Esophageal cancer (EC), which can be divided into
esophageal squamous cell carcinoma (ESCC) and esophageal
adenocarcinoma (EAC), is a common malignancy of the digestive tract
(1). In 2020, ~604,000 new cases
were reported and ~544,000 EC-associated mortalities were reported
worldwide (2). EAC occurs mostly in
North America and Western Europe, while ESCC occurs mostly in China
(3,4). ESCC is usually caused by smoking,
alcohol intake, poor diet, viral infection or genetic factors
(5). Surgical resection,
chemotherapy and radiotherapy are the primary therapeutic
strategies of ESCC. Since ESCC is mostly asymptomatic in the early
stages, patients often miss the optimal therapeutic opportunity
(6). In addition, the therapy for
ESCC is often ineffective due to metastasis and medication
resistance (7). Therefore, the
identification of promising diagnostic biomarkers and the
development of effective therapeutic strategies are crucial in
combatting ESCC.
Non-coding RNAs (ncRNAs) encompass a range of types
of RNA, with microRNAs (miRNAs/miRs), long ncRNAs (lncRNAs) and
circular RNAs (circRNAs) being the most well-known. They are
transcribed from DNA but lack the ability to encode proteins and
were previously considered to be ‘transcriptional waste’ (8–10).
However, with the advent of high-throughput sequencing technology,
recent research has revealed that ncRNAs constitute >90% of the
human genome (11). By contrast,
protein-coding genes make up <2% of the genome (12–14).
NcRNAs are widely involved in various cellular processes, such as
cell proliferation, differentiation, aging, apoptosis and immune
responses (15–18). Furthermore, extensive research has
highlighted the significant dysregulation of expression of numerous
ncRNAs in ESCC cells and tissues. This dysregulation is closely
linked to ESCC progression, drug resistance and unfavorable patient
prognosis (19–22). The present review provides a
comprehensive overview of the functions of miRNAs, lncRNAs and
circRNAs in ESCC, and investigates their roles as diagnostic
biomarkers and potential therapeutic targets for ESCC treatment.
This information highlights novel ideas and strategies for the
early diagnosis and potential therapeutic targets for ESCC.
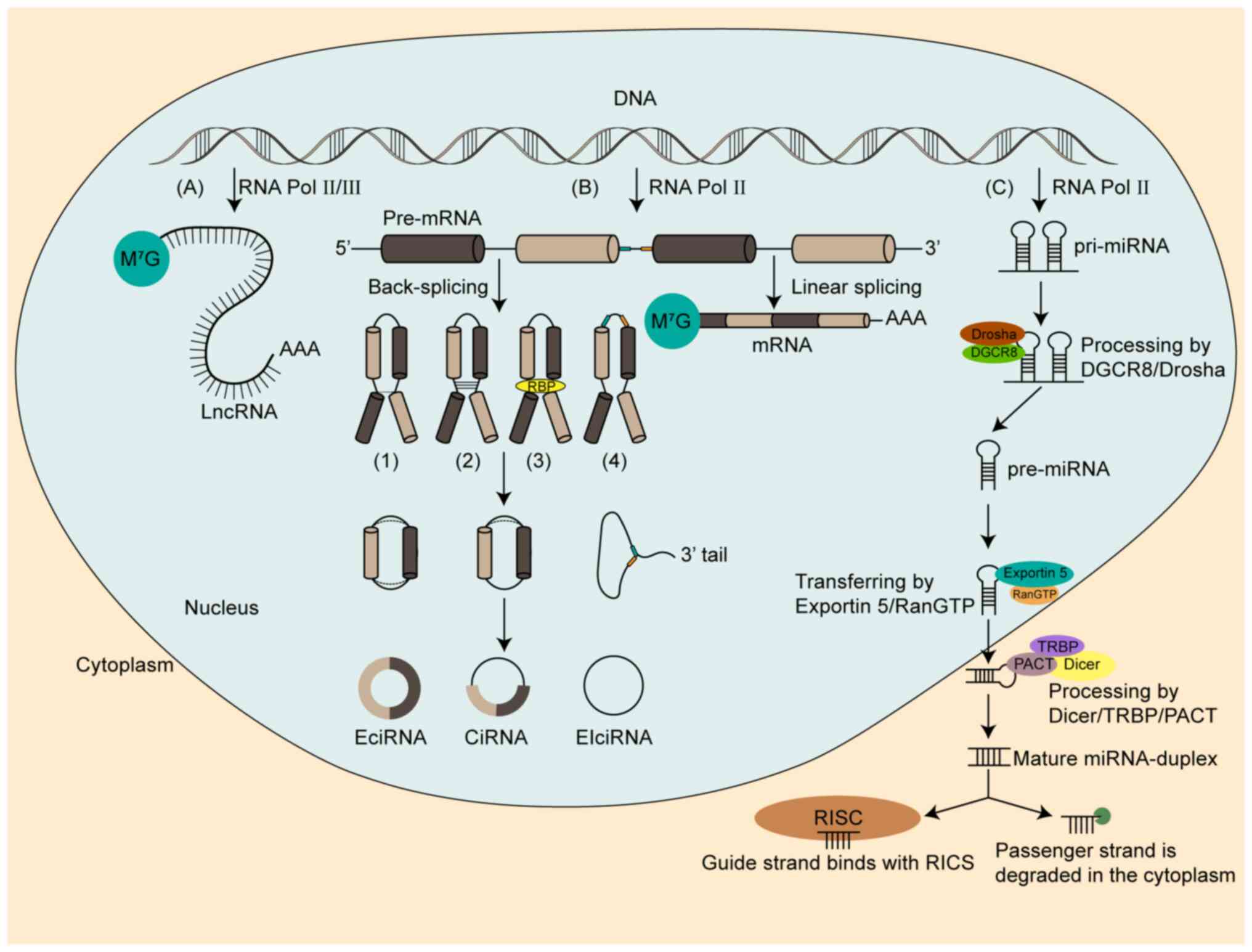
The generation of ncRNAs is a multifaceted and
intricate process that involves several sequential stages. In the
subsequent sections, we provide a comprehensive and detailed
explanation of the formation mechanisms that underlie miRNAs,
lncRNAs and circRNAs (Fig. 1).
MiRNAs are a category of endogenous and highly
conserved small ncRNAs that consist of ~22 nucleotides (23,24).
Processing involves two stages. In the first stage, RNA polymerase
II catalyzes DNA fragments in the nucleus to produce primary miRNA
(pri-miRNA) (25); subsequently, a
microprocessor complex, consisting of Drosha, an RNase type III
protein and DiGeorge syndrome critical region gene 8, recognizes
and cleaves the stem strand on the pri-miRNA hairpin to form
pre-miRNA (26). Thereafter, Ran
guanosine triphosphate-Exportin 5 facilitates pre-miRNA transfers
from the nucleus to the cytoplasm (27,28).
In the second stage, a mature double-stranded miRNA is generated
after pre-miRNA is identified by Dicer with trans-activation
response RNA-binding protein/protein activator of the
interferon-induced protein kinase (28,29).
The stem-loop structure is then cleaved and modified in the
cytoplasm (30,31). The guide strand attaches to the
RNA-induced silencing complex and targets the 3′-untranslated
Region (3′-UTR) of the mRNA via base complementary pairing when the
mature miRNA unwinds (32). The
passenger strand is degraded in the cytoplasm (33). Complete base-pair complementation
between miRNA and mRNA then causes cleavage of mRNA. An incomplete
match can inhibit protein translation of mRNA, which is the most
common mode of action of miRNA (34–37).
LncRNAs are a particular subtype of ncRNAs that are
longer than 200 nucleotides and are produced by RNA polymerase II
or RNA polymerase III (38,39). They are composed of multiple exons
and have a structure similar to mRNA. However, lncRNA transcripts
have fewer exons, lower overall expression levels and poorer
sequence conservation compared with mRNA (40,41).
LncRNAs can be divided into six types: i + ii) Bidirectional and
intergenic lncRNAs; iii) sense and iv) antisense exonic lncRNAs;
and v) sense and vi) antisense intronic lncRNAs, and they are able
to regulate different stages of transcription (42,43).
In pre-transcription, lncRNAs participate in the
regulation of histone acetylation and DNA methylation (44,45).
They also play a regulatory role during transcription (46). In addition, lncRNAs are directly
involved in post-transcriptional regulation of mRNA. Some lncRNAs
that contain miRNA complementary sites can change the effect of
miRNA by binding to the mRNA via a competing endogenous RNA (ceRNA)
mechanism (47). LncRNAs can also
regulate RNA editing (48). It is
clear that lncRNAs have multiple functions and are crucial for the
control of transcription.
CircRNAs are covalently closed, single-stranded
ncRNAs formed by back-splicing of precursor mRNA (pre-mRNA)
(49). CircRNAs lack 5′ caps and 3′
poly (A) tails and are classified into three types: i) Exonic
circRNAs (EciRNA); ii) intronic circRNAs (ciRNA); and iii)
exon-intron circRNAs (EIciRNA) (50,51).
