|
1
|
Llovet JM, Zucman-Rossi J, Pikarsky E,
Sangro B, Schwartz M, Sherman M and Gores G: Hepatocellular
carcinoma. Nat Rev Dis Primers. 2:160182016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Di Bisceglie AM, Rustgi VK, Hoofnagle JH,
Dusheiko GM and Lotze MT: NIH conference. Hepatocellular carcinoma.
Ann Intern Med. 108:390–401. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Villanueva A and Llovet JM: Targeted
therapies for hepatocellular carcinoma. Gastroenterology.
140:1410–1426. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rizzo A, Ricci AD and Brandi G: Systemic
adjuvant treatment in hepatocellular carcinoma: Tempted to do
something rather than nothing. Future Oncol. 16:2587–2589. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Santoni M, Rizzo A, Mollica V, Matrana MR,
Rosellini M, Faloppi L, Marchetti A, Battelli N and Massari F: The
impact of gender on The efficacy of immune checkpoint inhibitors in
cancer patients: The MOUSEION-01 study. Crit Rev Oncol Hematol.
170:1035962022. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rizzo A, Ricci AD and Brandi G:
Trans-arterial chemoembolization plus systemic treatments for
hepatocellular carcinoma: An update. J Pers Med. 12:17882022.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Rizzo A, Ricci AD and Brandi G:
Atezolizumab in advanced hepatocellular carcinoma: Good things come
to those who wait. Immunotherapy. 13:637–644. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bazzichetto C, Conciatori F, Pallocca M,
Falcone I, Fanciulli M, Cognetti F, Milella M and Ciuffreda L: PTEN
as a Prognostic/Predictive biomarker in cancer: An unfulfilled
promise? Cancers (Basel). 11:4352019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sengupta S and Parikh ND: Biomarker
development for hepatocellular carcinoma early detection: Current
and future perspectives. Hepat Oncol. 4:111–122. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ptolemy AS and Rifai N: What is a
biomarker? Research investments and lack of clinical integration
necessitate a review of biomarker terminology and validation
schema. Scand J Clin Lab Invest. 242:6–14. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tovar V, Cornella H, Moeini A, Vidal S,
Hoshida Y, Sia D, Peix J, Cabellos L, Alsinet C, Torrecilla S, et
al: Tumour initiating cells and IGF/FGF signalling contribute to
sorafenib resistance in hepatocellular carcinoma. Gut. 66:530–540.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sia D, Villanueva A, Friedman SL and
Llovet JM: Liver cancer cell of origin, molecular class, and
effects on patient prognosis. Gastroenterology. 152:745–761. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jing JS, Ye W, Jiang YK, Ma J, Zhu MQ, Ma
JM, Zhou H, Yu LQ, Yang YF and Wang SC: The value of GPC3 and GP73
in clinical diagnosis of hepatocellular carcinoma. Clin Lab.
63:1903–1909. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wongjarupong N, Negron-Ocasio GM,
Chaiteerakij R, Addissie BD, Mohamed EA, Mara KC, Harmsen WS,
Theobald JP, Peters BE, Balsanek JG, et al: Model combining
pre-transplant tumor biomarkers and tumor size shows more utility
in predicting hepatocellular carcinoma recurrence and survival than
the BALAD models. World J Gastroenterol. 24:1321–1331. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ono K, Kokubu S, Hidaka H, Watanabe M,
Nakazawa T and Saigenji K: Risk factors of delay in restoration of
hepatic reserve capacity and local recurrence after radiofrequency
ablation therapy for hepatocellular carcinoma (HCC). Hepatol Res.
31:172–177. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhu M, Zheng J, Wu F, Kang B, Liang J,
Heskia F, Zhang X and Shan Y: OPN is a promising serological
biomarker for hepatocellular carcinoma diagnosis. J Med Virol.
92:3596–3603. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Luo XY, Wu KM and He XX: Advances in drug
development for hepatocellular carcinoma: Clinical trials and
potential therapeutic targets. J Exp Clin Cancer Res. 40:1722021.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cancer Genome Atlas Research Network, .
Electronic address: simplewheeler@bcm.edu; Cancer
Genome Atlas Research Network: Comprehensive and integrative
genomic characterization of hepatocellular carcinoma. Cell.
