increase patient acceptance and
adherence in a future clinical application.
About 70% of urinary proteins are derived from
exosomes, secretory/excretory products and cells shed from
urogenital tract, including prostate gland. As urine is stored in
the bladder for hours, digestion of urinary proteins occurs prior
to voiding by the action of endogenous proteases (31). Therefore, the presence/abundance of
urinary peptides is altered according to the
physiological/pathological state of main urogenital tissues. Cancer
cells secret proteases that can act on proteins present in the
urine, ultimately leading to a differential abundance of urinary
peptides in individuals with cancer (18,32).
Previous studies have shown that endogenous urinary peptide
signatures have diagnostic value for PCa, especially in
discriminating between PCa and BPH, resulting in high sensitivity
(67.4 to 91.7%) and specificity (71.2 to 90.5%) (33–35).
Our urinary peptide panel outperformed PSA in discriminating
between PCa and BPH, showing high levels of sensitivity (87.88%)
and specificity (88%).
The urinary peptides found in our study were derived
from uromodulin (UMOD), also known as Tamm-Horsfall glycoprotein.
UMOD is the most abundant protein found in human urine under
physiological conditions (36).
Previous studies have identified UMOD peptides in urine under
physiological conditions, as part of urinary peptidome of healthy
individuals (19). Thus,
alterations in the presence and/or abundance of urinary UMOD
peptides may be associated with the individual's pathological
state. Urinary UMOD peptides identified across multiple studies are
derived from the C-terminal region of the protein, around 589–607
residues, which suggest that this region is more sensitive or
accessible to the action of endogenous proteases (20–22).
We identified five UMOD-derived urinary peptides with reduced
abundance in PCa samples. M'Koma et al (35) also identified urinary UMOD peptides
with reduced levels in patients with PCa. These results suggest
that PCa-related proteases may be acting on the further degradation
of UMOD peptides normally found in urine. From in silico
analyses of cleavage sites and expression data in PCa × normal
tissue, we identified proteases HTRA2, KLK3, KLK4, KLK14, and MMP25
as potentially acting on degradation of the identified UMOD
peptides.
Kallikrein-related peptidases (KLKs) are serine
proteases that are upregulated in PCa (37), including KLK2, KLK3 (also known as
PSA), KLK4, KLK11 and KLK14-15. These proteases have therefore been
proposed for use as PCa biomarkers (37,38).
KLKs are secreted by prostate cancer cells and many of them have
already been detected in human biofluids, including urine (23,37).
MMP25 (also known as MT6-MMP) was found upregulated in malignant
prostate tissue compared to benign prostate tissue (39). This metalloproteinase displays
intrinsic proteolytic activity towards extracellular matrix
components and therefore could play a direct role in prostate tumor
invasion. MMP25 is a GPI-anchored protein and its presence in
urinary exosomes has already been reported (40).
Although our results indicated the potential of
urinary peptides as biomarkers for PCa, they should be interpreted
considering the limitations of the study, including a relatively
small and age-biased cohort. Despite these limitations, the cohort
was appropriate to assess the feasibility of profiling endogenous
urine peptides and to estimate the potential of these peptides as
diagnostic biomarkers for PCa. The present study therefore
represents the first phase of the biomarker development pipeline
(the discovery phase), in which a small number of individual
samples are analyzed to identify biomarker candidates (10). Next, it will be necessary to further
evaluate the specificity of the identified peptides for use in PCa
diagnosis, since urinary peptides derived from UMOD have already
been described as altered in other pathophysiological conditions
(21–22,41).
It is noteworthy that, although the urine proteome is altered under
conditions of urinary tract infection or inflammation, no changes
were found in urinary levels of UMOD or enzymes potentially
involved in its proteolysis in samples from cases of urinary tract
infection or from animal models of prostatic inflammation (42,43).
Lastly, our findings require confirmatory studies using larger
age-matched cohorts to validate the pathophysiological relevance of
the identified urinary peptides and their potential use as PCa
biomarkers.
In conclusion, the profiling of urine by LC-MS
allowed the identification of endogenous peptides with potential
for use as PCa biomarkers. In addition, our peptide panel was able
to discriminate between individuals with PCa or BPH with high
sensitivity and specificity, overcoming an important limitation of
currently available biomarkers. We also identified
disease-associated proteases potentially involved in the
degradation of uromodulin peptides detected at low levels in the
urine of patients with PCa.