They are highly conserved, diverse and widely expressed in
different cells and tissues at various stages of development
(52–54). The closed-loop structure and the
absence of a 5′ cap or 3′ poly (A) tail in circRNAs also make them
less susceptible to exonuclease R compared with linear RNA
(55).
CircRNAs can competitively bind to proteins, serving
as miRNA decoys or protein scaffolds (56). EciRNAs can control the levels of
targeted genes by acting as miRNA sponges, due to their numerous
miRNA binding sites in the cytoplasm (57,58).
CiRNAs and EIciRNAs have few binding sites with miRNA in the
nucleus; however, they can regulate gene transcription in the
nucleus (59–61). Moreover, some circRNAs contain
extensive N6-methyladenosine (m6A) modification sites and can
translate proteins after m6A modification (62). Studies have shown that a single m6A
site is sufficient to drive the initiation of translation (63,64).
This m6A-driven translation requires the involvement of eukaryotic
translation initiation factor 4 gamma 2 (eIF4G2) and m6A reader YTH
m6A RNA binding protein (YTHDF)3. It is also enhanced by
methyltransferase like 3/14 methyltransferase, inhibited by fat
mass and obesity-associated protein (FTO) demethylase and
upregulated upon heat shock (65,66).
Insulin-like growth factor 2 mRNA-binding protein 1 facilitates
circMAP3K4 peptide translation and then promotes hepatocellular
carcinoma progression, which is driven by m6A modification
(67). In addition, accumulating
evidence suggests that several circRNAs are related to polysomes,
and they contain an AUG initiation codon and putative open reading
frames of optimal length (65,68).
This indicates an unexpected potential for protein coding by these
circRNAs.
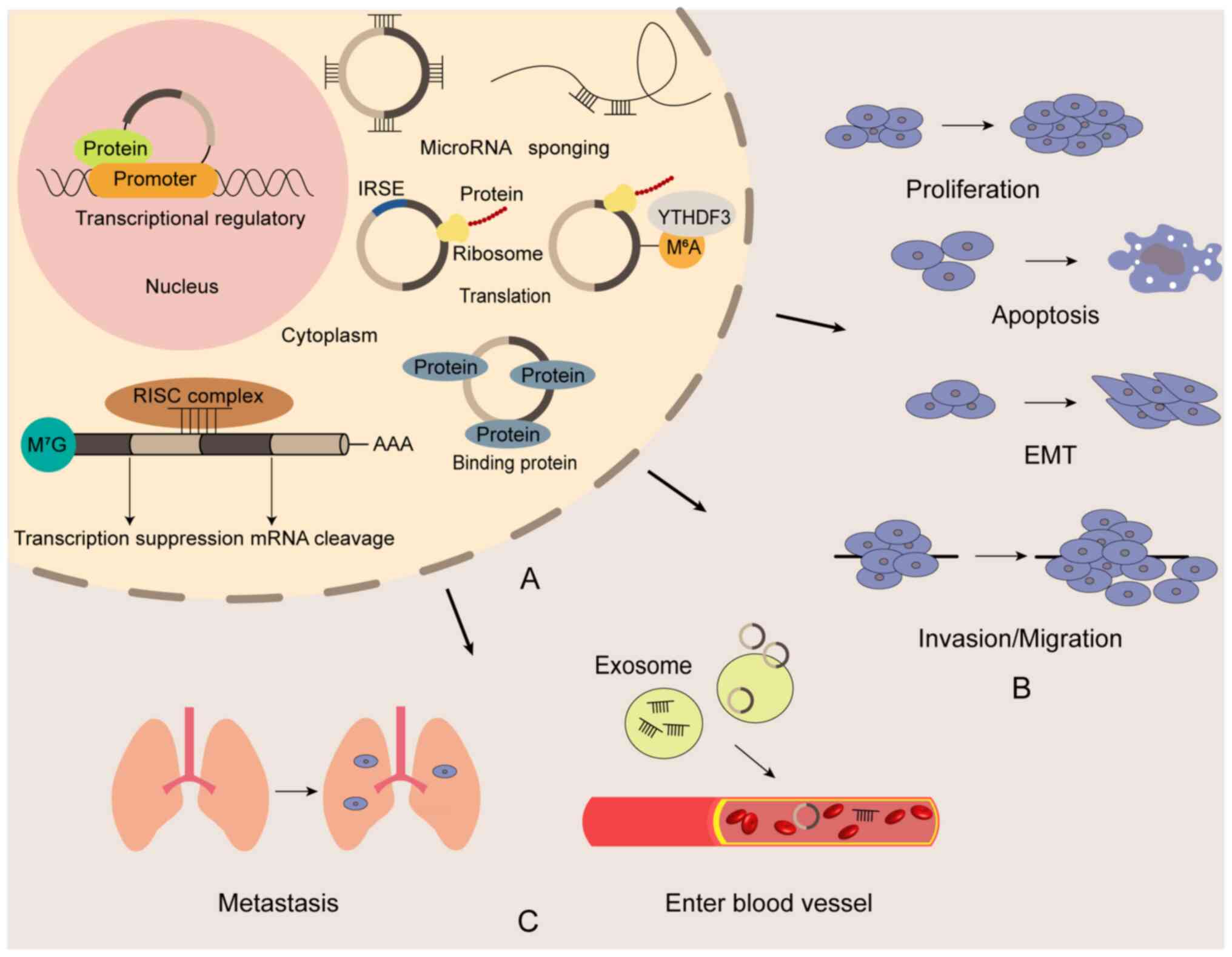
The pathogenesis of ESCC involves dysregulation,
activation, inactivation and mutation of multiple genes, with the
exact mechanisms still not fully understood (69). Recent studies emphasize the critical
role of ncRNAs in ESCC development (70–72)
(Fig. 2). Cui et al
(73) revealed that m6A-mediated
epigenetic modification of lncRNA is closely associated with ESCC
tumorigenesis. The FTO m6A demethylase can mediate the
demethylation of lncRNA LINC00022 in a YTHDF2-dependent manner,
promoting its accumulation in cells. Elevated levels of LINC00022
can bind specifically to the p21 protein and promote its
ubiquitination-mediated degradation, thereby upregulating the
levels of cyclin dependent kinase 2 and cyclin E1 proteins
(73). This leads to a shortened
G0/G1 phase and prolonged G2/M
phase in the cells, thus driving ESCC development (73). In addition, a study found that the
lncRNA HOX transcript antisense RNA can promote ESCC tumorigenesis
by binding to miR-1 and upregulating cyclin D1 expression, thereby
regulating changes in the cell cycle (74). These findings emphasize the critical
role of ncRNAs in ESCC pathogenesis. Current research predominantly
explores how ncRNAs promote ESCC progression by regulating cell
proliferation, metastasis and therapy resistance (21,75,76).
Therefore, this article focuses on elucidating the specific
mechanisms and current research status of ncRNAs in regulating ESCC
cell proliferation, metastasis, and therapy resistance, alongside
their potential as diagnostic markers for ESCC.
The miR-106b-5p oncogene is commonly upregulated in
various malignancies, such as ESCC (80,81).
In patients with ESCC, elevated miR-106b-5p levels are associated
with decreased expression of the 15-hydroxyprostaglandin
dehydrogenase (HPGD) tumor suppressor gene (82). Dual luciferase reporter assays have
confirmed HPGD as a direct target of miR-106b-5p, and miR-106b-5p
expression is inversely correlated with HPGD levels (82). Overexpression of miR-106b-5p
promotes tumor cell proliferation and inhibits apoptosis, while
upregulation of HPGD reverses these effects (82).
The levels of miR-497 have been found to be lower in
ESCC tissues and cells when compared with normal tissues (83). An increase in miR-497 expression
suppresses ESCC cell growth, while a reduction in miR-497
expression promotes ESCC cell proliferation (83). Furthermore, overexpression of
miR-497 in ESCC cells injected into BALB/c nude mice results in a
decreased level of proliferating cell nuclear antigen, increased
cleaved caspase-3 expression and inhibited tumor growth (83). Mechanistically, it has been revealed
that the YY1 protein can inhibit HIF2α expression by binding to the
HIF2α promoter region, and miR-497 upregulates YY1 protein
expression (83).
The regulatory impacts of lncRNAs on the progression
of ESCC have been shown in several studies. For example, the lncRNA
small nucleolar RNA host gene 16 (SNHG16) oncogene is upregulated
in the early stages of ESCC (85).
It can enhance the stability of Ras homologue family member U and
promote the proliferation of ESCC cells by recruiting the EIF4A3
RNA binding protein-eukaryotic translation initiation factor
(85). When cells that overexpress
SNHG16 are injected into mice, tumor growth is accelerated
(85). Similarly, lncRNA GK
intronic transcript 1 can competitively bind to mitogen-activated
protein kinase 1 (MAPK1). This prevents its interaction with dual
specificity phosphatase 6 and inhibits transmission of the
extracellular signal-regulated kinase/MAPK1 signaling pathway,
which is mediated by dual specificity phosphatase 6 (86). This facilitates growth and
suppresses apoptosis of ESCC cells (86).