169:1327–1341. e13232017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Schulze K, Imbeaud S, Letouze E,
Alexandrov LB, Calderaro J, Rebouissou S, Couchy G, Meiller C,
Shinde J, Soysouvanh F, et al: Exome sequencing of hepatocellular
carcinomas identifies new mutational signatures and potential
therapeutic targets. Nat Genet. 47:505–511. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hardy T and Mann DA: Epigenetics in liver
disease: From biology to therapeutics. Gut. 65:1895–1905. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Luo J, Dai X, Hu H, Chen J, Zhao L, Yang
C, Sun J, Zhang L, Wang Q, Xu S, et al: Fluzoparib increases
radiation sensitivity of non-small cell lung cancer (NSCLC) cells
without BRCA1/2 mutation, a novel PARP1 inhibitor undergoing
clinical trials. J Cancer Res Clin Oncol. 146:721–737. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang D, Zhao L, Wang D, Liu J, Yu X, Wei Y
and Ouyang Z: Transcriptome analysis and identification of key
genes involved in 1-deoxynojirimycin biosynthesis of mulberry
(Morus alba L.). PeerJ. 6:e54432018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang Y, Kang Z, Lv D, Zhang X, Liao Y, Li
Y, Liu R, Li P, Tong M, Tian J, et al: Longitudinal whole-genome
sequencing reveals the evolution of MPAL. Cancer Genet. 240:59–65.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Anwanwan D, Singh SK, Singh S, Saikam V
and Singh R: Challenges in liver cancer and possible treatment
approaches. Biochim Biophys Acta Rev Cancer. 1873:1883142020.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shi X, Chen G and Liu X, Qiu Y, Yang S,
Zhang Y, Fang X, Zhang C and Liu X: Scutellarein inhibits cancer
cell metastasis in vitro and attenuates the development of
fibrosarcoma in vivo. Int J Mol Med. 35:31–38. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cheng CY, Hu CC, Yang HJ, Lee MC and Kao
ES: Inhibitory effects of scutellarein on proliferation of human
lung cancer A549 cells through ERK and NFkappaB mediated by the
EGFR pathway. Chin J Physiol. 57:182–187. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Parajuli P, Joshee N, Rimando AM, Mittal S
and Yadav AK: In vitro antitumor mechanisms of various Scutellaria
extracts and constituent flavonoids. Planta Med. 75:41–48. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Guo F, Yang F and Zhu YH: Scutellarein
from Scutellaria barbata induces apoptosis of human colon cancer
HCT116 cells through the ROS-mediated mitochondria-dependent
pathway. Nat Prod Res. 33:2372–2375. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Goh D, Lee YH and Ong ES: Inhibitory
effects of a chemically standardized extract from Scutellaria
barbata in human colon cancer cell lines, LoVo. J Agric Food Chem.
53:8197–8204. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ha SE, Kim SM, Vetrivel P, Kim HH, Bhosale
PB, Heo JD, Lee HJ and Kim GS: Inhibition of cell proliferation and
metastasis by scutellarein regulating PI3K/Akt/NF-kappaB signaling
through PTEN activation in hepatocellular carcinoma. Int J Mol Sci.
22:88412021. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wiencke JK, Zheng S, Jelluma N, Tihan T,
Vandenberg S, Tamgüney T, Baumber R, Parsons R, Lamborn KR, Berger
MS, et al: Methylation of the PTEN promoter defines low-grade
gliomas and secondary glioblastoma. Neuro Oncol. 9:271–279. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tamguney T and Stokoe D: New insights into
PTEN. J Cell Sci. 120:4071–4079. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang L, Wang WL, Zhang Y, Guo SP, Zhang J
and Li QL: Epigenetic and genetic alterations of PTEN in
hepatocellular carcinoma. Hepatol Res. 37:389–396. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Pan L, Lu J, Wang X, Han L, Zhang Y, Han S
and Huang B: Histone deacetylase inhibitor trichostatin a
potentiates doxorubicin-induced apoptosis by up-regulating PTEN
expression. Cancer. 109:1676–1688. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kondo Y: Targeting histone
methyltransferase EZH2 as cancer treatment. J Biochem. 156:249–257.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Marino-Ramirez L, Kann MG, Shoemaker BA
and Landsman D: Histone structure and nucleosome stability. Expert
Rev Proteomics. 2:719–729. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Medrzycki M, Zhang Y, McDonald JF and Fan
Y: Profiling of linker histone variants in ovarian cancer. Front
Biosci (Landmrk Ed). 17:396–406. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chen ZH, Zhu M, Yang J, Liang H, He J, He
S, Wang P, Kang X, McNutt MA, Yin Y and Shen WH: PTEN interacts
with histone H1 and controls chromatin condensation. Cell Rep.