Supplementary Material
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Acknowledgements
The authors would like to thank Dr Ricardo Kaufmann
(Urology Service, Ernesto Dornelles Hospital, Porto Alegre, RS,
Brazil) for his assistance with the collection of urine and biopsy
specimens.
Funding
This work was supported by Programa de Pesquisa para o SUS:
Gestão Compartilhada em Saúde-PPSUS (grant no. 17/2551-0001 412-0),
Fundação de Amparo à Pesquisa do Estado do Rio Grande do Sul
(FAPERGS), Departamento de Ciência e Tecnologia da Secretaria de
Ciência, Tecnologia e Insumos Estratégicos do Ministério da Saúde
(DECIT/SCTIE/MS), Conselho Nacional de Desenvolvimento Científico e
Tecnológico (CNPq), and Secretaria da Saúde do Estado do Rio Grande
do Sul (SES/RS). CSD and DRK were supported by CNPq scholarship. MH
was supported by FAPERGS scholarship. DCS and RMP were supported by
Universidade Federal do Rio Grande do Sul (BIC/UFRGS)
scholarship.
Availability of data and materials
The mass spectrometry datasets generated during the
current study are available in the PRIDE repository with the
dataset identifier PXD037031 (https://www.ebi.ac.uk/pride/archive/projects/PXD037031).
Other datasets generated and/or analyzed during the current study
are available from the corresponding author on reasonable
request.
Authors' contributions
CDSD and KMM participated in the conception and
design of the study, and reviewed the literature. CDSD drafted the
manuscript. AZ, GB, DRK, DJB, HBF, KMM, LGP, RCD and MFB made
substantial contributions to the acquisition of data. CDSD, DDCS,
KMM, MH, RMP and NRM analyzed the data. HBF, AZ, KMM and LGP
critically revised the intellectual content of the manuscript prior
to submission. CDSD and KMM confirm the authenticity of all the raw
data. Each author participated sufficiently in the work to take
public responsibility for appropriate portions of the content. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
The study was approved by the Institutional Review
Boards of Universidade Federal do Rio Grande do Sul (UFRGS),
Universidade de Santa Cruz do Sul (UNISC), Hospital Ernesto
Dornelles (HED) and Hospital Ana Nery under the protocol CAAE
number 69852617.1.1001.5347. All subjects provided written informed
consent and the study was performed following the guidelines of The
Declaration of Helsinki.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global Cancer Statistics 2020:
GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36
Cancers in 185 Countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Heidenreich A, Bellmunt J, Bolla M, Joniau
S, Mason M, Matveev V, Mottet N, Schmid HP, Van Der Kwast T, Wiegel
T, et al: EAU guidelines on prostate cancer. Part 1: Screening,
diagnosis, and treatment of clinically localised disease. Eur Urol.
59:61–71. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gretzer MB and Partin AW: PSA levels and
the probability of prostate cancer on biopsy. Eur Urol. (Suppl
1):21–27. 2002. View Article : Google Scholar
|
|
4
|
Thompson IM, Pauler DK, Goodman PJ, Tangen
CM, Lucia MS, Parnes HL, Minasian LM, Ford LG, Lippman SM, Crawford
ED, et al: Prevalence of prostate cancer among men with a
prostate-specific antigen level < or =4.0 ng per milliliter. N
Engl J Med. 350:2239–2246. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Oshi M, Murthy V, Takahashi H, Huyser M,
Okano M, Tokumaru Y, Rashid OM, Matsuyama R, Endo I and Takabe K:
Urine as a source of liquid biopsy for cancer. Cancers (Basel).
13:26522021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wood SL, Knowles MA, Thompson D, Selby PJ
and Banks RE: Proteomic studies of urinary biomarkers for prostate,
bladder and kidney cancers. Nat Rev Urol. 10:206–218. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jedinak A, Loughlin KR and Moses MA:
Approaches to the discovery of non-invasive urinary biomarkers of
prostate cancer. Oncotarget. 9:32534–32550. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Koo KM, Mainwaring PN, Tomlins SA and Trau
M: Merging new-age biomarkers and nanodiagnostics for precision
prostate cancer management. Nat Rev Urol. 16:302–317. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tonry C, Finn S, Armstrong J and
Pennington SR: Clinical proteomics for prostate cancer:
Understanding prostate cancer pathology and protein biomarkers for
improved disease management. Clin Proteomics. 17:412020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Parker BL, Burchfield JG, Clayton D,
Geddes TA, Payne RJ, Kiens B, Wojtaszewski JFP, Richter EA and
James DE: Multiplexed temporal quantification of the
exercise-regulated plasma peptidome. Mol Cell Proteomics.