CircRNAs that are aberrantly expressed have been
found to facilitate the proliferative ability of ESCC cells. Xiong
et al (88) demonstrated
that circNELL2 can activate the translation of cell division
cyclin-6 by acting as a sponge for miR-127-5p, which in turn
encourages the growth of ESCC cells. MiR-124 has been demonstrated
to be an anti-oncogene and shows markedly declined levels in ESCC
(89,90). The levels of circ2646 are elevated
and inversely correlated with miR-124 expression in ESCC (91). Increased circ2646 reduces miR-124
levels and promotes tumor cell proliferation (91). Yao et al (92) report elevated circHIPK3 levels in
ESCC tissues when compared with normal tissues, which correlates
with lymph node metastasis (LNM) and poor tumor differentiation.
Knockdown of circHIPK3 inhibits ESCC cell proliferation by acting
as a sponge for miR-124 (92).
These findings suggest that circRNAs can boost ESCC cell
proliferation and tumor formation. Moreover, some circRNAs have
been shown to increase apoptosis of ESCC cells (93,94).
Jiang et al (95) found that
low circ0086414 expression correlates with poor prognosis in
patients with ESCC. They observed that circ0086414 inhibits tumor
cell proliferation, suppresses glycolysis and increases apoptosis
in ESCC cells. These findings highlight the importance of circRNAs
in ESCC development and suggest potential therapeutic strategies
that target circRNAs.
The aforementioned studies highlight the involvement
of ncRNAs in the regulation of tumor cell proliferation via
intricate mechanisms. Future research should aim for a deeper
understanding of these mechanisms, to distinguish between ncRNAs
that promote or inhibit cancer progression. Such insights can guide
clinical approaches to treating ESCC.
The two main factors that contribute to therapy
failure in individuals with advanced-stage ESCC are local invasion
and distant metastases (96). It is
crucial to understand the molecular processes behind the spread of
ESCC and to investigate prospective treatment targets. The present
review provides a summary of current research on the connection
between ncRNAs and ESCC metastasis.
The levels of miR-17-5p and miR-4443 are elevated in
ESCC cells and correlate with the tumor node metastasis (TNM)
stage. Both miR-17-5p and miR-4443 promote ESCC cell invasion and
migration by targeting metallopeptidase-2 (97). Exosomes, which transport proteins,
lipids and nucleic acids, are vital mediators of intercellular
communication (98). Exosomes
secreted by tumor cells are crucial for the development and spread
of malignancies (99). Liu et
al (100) revealed a positive
correlation between the LNM of patients with ESCC and miR-320b
levels in ESCC cell-derived exosomes. MiR-320b is transferred to
human lymphatic endothelial cells via exosomes, inhibiting
programmed cell death 4, activating protein kinase B (AKT)
phosphorylation and promoting peritumoral lymphangiogenesis
(100). It also enhances ESCC cell
invasion and migration through these pathways (100). In addition, certain miRNAs have
been shown to prevent ESCC cell invasion and migration. For
instance, miR-338-5p targets epidermal growth factor receptor
(EGFR) and the hepatocyte growth factor receptor (cMET) (101). Increasing miR-338-5p levels
reduces cMET and EGFR expression and blocks their signaling
pathways (101). Consequently,
activation of growth factor receptor-bound protein 2-associated
binding protein 1, AKT and extracellular signal-regulated kinase
are inhibited (101). This
prevents the extracellular matrix from remodeling and impedes ESCC
cell invasion and migration (101). Moreover, it has been discovered
that miR-485-5p significantly inhibits the mRNA and protein
expression levels of Flotillin-1 by binding to its 3′-UTR,
resulting in increased E-cadherin expression in ESCC cells, whereas
the expression levels of Vimentin, N-cadherin, and zinc finger
E-box-binding homeobox 1 were decreased (102). This impedes the invasion and
migration of ESCC cells by inhibiting the epithelial-mesenchymal
transition (EMT) (102). These
results underline the critical role that miRNAs play in controlling
ESCC cell invasion and migration.
Recently, several lncRNAs have been revealed to act
as oncogenic genes and promote ESCC invasion and migration
(103,104). The lncRNA LINC02820 correlates
with poor prognosis in patients with ESCC. While its overexpression
does not impact ESCC cell proliferation, it enhances cell invasion
and migration (105). It also
causes an increase in levels of the cortactin and phosphorylated
Neural Wiskott-Aldrich syndrome protein invadopodia markers
(105). A number of malignancies
show high levels of expression of the GTPase-activating protein
(SH3 domain)-binding protein 2 (G3BP2) oncogene (106,107). Zheng et al (108) found that G3BP2 can promote ESCC
invasion and migration by upregulating the levels of
hepatoma-derived growth factor mRNA, and the poor prognosis of
patients with ESCC has a positive relationship with G3BP2
expression. The lncRNA LINC01554 binds to the RNA recognition motif
of G3BP2 and prolongs its half-life by reducing the ubiquitination
level of G3BP2. This indicates that LINC01554 is essential for the
oncogenic effect of G3BP2 (108).
The lncRNA BAALC-AS1 can also directly interact with G3BP2 to block
G3BP2-mediated c-Myc degradation, and to enable ESCC invasion and
migration (109). A study has
revealed a correlation between the lncRNA PTPRG-AS1 and pyruvate
dehydrogenase kinase 1 (PDK1) expression in tumor cells (110). Notably, PTPRG-AS1 has been
revealed to regulate PDK1 expression by acting as a sponge for
miR-599. Silencing PTPRG-AS1 has been shown to inhibit ESCC
glycolysis, invasion and migration (110).
CircRNAs have been shown to regulate ESCC invasion
and migration through their ability to act as a sponge for various
types of miRNAs. For example, Shi et al (111) revealed that circLPAR3 upregulates
MET gene expression and activates the RAS protein/MAPK and
phosphoinositide 3-kinase/AKT pathways by acting as a sponge for
miR-198. This promotes ESCC cell migration, invasion and
metastasis. Meanwhile, Xu et al (112) demonstrated that circ0000654
promotes interleukin-6 secretion by acting as a sponge for
miR-149-5p, which enhances ESCC cell migration by activating signal
transducer and activator of transcription 3 (STAT3) signaling. This
leads to malignant progression of the disease. Wang et al
(113) demonstrated that
circ0087378 correlates with poor overall survival (OS) in patients
with ESCC and is highly expressed in ESCC cells. The authors
observed a negative correlation between circ0087378 expression and
miR-140-3p levels. Through binding to miR-140-3p, circ0087378
alleviates the suppressive effect of miR-140-3p on its target gene,
E2F transcription factor 3 (E2F3). This circ0087378/miR-140-3p/E2F3
axis is crucial for promoting ESCC cell invasion and migration
(114). The EMT facilitates tumor
progression by altering cell-cell adhesion, polarity and
morphology, which enhances tumor cell invasion and metastasis
(114,115).
Numerous studies have demonstrated that, through
affecting the EMT process, circRNAs may regulate the capacity of
ESCC to invade and migrate. For instance, circLONP2 can promote EMT
and lead to ESCC metastasis by regulating the miR-27b-3p/zinc
finger E-box binding homeobox 1 axis (116). Chen et al (117) revealed that by reducing
circ0004370, EMT-related protein expression can be decreased and
ESCC cell invasion and migration can be prevented. Mechanically,
collagen type I α 1 expression is upregulated by circ0004370 acting
as a sponge for miR-1301-3p (117). Moreover, circARAP2 knockdown
elevates E-Cadherin expression, while suppressing N-Cadherin
expression. However, these effects are reversed when miR-761 is
reduced or Forkhead Box M1 is upregulated (118). The downregulation of circARAP2
results in decreased expression of ESCC tumor stem cell-related
genes and inhibits ESCC progression (118). Thus, targeting circRNAs in ESCC
may represent a novel strategy to inhibit metastasis (118).
In summary, ncRNAs are pivotal regulators of ESCC
metastasis, offering potential as therapeutic targets to curb tumor
migration and recurrence. Further research is warranted to
elucidate their precise mechanisms and optimize their utilization
in controlling metastasis.
While chemotherapy and radiotherapy remain the
primary treatment modalities for ESCC, frequent drug resistance is
a major contributing factor to poor treatment outcomes. In recent
years, several reports have indicated a significant association
between ncRNAs and resistance to chemotherapy and radiotherapy in
ESCC (119–121).