8:2003–2014. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Martin M: CUTADAPT removes adapter
sequences from high-throughput sequencing reads. EMBnet J.
17:10–12. 2011. View Article : Google Scholar
|
|
40
|
Dobin A, Davis CA, Schlesinger F, Drenkow
J, Zaleski C, Jha S, Batut P, Chaisson M and Gingeras TR: STAR:
Ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li B and Dewey CN: RSEM: Accurate
transcript quantification from RNA-Seq data with or without a
reference genome. BMC Bioinformatics. 12:3232011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Sun J, Nishiyama T, Shimizu K and Kadota
K: TCC: An R package for comparing tag count data with robust
normalization strategies. BMC Bioinformatics. 14:2192013.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Love MI, Huber W and Anders S: Moderated
estimation of fold change and dispersion for RNA-seq data with
DESeq2. Genome Biol. 15:5502014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Robinson MD, McCarthy DJ and Smyth GK:
edgeR: A Bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Liao Y, Wang J, Jaehnig EJ, Shi Z and
Zhang B: WebGestalt 2019: Gene set analysis toolkit with revamped
UIs and APIs. Nucleic Acids Res. 47:W199–W205. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Young MD, Wakefield MJ, Smyth GK and
Oshlack A: Gene ontology analysis for RNA-seq: Accounting for
selection bias. Genome Biol. 11:R142010. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Pettersen EF, Goddard TD, Huang CC, Couch
GS, Greenblatt DM, Meng EC and Ferrin TE: UCSF Chimera-a
visualization system for exploratory research and analysis. J
Comput Chem. 25:1605–1612. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Boosani CS and Agrawal DK: PTEN
modulators: A patent review. Expert Opin Ther Pat. 23:569–580.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhu AX, Rosmorduc O, Evans TR, Ross PJ,
Santoro A, Carrilho FJ, Bruix J, Qin S, Thuluvath PJ, Llovet JM, et
al: SEARCH: A phase III, randomized, double-blind,
placebo-controlled trial of sorafenib plus erlotinib in patients
with advanced hepatocellular carcinoma. J Clin Oncol. 33:559–566.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Byron SA, Van Keuren-Jensen KR,
Engelthaler DM, Carpten JD and Craig DW: Translating RNA sequencing
into clinical diagnostics: opportunities and challenges. Nat Rev
Genet. 17:257–271. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Garber ME, Troyanskaya OG, Schluens K,
Petersen S, Thaesler Z, Pacyna-Gengelbach M, van de Rijn M, Rosen
GD, Perou CM, Whyte RI, et al: Diversity of gene expression in
adenocarcinoma of the lung. Proc Natl Acad Sci USA. 98:13784–13789.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Cajigas-Du Ross CK, Martinez SR,
Woods-Burnham L, Durán AM, Roy S, Basu A, Ramirez JA,
Ortiz-Hernández GL, Ríos-Colón L, Chirshev E, et al: RNA sequencing
reveals upregulation of a transcriptomic program associated with
stemness in metastatic prostate cancer cells selected for taxane
resistance. Oncotarget. 9:30363–30384. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Sos ML, Koker M, Weir BA, Heynck S,
Rabinovsky R, Zander T, Seeger JM, Weiss J, Fischer F, Frommolt P,
et al: PTEN loss contributes to erlotinib resistance in EGFR-mutant
lung cancer by activation of Akt and EGFR. Cancer Res.