16:2055–2068. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Perez-Riverol Y, Bai J, Bandla C,
García-Seisdedos D, Hewapathirana S, Kamatchinathan S, Kundu DJ,
Prakash A, Frericks-Zipper A, Eisenacher M, et al: The PRIDE
database resources in 2022: A Hub for mass spectrometry-based
proteomics evidences. Nucleic Acids Res. 50((D1)): D543–D552. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Schilling B, Rardin MJ, MacLean BX,
Zawadzka AM, Frewen BE, Cusack MP, Sorensen DJ, Bereman MS, Jing E,
Wu CC, et al: Platform-independent and label-free quantitation of
proteomic data using MS1 extracted ion chromatograms in skyline:
Application to protein acetylation and phosphorylation. Mol Cell
Proteomics. 11:202–214. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Willforss J, Chawade A and Levander F:
NormalyzerDE: Online tool for improved normalization of omics
expression data and high-sensitivity differential expression
analysis. J Proteome Res. 18:732–740. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Klein J, Eales J, Zürbig P, Vlahou A,
Mischak H and Stevens R: Proteasix: A tool for automated and
large-scale prediction of proteases involved in naturally occurring
peptide generation. Proteomics. 13:1077–1082. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and
Varambally S: UALCAN: A portal for facilitating tumor subgroup gene
expression and survival analyses. Neoplasia. 19:649–658. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Loeb S and Partin AW: Review of the
Literature: PCA3 for prostate cancer risk assessment and
prognostication. Rev Urol. 13:e191–e195. 2011.PubMed/NCBI
|
|
17
|
Fujita K and Nonomura N: Urinary
biomarkers of prostate cancer. Int J Urol. 25:770–779. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zheng B, Zhang P, Wang H, Wang J, Liu ZH
and Zhang DH: Advances in research on bladder cancer targeting
peptides: A review. Cell Biochem Biophys. 79:711–718. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mavrogeorgis E, Mischak H, Latosinska A,
Siwy J, Jankowski V and Jankowski J: Reproducibility evaluation of
urinary peptide detection using CE-MS. Molecules. 26:72602021.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Van JAD, Clotet-Freixas S, Zhou J, Batruch
I, Sun C, Glogauer M, Rampoldi L, Elia Y, Mahmud FH, Sochett E, et
al: Peptidomic analysis of urine from youths with early type 1
diabetes reveals novel bioactivity of uromodulin peptides in vitro.
Mol Cell Proteomics. 19:501–517. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nkuipou-Kenfack E, Bhat A, Klein J,
Jankowski V, Mullen W, Vlahou A, Dakna M, Koeck T, Schanstra JP,
Zürbig P, et al: Identification of ageing-associated naturally
occurring peptides in human urine. Oncotarget. 6:34106–34117. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Good DM, Zürbig P, Argilés À, Bauer HW,
Behrens G, Coon JJ, Dakna M, Decramer S, Delles C, Dominiczak AF,
et al: Naturally occurring human urinary peptides for use in
diagnosis of chronic kidney disease. Mol Cell Proteomics.
9:2424–2437. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Principe S, Kim Y, Fontana S, Ignatchenko
V, Nyalwidhe JO, Lance RS, Troyer DA, Alessandro R, Semmes OJ,
Kislinger T, et al: Identification of prostate-enriched proteins by
in-depth proteomic analyses of expressed prostatic secretions in
urine. J Proteome Res. 11:2386–2396. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Haffner MC, Zwart W, Roudier MP, True LD,
Nelson WG, Epstein JI, De Marzo AM, Nelson PS and Yegnasubramanian
S: Genomic and phenotypic heterogeneity in prostate cancer. Nat Rev
Urol. 18:79–92. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xiao K, Guo J, Zhang X, Feng X, Zhang H,
Cheng Z, Johnson H, Persson JL and Chen L: Use of two gene panels
for prostate cancer diagnosis and patient risk stratification.
Tumour Biol. 37:10115–10122. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Alarcón-Zendejas AP, Scavuzzo A,
Jiménez-Ríos MA, Álvarez-Gómez RM, Montiel-Manríquez R,
Castro-Hernández C, Jiménez-Dávila MA, Pérez-Montiel D,
González-Barrios R, Jiménez-Trejo F, et al: The promising role of
new molecular biomarkers in prostate cancer: From coding and
non-coding genes to artificial intelligence approaches. Prostate
Cancer Prostatic Dis. 5:431–443. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lone SN, Nisar S, Masoodi T, Singh M,
Rizwan A, Hashem S, El-Rifai W, Bedognetti D, Batra SK, Haris M, et
al: Liquid biopsy: A step closer to transform diagnosis, prognosis
and future of cancer treatments. Mol Cancer. 21:792022. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hessels D and Schalken JA: Urinary
biomarkers for prostate cancer: A review. Asian J Androl.