Recent studies have shown that miRNAs are involved
in regulating drug resistance in ESCC. For example, Zuo et
al (122) demonstrated that
overexpression of miR-153-3p inhibits the growth of ESCC cells and
increases the level of cleaved caspase-3 after treatment with
cisplatin. This indicates that upregulation of miR-153-3p may
improve cisplatin resistance in ESCC cells. Previous studies have
demonstrated that cancer-associated fibroblasts (CAFs) contribute
to cisplatin resistance in ESCC (123,124). Another recent study further
revealed the critical role of miRNAs in CAFs-induced cisplatin
resistance (125,126). Exosomes secreted by CAFs, enriched
with miR-21, inhibit phosphatase and tensin homolog (PTEN)
activation in monocytes, which leads to STAT3 activation. This
promotes monocyte differentiation into myeloid-derived suppressor
cells, ultimately inducing cisplatin resistance (127). Gou et al (128) revealed that patients with ESCC
with low miR-29b expression demonstrate increased resistance to
radiotherapy. The authors identified a miR-29b binding site in the
3′-UTR of BTG anti-proliferation factor 2 (BTG2) mRNA.
Downregulation of miR-29b increases BTG2 expression, decreases
cyclin D1 expression and leads to cell cycle arrest in the
G0/G1 phase of radiotherapy resistant ESCC
cells (128). These findings
suggest that targeting miRNAs could be a viable strategy to
mitigate resistance to therapy in ESCC (128).
LncRNAs play a crucial role in drug resistance in
ESCC. The ALKBH5-mediated m6A demethylation process upregulates
lnc-cancer susceptibility 8 (lncCASC8) expression, which induces
drug resistance in ESCC cells. LncCASC8 targets heterogeneous
nuclear ribonucleoprotein L, preventing its ubiquitination and
stabilizing its expression. This leads to increased Bcl2 production
and decreased cleaved caspase-3 levels, ultimately resulting in
ESCC drug resistance (129). The
lnc-non-coding RNA activated by DNA damage (NORAD) is associated
with cisplatin resistance and poor prognosis in patients with ESCC
(130). Upregulation of lncNORAD
increases the IC50 value of cisplatin and reduces
apoptosis in ESCC cells treated with cisplatin. Mechanistically,
lncNORAD acts as a sponge for miR-224-3p and represses miR-224-3p
expression (130). Downregulation
of miR-224-3p decreases the expression of its target gene,
metadherin, and inhibits nuclear accumulation of β-catenin, which
results in cisplatin resistance in ESCC (130). The lncPOU3F3 from tumor cell
exosomes induces cisplatin resistance in ESCC by promoting the
conversion of normal fibroblasts to CAFs (131). In addition, Wang et al
(132) demonstrated that
lnc-Taurine upregulated 1 (lncTUG1) knockdown, combined with
radiotherapy, significantly inhibits ESCC cell growth, when
compared to radiotherapy alone. Further investigation revealed that
lncTUG1 acts as a sponge for miR-144-3p, promoting MET expression.
Decreased lncTUG1 levels are associated with reduced MET and AKT
phosphorylation, enhancing the efficacy of radiotherapy (132). These findings highlight the
regulatory role of the lncTUG1/miR-144-3p/MET axis in ESCC
radiotherapy resistance (132).
CircRNAs are also closely associated with ESCC drug
resistance. The circ0014879 increases in radiotherapy resistant
ESCC cells and controls the miR-519-3p/cell division cycle 25A axis
to cause drug resistance (133).
The upregulation of circ0000277 in cisplatin-resistant ESCC
enhances drug resistance by modulating miR-873-5p and activating
the SRY-box transcription factor 4/Wnt/β-catenin axis (134). Knockdown of circ0000277 promotes
ESCC cell apoptosis and reduces cisplatin resistance, as evidenced
by increased proapoptotic protein levels (PARP and caspase-3) and
decreased IC50 values, following cisplatin treatment
(134). Zhou et al
(135) revealed that
circ-glutamic-oxaloacetic transaminase 1 (circGOT1) is highly
expressed in cisplatin-resistant ESCC cells, while miR-606
expression is reduced. This circRNA upregulates the expression of
its host gene, GOT1, by acting as a sponge for miR-606. Inhibition
of circGOT1 decreases ESCC cell glycolysis and enhances cisplatin
sensitivity (135). These effects
are reversed by miR-606 knockdown or GOT1 overexpression. It is
hypothesized that circGOT1 promotes ESCC resistance via the
circGOT1/miR-606/GOT1 axis (135).
Certain circRNAs, such as circDOPEY2, inhibit ESCC
resistance and circDOPEY2 is expressed at low levels in
cisplatin-resistant ESCC cells (136). It acts as a protein scaffold to
enhance the binding between cytoplasmic polyadenylation element
binding protein (CPEB4) and TRIM25 E3 ligase. This promotes the
formation of the CPEB4/circDOPEY2/TRIM25 complex, accelerating
CPEB4 degradation by promoting its ubiquitination and blocking
CPEB4-mediated Mcl-1 translation (136). An in vivo study
demonstrated that combining circDOPEY2 with cisplatin significantly
suppresses cisplatin resistance (136). Zhu et al (137) indicated that circPSMC3 is
downregulated in gefitinib-resistant ESCC cells. Increasing
circPSMC3 expression significantly reduces ESCC resistance to
gefitinib, inhibits cell proliferation and promotes apoptosis
(137). A negative correlation has
been found between circPSMC3 and miR-10a-5p expression, but
circPSMC3 is positively correlated with PTEN expression, downstream
of miR-10a-5p (137). Modulating
miR-10a-5p or PTEN expression alters the effect of circPSMC3 on
gefitinib resistance in ESCC. These findings suggest that circPSMC3
inhibits gefitinib resistance in ESCC by modulating the
miR-10a-5p/PTEN axis (137).
Programmed cell death receptor 1 (PD-1)/programmed cell
death-ligand (PD-L1) inhibitors can act to slow tumor development,
according to recent research (138). However, due to the emergence of
resistance, only a small percentage of individuals benefit from
this medication (139).
It has been demonstrated that circRNAs regulate drug
resistance in PD-1/PD-L1 therapy (140,141). A study has reported elevated
levels of circ-vimentin (circVIM) in ESCC cells and showed its
ability to suppress miR-124 expression (142). Knockdown of circVIM in ESCC cells
results in upregulation of miR-124 expression, a reduction in PD-L1
expression and a mitigation of tumor escape (142). Furthermore, silencing circVIM can
be synergized with sevoflurane to inhibit multiple oncogenic
activities and counter immune escape in ESCC by targeting the
miR-124/PD-L1 axis (142). These
findings suggest that circRNAs are an important regulator of drug
resistance in ESCC therapy. If the efficacy and application plan
can be further clarified, circRNA could have a high impact on the
treatment of numerous patients and could reduce the recurrence rate
of ESCC.
The aforementioned studies reveal a strong link
between ncRNAs and tumor drug resistance, which highlights their
potential in overcoming this challenge and guiding future
therapeutic strategies.
Due to the subtle onset and rapid progression of
ESCC, early detection and diagnosis are crucial for the treatment
of this disease. Several studies have validated the use of ncRNAs
as reliable biomarkers for the diagnosis of ESCC.
Researchers have shown that patients with ESCC with
low miR-378 expression have a short OS and a poor prognosis. Low
expression of miR-378 is correlated with TNM stage and LNM of
patients (143). These findings
indicate that miR-378 serves as an independent prognostic factor
for the prediction of OS in patients diagnosed with ESCC (143). MiR-205-5-5p and miR-429 show
substantial increases in plasma exosomes among patients with ESCC,
while miR-375-3p exhibits a marked decrease (144).
Researchers have evaluated whether these miRNAs in
plasma exosomes can be potential biomarkers for ESCC diagnosis
using receiver operating characteristic (ROC) curves (144). It has been revealed that the area
under the curve (AUC) of miR-205-5p, miR-429 and miR-375-3p are
0.770, 0.699 and 0.741, respectively (144). The sensitivities of miR-205-5-5p,
miR-429 and miR-375-3p in diagnosing ESCC were 72.5, 60.0 and
65.0%, respectively, while the diagnostic specificities were 70.0,
60.0 and 65.0%, respectively (144). Wen et al (145) has identified miR-135b-5p,
miR-139-5p, miR-29c-5p and miR-338-3p as potential predictors of OS
in patients with ESCC with local metastasis, post surgical
resection. These four miRNAs exhibit predictive capabilities that
are comparable to established clinical prognostic indicators, such
as pathological tumor-stage, pathological node-stage and lymph node
examination count. Notably, the four-miRNA-based classifier has
achieved an AUC of 0.597, underscoring its significance as a highly
valuable diagnostic tool.
In addition, miRNAs in urine can be used for the
diagnosis of ESCC. A study has revealed that the urine of patients
contains elevated levels of certain miRNAs, such as miR-1273f,
miR-619-5p, miR-150-3p, miR-4327 and miR-3135b. Urinary miR-1273f
and miR-619-5p sensitivity and specificity for diagnosing all
stages of ESCC were 82.5 and 59.0%, and 80.0 and 65.3%,
respectively. These findings suggest that miRNAs have potential as
biomarkers for ESCC (146).