69:3256–3261. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Martire S and Banaszynski LA: The roles of
histone variants in fine-tuning chromatin organization and
function. Nat Rev Mol Cell Biol. 21:522–541. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Malik HS and Henikoff S: Phylogenomics of
the nucleosome. Nat Struct Biol. 10:882–891. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Thatcher TH and Gorovsky MA: Phylogenetic
analysis of the core histones H2A, H2B, H3, and H4. Nucleic Acids
Res. 22:174–179. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Freeman L, Kurumizaka H and Wolffe AP:
Functional domains for assembly of histones H3 and H4 into the
chromatin of Xenopus embryos. Proc Natl Acad Sci USA.
93:12780–12785. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Liu Z, Zhu Y, Gao J, Yu F, Dong A and Shen
WH: Molecular and reverse genetic characterization of NUCLEOSOME
ASSEMBLY PROTEIN1 (NAP1) genes unravels their function in
transcription and nucleotide excision repair in Arabidopsis
thaliana. Plant J. 59:27–38. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Ong J, van den Berg A, Faiz A, Boudewijn
IM, Timens W, Vermeulen CJ, Oliver BG, Kok K, Terpstra MM, van den
Berge M, et al: Current smoking is associated with decreased
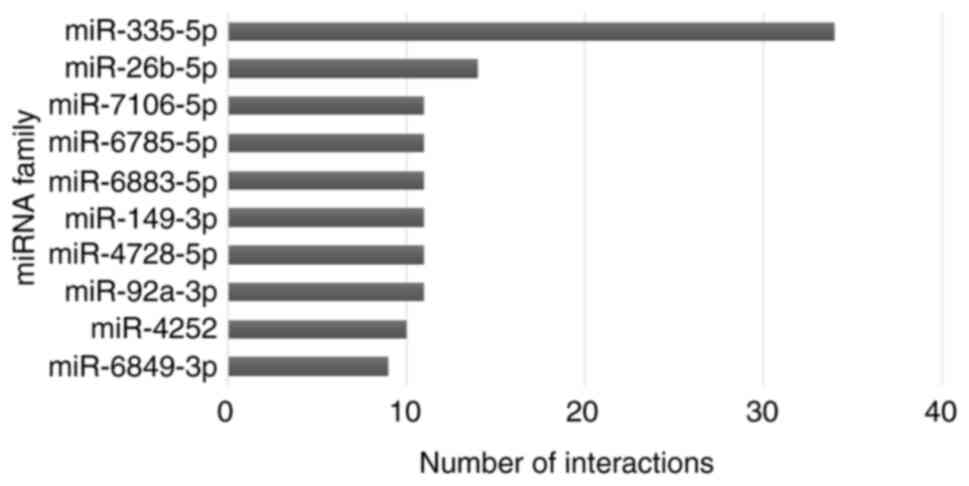
expression of miR-335-5p in parenchymal lung fibroblasts. Int J Mol
Sci. 20:51762019. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Li LM, Liu ZX and Cheng QY: Exosome plays
an important role in the development of hepatocellular carcinoma.
Pathol Res Pract. 215:1524682019. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Xiao Y, Zheng S, Duan N, Li X and Wen J:
MicroRNA-26b-5p alleviates cerebral ischemia-reperfusion injury in
rats via inhibiting the N-myc/PTEN axis by downregulating KLF10
expression. Hum Exp Toxicol. 40:1250–1262. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Wang Y, Sun B, Sun H, Zhao X, Wang X, Zhao
N, Zhang Y, Li Y, Gu Q, Liu F, et al: Regulation of proliferation,
angiogenesis and apoptosis in hepatocellular carcinoma by
miR-26b-5p. Tumour Biol. 37:10965–10979. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Fan X, Kraynak J, Knisely JPS, Formenti SC
and Shen WH: PTEN as a guardian of the genome: Pathways and
targets. Cold Spring Harb Perspect Med. 10:a0361942020. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Chew A, Salama P, Robbshaw A, Klopcic B,
Zeps N, Platell C and Lawrance IC: SPARC, FOXP3, CD8 and CD45
correlation with disease recurrence and long-term disease-free
survival in colorectal cancer. PLoS One. 6:e220472011. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Hafsi S, Pezzino FM, Candido S, Ligresti
G, Spandidos DA, Soua Z, McCubrey JA, Travali S and Libra M: Gene
alterations in the PI3K/PTEN/AKT pathway as a mechanism of
drug--resistance (review). Int J Oncol. 40:639–644. 2012.PubMed/NCBI
|