15:333–339. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Padoan A, Basso D, Zambon CF,
Prayer-Galetti T, Arrigoni G, Bozzato D, Moz S, Zattoni F, Bellocco
R and Plebani M: MALDI-TOF peptidomic analysis of serum and
post-prostatic massage urine specimens to identify prostate cancer
biomarkers. Clin Proteomics. 15:232018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Clarke RA, Schirra HJ, Catto JW, Lavin MF
and Gardiner RA: Markers for detection of prostate cancer. Cancers
(Basel). 2:1125–1154. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Good DM, Thongboonkerd V, Novak J,
Bascands JL, Schanstra JP, Coon JJ, Dominiczak A and Mischak H:
Body fluid proteomics for biomarker discovery: Lessons from the
past hold the key to success in the future. J Proteome Res.
6:4549–4555. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Schönemeier B, Metzger J, Klein J, Husi H,
Bremer B, Armbrecht N, Dakna M, Schanstra JP, Rosendahl J, Wiegand
J, et al: Urinary peptide analysis differentiates pancreatic cancer
from chronic pancreatitis. Pancreas. 45:1018–1026. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Theodorescu D, Fliser D, Wittke S, Mischak
H, Krebs R, Walden M, Ross M, Eltze E, Bettendorf O, Wulfing C and
Semjonow A: Pilot study of capillary electrophoresis coupled to
mass spectrometry as a tool to define potential prostate cancer
biomarkers in urine. Electrophoresis. 26:2797–2808. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Okamoto A, Yamamoto H, Imai A, Hatakeyama
S, Iwabuchi I, Yoneyama T, Hashimoto Y, Koie T, Kamimura N, Mori K,
et al: Protein profiling of post-prostatic massage urine specimens
by surface-enhanced laser desorption/ionization time-of-flight mass
spectrometry to discriminate between prostate cancer and benign
lesions. Oncol Rep. 21:73–79. 2009.PubMed/NCBI
|
|
35
|
M'Koma AE, Blum DL, Norris JL, Koyama T,
Billheimer D, Motley S, Ghiassi M, Ferdowsi N, Bhowmick I, Chang
SS, et al: Detection of pre-neoplastic and neoplastic prostate
disease by MALDI profiling of urine. Biochem Biophys Res Commun.
353:829–834. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Serafini-Cessi F, Malagolini N and
Cavallone D: Tamm-Horsfall glycoprotein: Biology and clinical
relevance. Am J Kidney Dis. 42:658–676. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Fuhrman-Luck RA, Loessner D and Clements
JA: Kallikrein-Related peptidases in prostate cancer: From
molecular function to clinical application. EJIFCC.
25:2692014.PubMed/NCBI
|
|
38
|
Hong SK: Kallikreins as biomarkers for
prostate cancer. Biomed Res Int. 2014:5263412014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Riddick ACP, Shukla CJ, Pennington CJ,
Bass R, Nuttall RK, Hogan A, Sethia KK, Ellis V, Collins AT,
Maitland NJ, et al: Identification of degradome components
associated with prostate cancer progression by expression analysis
of human prostatic tissues. Br J Cancer. 92:2171–2180. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Shimoda M and Khokha R: Metalloproteinases
in extracellular vesicles. Biochim Biophys Acta Mol Cell Res.
1864((11 Pt A)): 1989–2000. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Mary S, Small HY, Siwy J, Mullen W, Giri A
and Delles C: Polymerization-incompetent uromodulin in the pregnant
stroke-prone spontaneously hypertensive rat. Hypertens. 69:910–918.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yu Y, Sikorski P, Smith M, Bowman-Gholston
C, Cacciabeve N, Nelson KE and Pieper R: Comprehensive
metaproteomic analyses of urine in the presence and absence of
neutrophil-associated inflammation in the urinary tract.
Theranostics. 7:238–252. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wei P, Hao L, Ma F, Yu Q, Buchberger AR,
Lee S, Bushman W and Li L: Urinary metabolomic and proteomic
analyses in a mouse model of prostatic inflammation. Urine (Amst).
1:17–23. 2019. View Article : Google Scholar : PubMed/NCBI
|