LncRNAs have been shown to play a critical role in
the diagnosis and prognosis of ESCC. Research suggests that the
prognosis of patients with ESCC is inversely related to the risk
score associated with an 8-lncRNA signature. This comprises
ADAMTS9-AS1, DLX6-AS1, LINC00470, LINC00520, LINC01497, LINC01749,
MAMDC2-AS1 and SSTR5-AS1 (147).
In addition, six differentially expressed lncRNAs (6-DElncRNAs),
namely AP000696.2, LINC01711, RP11-70C1.3, AP000487.5, AC011997.1
and RP11-225N10.1, have been identified as potential prognostic
indicators for patients with ESCC. A 6-DElncRNA model has also been
developed to classify patients with ESCC into high-risk and
low-risk groups, with the former exhibiting lower OS rates. A ROC
curve analysis showed that the AUCs for 6-DElncRNAs for predicting
1-, 3- and 5-year survival rates of patients with ESCC are 0.768,
0.825 and 0.822, respectively. As the high-risk group has higher
gefitinib and lapatinib IC50 values, 6-DElncRNAs may
also be able to predict drug susceptibility (148). Liu et al (149) demonstrated that a 6-lncRNA
signature (AC005091.1, SNHG6, AC091544.4, DNAJB5-DT, HTT-AS and
ANKRD10-IT1) has a high predictive value for ESCC. The ROC curve
analysis gave AUCs of 0.785 and 0.786 for the 6-lncRNA signature
for predicting 3- and 5-year survival rates, respectively. The AUCs
of the 6-lncRNA signature for predicting 3- and 5-year
recurrence-free survival are 0.795 and 0.783, respectively. These
findings provide additional perspectives for the development and
research of ESCC biomarkers.
There is growing evidence to suggest that circRNAs
can serve as biomarkers for ESCC. Huang et al (151) revealed a substantial correlation
between the level of circ0004771, vascular invasion and a worse OS
in patients with ESCC. Furthermore, the authors observed that
circ0004771 levels increase in an extracellular medium in a
time-dependent manner and can be downregulated by the GW4869
exosome blocker, without affecting its expression within the cells.
A ROC curve analysis provided an AUC for circ0004771 of 0.816,
which indicates that circ0004771 has potential as a diagnostic
biomarker for ESCC (151). Wang
et al (152) identified
1,202 circRNAs related to ESCC prognosis through bioinformatics.
The authors also revealed four circRNAs (circ0000005, circ0007541,
circ0008199 and circ0077536) that are primarily found in the
nucleus and can be used to divide patients with ESCC into two
groups with noticeably different survival rates. This four-circRNA
signature (AUC=0.839) performs better for ESCC prediction compared
with using the TNM stage (AUC=0.657). When the four-circRNA
signature and TNM are combined, the predictive value is higher
(AUC=0.874).
The aforementioned studies show the significant
dysregulation of specific ncRNAs in ESCC, which facilitates early
diagnosis. Given their widespread expression and easy detection,
ncRNAs serve as promising and cost-effective biomarkers for
ESCC.
Since the late 1990s to the present day, an
increasing number of ncRNAs have been discovered. Initially, these
non-coding fragments of RNA were considered to be useless
byproducts (158). However, with
the advent of RNA-sequencing technology, ncRNAs have gained growing
attention and have emerged as a novel research paradigm within RNA
biology (159). Furthermore, an
expanding body of research has indicated that ncRNAs play pivotal
roles in the onset and progression of various diseases, such as
cancer, cardiovascular diseases and neurological disorders. This
field of study has evolved into a crucial component of life
sciences (160,161).
The present review provides an overview of the
biogenesis, characteristics and functions of ncRNA family members,
which include miRNAs, lncRNAs and circRNAs, along with the latest
developments in the treatment of ESCC. The present study also
summarizes, in Table I, studies on
the regulation of ESCC progression by ncRNAs that are not detailed
in this article. Aberrant expression of ncRNAs has been observed in
ESCC cells, tissues and peripheral blood of patients with ESCC,
which suggests their potential involvement in the disease and their
utility as biomarkers (162).
Furthermore, ncRNAs have been found to impact the progression of
ESCC through multiple pathways, such as acting as molecular sponges
for miRNA, participating in transcriptional regulation and
influencing protein coding (163–165). These findings underscore the
potential of ncRNAs as biomarkers and therapeutic targets for ESCC,
with implications for early diagnosis, treatment and prognosis
assessment (162,166).
Although the critical regulatory role of ncRNAs in
the occurrence and progression of ESCC has received increasing
attention, further in-depth research is still required. It is well
known that ncRNAs exhibit rich diversity in terms of quantity and
types. Future investigations should focus more on the interactions
among ncRNAs, to elucidate their regulatory roles in ESCC
progression by constructing and depicting ncRNA regulatory
networks. In addition, the subcellular localization of ncRNAs is
closely associated with their specific functions (167). Therefore, future studies should
explore whether changes in the localization of ncRNAs during
disease progression result in alterations to their structure,
function and downstream target molecules. Most current studies
focus primarily on the functional aspects of aberrantly expressed
ncRNAs, in vivo, but lack investigation into the causes of
their aberrant expression (168,169). Consequently, future research
should further elucidate the specific mechanisms that underlie the
aberrant expression of ncRNAs in cells or peripheral blood.
Moreover, the occurrence and progression of various malignant
tumors, such as ESCC, are closely related to the status of the
tumor microenvironment. Therefore, subsequent research should not
only focus on the role of ncRNAs in tumor cells but should also
clarify their effects on the tumor microenvironment. For instance,
it is crucial to elucidate the specific mechanisms by which ncRNAs,
mediated by exosomes, enter other cells, as this leads to cellular
dysfunction and disruption of the tumor microenvironment
homeostasis.
Targeted therapies for ESCC have shown promising
efficacy, with targeted drugs against key genes and signaling
pathways implicated in ESCC now available. Given that ncRNAs can
regulate the expression of downstream target genes through multiple
mechanisms, future considerations may involve combining
ncRNA-targeted therapy with existing targeted drugs for ESCC. This
approach could involve the synergistic regulation of key genes in
ESCC progression, such as Notch1, EGFR and human epidermal growth
factor receptor 2 (170–172). In addition, the regulatory effects
of ncRNAs on multiple immune checkpoints have been confirmed and a
potential combination of ncRNA-targeted therapy with immunotherapy
could be trialed for ESCC in future studies (173,174). Increasing attention has been paid
to the protein-coding ability of circRNAs. Peptide or protein-based
drugs exhibit high specificity and efficiency in the clinical
treatment of ESCC, which suggests that proteins encoded by circRNAs
may serve as potential sources for screening therapeutic drugs for
ESCC (175–177). Indeed, studies have examined the
potential of ncRNAs as target in the treatment of ESCC, such as
miR-877-3p, miRNA-20b-5p and circFoxo3 (178–180). However, the therapeutic efficacy
of these ncRNA for ESCC in the clinic still requires in-depth
study.
In summary, ncRNAs have potential as non-invasive
diagnostic markers and specific therapeutic targets in clinical
diagnosis and treatment. However, there is still a long way to go
from basic research to clinical application. Extensive clinical
studies are needed to demonstrate the safety of ncRNAs and
gradually reduce the risks associated with their use as therapeutic
targets, as well as to minimize treatment-related side effects.
Moreover, continuous improvement and refinement of high-throughput
sequencing technologies are necessary to ensure the specificity and
integrity of ncRNA detection.
Not applicable.
This research was funded by the National Natural Science
Foundation of China (grant nos. 32270848 and 82360044), the
Collaborative Innovation Center of Chinese Ministry of Education
(grant no. 2020-39), the Guizhou Provincial Natural Science
Foundation [grant nos. ZK(2022)-607 and ZK(2024)-269], the Science
and Technology Plan Project of Guizhou Province (grant no.
2020-032) and the Guizhou Provincial Traditional Chinese Medicine
Project (grant no. QZYY-2022-027).
Not applicable.
LZ, XW and ZH designed and organized this
manuscript. LZ, YW and XZ contributed to the first draft of the
manuscript. LZ, YW and MH designed the tables and figures. XW, JG
and ZH reviewed the manuscript critically for important
intellectual content. JG, MH and XZ revised the manuscript. YW, XZ
and MH investigated and resolved any parts of the work
appropriately. XW and ZH approved the final version to be
published. Data authentication is not applicable. All authors read
and approved the final version of the manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Rogers JE, Sewastjanow-Silva M, Waters RE
and Ajani JA: Esophageal cancer: Emerging therapeutics. Expert Opin
Ther Targets. 26:107–117. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Morgan E, Soerjomataram I, Rumgay H,
Coleman HG, Thrift AP, Vignat J, Laversanne M, Ferlay J and Arnold
M: The global landscape of esophageal squamous cell carcinoma and
esophageal adenocarcinoma incidence and mortality in 2020 and
projections to 2040: New estimates from GLOBOCAN 2020.
Gastroenterology. 163:649–658.e2. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
He S, Xu J, Liu X and Zhen Y: Advances and
challenges in the treatment of esophageal cancer. Acta Pharm Sin B.
11:3379–3392. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rumgay H, Arnold M, Laversanne M, Whiteman
DC, Thrift AP, Wei W, Lemmens VEPP and Soerjomataram I:
International trends in esophageal squamous cell carcinoma and
adenocarcinoma incidence. Am J Gastroenterol. 116:1072–1076. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li J, Xu J, Zheng Y, Gao Y, He S, Li H,
Zou K, Li N, Tian J, Chen W and He J: Esophageal cancer:
Epidemiology, risk factors and screening. Chin J Cancer Res.
33:535–547. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bhat AA, Nisar S, Maacha S, Carneiro-Lobo
TC, Akhtar S, Siveen KS, Wani NA, Rizwan A, Bagga P, Singh M, et
al: Cytokine-chemokine network driven metastasis in esophageal
cancer; promising avenue for targeted therapy. Mol Cancer.
20:22021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Huang FL and Yu SJ: Esophageal cancer:
Risk factors, genetic association, and treatment. Asian J Surg.
41:210–215. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mattick JS and Makunin IV: Non-coding RNA.
Hum Mol Genet. 15:R17–R29. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu Y, Liu X, Lin C, Jia X, Zhu H, Song J
and Zhang Y: Noncoding RNAs regulate alternative splicing in
cancer. J Exp Clin Cancer Res. 40:112021. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mohapatra S, Pioppini C, Ozpolat B and
Calin GA: Non-coding RNAs regulation of macrophage polarization in
cancer. Mol Cancer. 20:242021. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Poller W, Sahoo S, Hajjar R, Landmesser U
and Krichevsky AM: Exploration of the noncoding genome for
human-specific therapeutic targets-recent insights at molecular and
cellular level. Cells. 12:26602023. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Beermann J, Piccoli MT, Viereck J and Thum
T: Non-coding RNAs in development and disease: Background,
mechanisms, and therapeutic approaches. Physiol Rev. 96:1297–1325.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhou B, Yang H, Yang C, Bao YL, Yang SM,
Liu J and Xiao YF: Translation of noncoding RNAs and cancer. Cancer
Lett. 497:89–99. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Slack FJ and Chinnaiyan AM: The role of
non-coding RNAs in oncology. Cell. 179:1033–1055. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ran H, Yang Y, Luo M, Liu X, Yue B, Chai
Z, Zhong J and Wang H: Molecular regulation of yak preadipocyte
differentiation and proliferation by LncFAM200B and ceRNA
regulatory network analysis. Cells. 11:23662022. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Min X, Cai MY, Shao T, Xu ZY, Liao Z, Liu
DL, Zhou MY, Wu WP, Zhou YL, Mo MH, et al: A circular intronic RNA
ciPVT1 delays endothelial cell senescence by regulating the
miR-24-3p/CDK4/pRb axis. Aging Cell. 21:e135292022. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Qu J, Xiong X, Hujie G, Ren J, Yan L and
Ma L: MicroRNA-132-3p alleviates neuron apoptosis and impairments
of learning and memory abilities in Alzheimer's disease by
downregulation of HNRNPU stabilized BACE1. Cell Cycle.
20:2309–2320. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu CX, Guo SK, Nan F, Xu YF, Yang L and
Chen LL: RNA circles with minimized immunogenicity as potent PKR
inhibitors. Mol Cell. 82:420–434.e6. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Feng Q, Zhang H, Yao D, Chen WD and Wang
YD: Emerging role of non-coding RNAs in esophageal squamous cell
carcinoma. Int J Mol Sci. 21:2582019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Miyoshi J, Zhu Z, Luo A, Toden S, Zhou X,
Izumi D, Kanda M, Takayama T, Parker IM, Wang M, et al: A
microRNA-based liquid biopsy signature for the early detection of
esophageal squamous cell carcinoma: A retrospective, prospective
and multicenter study. Mol Cancer. 21:442022. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sharma U, Murmu M, Barwal TS, Tuli HS,
Jain M, Prakash H, Kaceli T, Jain A and Bishayee A: A pleiotropic
role of long non-coding RNAs in the modulation of Wnt/β-catenin and
PI3K/Akt/mTOR signaling pathways in esophageal squamous cell
carcinoma: Implication in chemotherapeutic drug response. Curr
Oncol. 29:2326–2349. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang C, Liu WR, Tan S, Zhou JK, Xu X, Ming
Y, Cheng J, Li J, Zeng Z, Zuo Y, et al: Characterization of
distinct circular RNA signatures in solid tumors. Mol Cancer.
21:632022. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jansson MD and Lund AH: MicroRNA and
cancer. Mol Oncol. 6:590–610. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Acunzo M and Croce CM: MicroRNA in cancer
and cachexia-a mini-review. J Infect Dis. 212 (Suppl 1):S74–S77.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mengistu AA and Tenkegna TA: The role of
miRNA in plant-virus interaction: A review. Mol Biol Rep.
48:2853–2861. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Peng Y and Croce CM: The role of MicroRNAs
in human cancer. Signal Transduct Target Ther. 1:150042016.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wahid F, Shehzad A, Khan T and Kim YY:
MicroRNAs: Synthesis, mechanism, function, and recent clinical
trials. Biochim Biophys Acta. 1803:1231–1243. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Budakoti M, Panwar AS, Molpa D, Singh RK,
Büsselberg D, Mishra AP, Coutinho HDM and Nigam M: Micro-RNA: The
darkhorse of cancer. Cell Signal. 83:1099952021. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Krol J, Loedige I and Filipowicz W: The
widespread regulation of microRNA biogenesis, function and decay.
Nat Rev Genet. 11:597–610. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wilson RC, Tambe A, Kidwell MA, Noland CL,
Schneider CP and Doudna JA: Dicer-TRBP complex formation ensures
accurate mammalian microRNA biogenesis. Mol Cell. 57:397–407. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
He B, Zhao Z, Cai Q, Zhang Y, Zhang P, Shi
S, Xie H, Peng X, Yin W, Tao Y and Wang X: miRNA-based biomarkers,
therapies, and resistance in cancer. Int J Biol Sci. 16:2628–2647.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ha M and Kim VN: Regulation of microRNA
biogenesis. Nat Rev Mol Cell Biol. 15:509–524. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ho PTB, Clark IM and Le LTT:
MicroRNA-based diagnosis and therapy. Int J Mol Sci. 23:71972022.
View Article : Google Scholar
|
|
34
|
Iqbal MA, Arora S, Prakasam G, Calin GA
and Syed MA: MicroRNA in lung cancer: Role, mechanisms, pathways
and therapeutic relevance. Mol Aspects Med. 70:3–20. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
O'Brien J, Hayder H, Zayed Y and Peng C:
Overview of MicroRNA biogenesis, mechanisms of actions, and
circulation. Front Endocrinol (Lausanne). 9:4022018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Medley JC, Panzade G and Zinovyeva AY:
microRNA strand selection: Unwinding the rules. Wiley Interdiscip
Rev RNA. 12:e16272021. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Bartel DP: Metazoan MicroRNAs. Cell.
173:20–51. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Sun Q, Hao Q and Prasanth KV: Nuclear long
noncoding RNAs: Key regulators of gene expression. Trends Genet.
34:142–157. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Cao Q, Guo Z, Yan Y, Wu J and Song C:
Exosomal long noncoding RNAs in aging and age-related diseases.
IUBMB Life. 71:1846–1856. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kour S and Rath PC: Long noncoding RNAs in
aging and age-related diseases. Ageing Res Rev. 26:1–21. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Statello L, Guo CJ, Chen LL and Huarte M:
Gene regulation by long non-coding RNAs and its biological
functions. Nat Rev Mol Cell Biol. 22:96–118. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
He Z, Yang D, Fan X, Zhang M, Li Y, Gu X
and Yang M: The roles and mechanisms of lncRNAs in liver fibrosis.
Int J Mol Sci. 21:14822020. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Aboudehen K: Regulation of mTOR signaling
by long non-coding RNA. Biochim Biophys Acta Gene Regul Mech.
1863:1944492020. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Jain AK, Xi Y, McCarthy R, Allton K,
Akdemir KC, Patel LR, Aronow B, Lin C, Li W, Yang L, et al:
LncPRESS1 Is a p53-regulated LncRNA that safeguards pluripotency by
disrupting SIRT6-mediated de-acetylation of histone H3K56. Mol
Cell. 64:967–981. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Du Z, Wen X, Wang Y, Jia L, Zhang S, Liu
Y, Zhou L, Li H, Yang W, Wang C, et al: Chromatin lncRNA Platr10
controls stem cell pluripotency by coordinating an intrachromosomal
regulatory network. Genome Biol. 22:2332021. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhang Y, Huang YX, Wang DL, Yang B, Yan
HY, Lin LH, Li Y, Chen J, Xie LM, Huang YS, et al: LncRNA DSCAM-AS1
interacts with YBX1 to promote cancer progression by forming a
positive feedback loop that activates FOXA1 transcription network.
Theranostics. 10:10823–10837. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Su K, Wang N, Shao Q, Liu H, Zhao B and Ma
S: The role of a ceRNA regulatory network based on lncRNA MALAT1
site in cancer progression. Biomed Pharmacother. 137:1113892021.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Huang W, Sun YM, Pan Q, Fang K, Chen XT,
Zeng ZC, Chen TQ, Zhu SX, Huang LB, Luo XQ, et al: The snoRNA-like
lncRNA LNC-SNO49AB drives leukemia by activating the RNA-editing
enzyme ADAR1. Cell Discov. 8:1172022. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Yang X, Ye T, Liu H, Lv P, Duan C, Wu X,
Jiang K, Lu H, Xia D, Peng E, et al: Expression profiles,
biological functions and clinical significance of circRNAs in
bladder cancer. Mol Cancer. 20:42021. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Hu Q and Zhou T: EIciRNA-mediated gene
expression: Tunability and bimodality. FEBS Lett. 592:3460–3471.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zhou X and Du J: CircRNAs: Novel
therapeutic targets in multiple myeloma. Mol Biol Rep.
49:10667–10676. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Vahabi A, Rezaie J, Hassanpour M, Panahi Y
and Nemati M, Rasmi Y and Nemati M: Tumor cells-derived exosomal
CircRNAs: Novel cancer drivers, molecular mechanisms, and clinical
opportunities. Biochem Pharmacol. 200:1150382022. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Amicone L, Marchetti A and Cicchini C:
Exosome-associated circRNAs as key regulators of EMT in cancer.
Cells. 11:17162022. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Guo X, Chang X, Wang Z, Jiang C and Wei Z:
CircRNAs: Promising factors for regulating angiogenesis in
colorectal cancer. Clin Transl Oncol. 24:1673–1681. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Li X, Yang L and Chen LL: The biogenesis,
functions, and challenges of circular RNAs. Mol Cell. 71:428–442.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Zhang LX, Gao J, Long X, Zhang PF, Yang X,
Zhu SQ, Pei X, Qiu BQ, Chen SW, Lu F, et al: The circular RNA
circHMGB2 drives immunosuppression and anti-PD-1 resistance in lung
adenocarcinomas and squamous cell carcinomas via the
miR-181a-5p/CARM1 axis. Mol Cancer. 21:1102022. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Hollensen AK, Thomsen HS, Lloret-Llinares
M, Kamstrup AB, Jensen JM, Luckmann M, Birkmose N, Palmfeldt J,
Jensen TH, Hansen TB and Damgaard CK: circZNF827 nucleates a
transcription inhibitory complex to balance neuronal
differentiation. Elife. 9:e584782020. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Misir S, Wu N and Yang BB: Specific
expression and functions of circular RNAs. Cell Death Differ.
29:481–491. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Li Z, Huang C, Bao C, Chen L, Lin M, Wang
X, Zhong G, Yu B, Hu W, Dai L, et al: Exon-intron circular RNAs
regulate transcription in the nucleus. Nat Struct Mol Biol.
22:256–264. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Wang X, Ma R, Zhang X, Cui L, Ding Y, Shi
W, Guo C and Shi Y: Crosstalk between N6-methyladenosine
modification and circular RNAs: Current understanding and future
directions. Mol Cancer. 20:1212021. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Lin H, Wang Y, Wang P, Long F and Wang T:
Mutual regulation between N6-methyladenosine (m6A) modification and
circular RNAs in cancer: Impacts on therapeutic resistance. Mol
Cancer. 21:1482022. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Meyer KD, Patil DP, Zhou J, Zinoviev A,
Skabkin MA, Elemento O, Pestova TV, Qian SB and Jaffrey SR: 5′ UTR
m(6)A promotes cap-independent translation. Cell. 163:999–1010.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Li R, Jiang J, Shi H, Qian H, Zhang X and
Xu W: CircRNA: A rising star in gastric cancer. Cell Mol Life Sci.
77:1661–1680. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Yang Y, Fan X, Mao M, Song X, Wu P, Zhang
Y, Jin Y, Yang Y, Chen LL, Wang Y, et al: Extensive translation of
circular RNAs driven by N6-methyladenosine. Cell Res.
27:626–641. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Duan JL, Chen W, Xie JJ, Zhang ML, Nie RC,
Liang H, Mei J, Han K, Xiang ZC, Wang FW, et al: A novel peptide
encoded by N6-methyladenosine modified circMAP3K4 prevents
apoptosis in hepatocellular carcinoma. Mol Cancer. 21:932022.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Lei M, Zheng G, Ning Q, Zheng J and Dong
D: Translation and functional roles of circular RNAs in human
cancer. Mol Cancer. 19:302020. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Song Y, Li L, Ou Y, Gao Z, Li E, Li X,
Zhang W, Wang J, Xu L, Zhou Y, et al: Identification of genomic
alterations in oesophageal squamous cell cancer. Nature. 509:91–95.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Yu XM, Li SJ, Yao ZT, Xu JJ, Zheng CC, Liu
ZC, Ding PB, Jiang ZL, Wei X, Zhao LP, et al: N4-acetylcytidine
modification of lncRNA CTC-490G23.2 promotes cancer metastasis
through interacting with PTBP1 to increase CD44 alternative
splicing. Oncogene. 42:1101–1116. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Huang X, Liu C, Li H, Dai T, Luo G, Zhang
C, Li T and Lü M: Hypoxia-responsive lncRNA G077640 promotes ESCC
tumorigenesis via the H2AX-HIF1α-glycolysis axis. Carcinogenesis.
44:383–393. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Xiao Z, Feng X, Zhou Y, Li P, Luo J, Zhang
W, Zhou J, Zhao J, Wang D, Wang Y, et al: Exosomal miR-10527-5p
inhibits migration, invasion, lymphangiogenesis and lymphatic
metastasis by affecting Wnt/β-catenin signaling via Rab10 in
esophageal squamous cell carcinoma. Int J Nanomedicine. 18:95–114.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Cui Y, Zhang C, Ma S, Li Z, Wang W, Li Y,
Ma Y, Fang J, Wang Y, Cao W and Guan F: RNA m6A demethylase
FTO-mediated epigenetic up-regulation of LINC00022 promotes
tumorigenesis in esophageal squamous cell carcinoma. J Exp Clin
Cancer Res. 40:2942021. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Ren K, Li Y, Lu H, Li Z, Li Z, Wu K, Li Z
and Han X: Long noncoding RNA HOTAIR controls cell cycle by
functioning as a competing endogenous RNA in esophageal squamous
cell carcinoma. Transl Oncol. 9:489–497. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Xu T, Hu Y, Zhao Y, Qi Y, Zhang S and Li
P: Hsa_circ_0046534 accelerates esophageal squamous cell carcinoma
proliferation and metastasis via regulating MMP2 expression by
sponging miR-339-5p. Cell Signal. 112:1109062023. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Xian D, Yang S, Liu Y, Liu Q, Huang D and
Wu Y: MicroRNA-196a-5p facilitates the onset and progression via
targeting ITM2B in esophageal squamous cell carcinoma. Pathol Int.
74:129–138. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Zhang Y, Dong X, Guo X, Li C, Fan Y, Liu
P, Yuan D, Ma X, Wang J, Zheng J, et al: LncRNA-BC069792 suppresses
tumor progression by targeting KCNQ4 in breast cancer. Mol Cancer.
22:412023. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Liu QL, Zhang Z, Wei X and Zhou ZG:
Noncoding RNAs in tumor metastasis: Molecular and clinical
perspectives. Cell Mol Life Sci. 78:6823–6850. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Enkhnaran B, Zhang GC, Zhang NP, Liu HN,
Wu H, Xuan S, Yu XN, Song GQ, Shen XZ, Zhu JM, et al:
microRNA-106b-5p promotes cell growth and sensitizes
chemosensitivity to sorafenib by targeting the BTG3/Bcl-xL/p27
signaling pathway in hepatocellular carcinoma. J Oncol.
2022:19715592022. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Wang H, Peng D, Gan M, He Z and Kuang Y:
CPEB3 overexpression caused by miR-106b-5p inhibition inhibits
esophageal carcinoma in-vitro progression and metastasis.
Anticancer Drugs. 33:335–351. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Yang F, Sun Z, Wang D and Du T:
MiR-106b-5p regulates esophageal squamous cell carcinoma
progression by binding to HPGD. BMC Cancer. 22:3082022. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Deng W, Fan W, Li P, Yao J, Qi J, Chi H,
Ji G and Zhao J: microRNA-497-mediated Smurf2/YY1/HIF2α axis in
tumor growth and metastasis of esophageal squamous cell carcinoma.
J Biochem Mol Toxicol. 36:e231822022. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
He Z, Chen J, Chen X, Wang H, Tang L and
Han C: microRNA-377 acts as a suppressor in esophageal squamous
cell carcinoma through CBX3-dependent P53/P21 pathway. J Cell
Physiol. 236:107–120. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Ren L, Fang X, Shrestha SM, Ji Q, Ye H,
Liang Y, Liu Y, Feng Y, Dong J and Shi R: LncRNA SNHG16 promotes
development of oesophageal squamous cell carcinoma by interacting
with EIF4A3 and modulating RhoU mRNA stability. Cell Mol Biol Lett.
27:892022. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Yang X, Zeng T, Liu Z, He W, Hu M, Tang T,
Chen L and Xing L: Long noncoding RNA GK-IT1 promotes esophageal
squamous cell carcinoma by regulating MAPK1 phosphorylation. Cancer
Med. 11:4555–4574. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Xue ST, Zheng B, Cao SQ, Ding JC, Hu GS,
Liu W and Chen C: Long non-coding RNA LINC00680 functions as a
ceRNA to promote esophageal squamous cell carcinoma progression
through the miR-423-5p/PAK6 axis. Mol Cancer. 21:692022. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Xiong G, Diao D, Lu D, Liu X, Liu Z, Mai
S, Feng S, Dong X and Cai K: Circular RNA circNELL2 acts as the
sponge of miR-127-5p to promote esophageal squamous cell carcinoma
progression. Onco Targets Ther. 13:9245–9255. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Li Z, Qin X, Bian W, Li Y, Shan B, Yao Z
and Li S: Exosomal lncRNA ZFAS1 regulates esophageal squamous cell
carcinoma cell proliferation, invasion, migration and apoptosis via
microRNA-124/STAT3 axis. J Exp Clin Cancer Res. 38:4772019.
View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Tian Z, Li Z, Zhu Y, Meng L, Liu F, Sang M
and Wang G: Hypermethylation-mediated inactivation of miR-124
predicts poor prognosis and promotes tumor growth at least
partially through targeting EZH2/H3K27me3 in ESCC. Clin Exp
Metastasis. 36:381–391. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Zeng B, Liu Z, Zhu H, Zhang X, Yang W, Li
X and Cheng C: CircRNA_2646 functions as a ceRNA to promote
progression of esophageal squamous cell carcinoma via inhibiting
miR-124/PLP2 signaling pathway. Cell Death Discov. 7:992021.
View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Yao D, Lin S, Chen S and Wang Z: circHIPK3
regulates cell proliferation and migration by sponging microRNA-124
and regulating serine/threonine kinase 3 expression in esophageal
squamous cell carcinoma. Bioengineered. 13:9767–9780. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Cheng J, Ma H, Yan M, Zhang Z and Xing W:
Circ_0007624 suppresses the development of esophageal squamous cell
carcinoma via targeting miR-224-5p/CPEB3 to inactivate the
EGFR/PI3K/AKT signaling. Cell Signal. 99:1104482022. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Chen Z, Yao N, Gu H, Song Y, Ye Z, Li L,
Lu P and Shao Q: Circular RNA_LARP4 sponges miR-1323 and hampers
progression of esophageal squamous cell carcinoma through
modulating PTEN/PI3K/AKT pathway. Dig Dis Sci. 65:2272–2283. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Jiang Q, Wang H, Yuan D, Qian X, Ma X, Yan
M and Xing W: Circular_0086414 induces SPARC like 1 (SPARCL1)
production to inhibit esophageal cancer cell proliferation,
invasion and glycolysis and induce cell apoptosis by sponging
miR-1290. Bioengineered. 13:12099–12114. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Zheng S, Liu B and Guan X: The role of
tumor microenvironment in invasion and metastasis of esophageal
squamous cell carcinoma. Front Oncol. 12:9112852022. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Wang X, Han J, Liu Y, Hu J, Li M, Chen X
and Xu L: miR-17-5p and miR-4443 promote esophageal squamous cell
carcinoma development by targeting TIMP2. Front Oncol.
11:6058942021. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Liang Y, Duan L, Lu J and Xia J:
Engineering exosomes for targeted drug delivery. Theranostics.
11:3183–3195. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Paskeh MDA, Entezari M, Mirzaei S,
Zabolian A, Saleki H, Naghdi MJ, Sabet S, Khoshbakht MA, Hashemi M,
Hushmandi K, et al: Emerging role of exosomes in cancer progression
and tumor microenvironment remodeling. J Hematol Oncol. 15:832022.
View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Liu T, Li P, Li J, Qi Q, Sun Z, Shi S, Xie
Y, Liu S, Wang Y, Du L and Wang C: Exosomal and intracellular
miR-320b promotes lymphatic metastasis in esophageal squamous cell
carcinoma. Mol Ther Oncolytics. 23:163–180. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Cui D, Zhu Y, Yan D, Lee NPY, Han L, Law
S, Tsao GSW and Cheung ALM: Dual inhibition of cMET and EGFR by
microRNA-338-5p suppresses metastasis of esophageal squamous cell
carcinoma. Carcinogenesis. 42:995–1007. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Zhao R, Shan Y, Zhou X, Zhang C, Zhao R,
Zhao L and Shan B: MicroRNA-485-5p suppresses the progression of
esophageal squamous cell carcinoma by targeting flotillin-1 and
inhibits the epithelial-mesenchymal transition. Oncol Rep.
45:932021. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Cheng W, Yang F and Ma Y: lncRNA TPT1-AS1
promotes cell migration and invasion in esophageal squamous-cell
carcinomas by regulating the miR-26a/HMGA1 axis. Open Med (Wars).
18:202205332023. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Yang R, Wan J, Ma L, Zhou F, Yang Z, Li Z,
Zhang M and Ming L: TMEM44-AS1 promotes esophageal squamous cell
carcinoma progression by regulating the IGF2BP2-GPX4 axis in
modulating ferroptosis. Cell Death Discov. 9:4312023. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Wang J, Huang TJ, Mei Y, Luo FF, Xie DH,
Peng LX, Liu BQ, Fan ML, Zhang JB, Zheng ST, et al: Novel long
noncoding RNA LINC02820 augments TNF signaling pathway to remodel
cytoskeleton and potentiate metastasis in esophageal squamous cell
carcinoma. Cancer Gene Ther. 30:375–387. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Li H, Lin PH, Gupta P, Li X, Zhao SL, Zhou
X, Li Z, Wei S, Xu L, Han R, et al: MG53 suppresses tumor
progression and stress granule formation by modulating G3BP2
activity in non-small cell lung cancer. Mol Cancer. 20:1182021.
View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Zhang Y, Yue C, Krichevsky AM and
Garkavtsev I: Repression of the stress granule protein G3BP2
inhibits immune checkpoint molecule PD-L1. Mol Oncol. Feb
1–2021.(Epub ahead of print).
|
|
108
|
Zheng Y, Wu J, Deng R, Lin C, Huang Y,
Yang X, Wang C, Yang M, He Y, Lu J, et al: G3BP2 regulated by the
lncRNA LINC01554 facilitates esophageal squamous cell carcinoma
metastasis through stabilizing HDGF transcript. Oncogene.
41:515–526. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Zhang H, Wang Y, Zhang W, Wu Q, Fan J and
Zhan Q: BAALC-AS1/G3BP2/c-Myc feedback loop promotes cell
proliferation in esophageal squamous cell carcinoma. Cancer Commun
(Lond). 41:240–257. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Wu K, Wang Z, Huang Y, Yao L, Kang N, Ge
W, Zhang R and He W: LncRNA PTPRG-AS1 facilitates glycolysis and
stemness properties of esophageal squamous cell carcinoma cells
through miR-599/PDK1 axis. J Gastroenterol Hepatol. 37:507–517.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Shi Y, Fang N, Li Y, Guo Z, Jiang W, He Y,
Ma Z and Chen Y: Circular RNA LPAR3 sponges microRNA-198 to
facilitate esophageal cancer migration, invasion, and metastasis.
Cancer Sci. 111:2824–2836. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Xu Z, Tie X, Li N, Yi Z, Shen F and Zhang
Y: Circular RNA hsa_circ_0000654 promotes esophageal squamous cell
carcinoma progression by regulating the miR-149-5p/IL-6/STAT3
pathway. IUBMB Life. 72:426–439. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Wang J, Wang Q, Gong Y, Hu Q, Zhang H, Ke
S and Chen Y: Knockdown of circRNA circ_0087378 represses the
tumorigenesis and progression of esophageal squamous cell carcinoma
through modulating the miR-140-3p/E2F3 axis. Front Oncol.
10:6072312021. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Brown MS, Muller KE and Pattabiraman DR:
Quantifying the epithelial-to-mesenchymal transition (EMT) from
bench to bedside. Cancers (Basel). 14:11382022. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Huang Y, Hong W and Wei X: The molecular
mechanisms and therapeutic strategies of EMT in tumor progression
and metastasis. J Hematol Oncol. 15:1292022. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Zhu C, Bi W, Li H and Wang W: CircLONP2
accelerates esophageal squamous cell carcinoma progression via
direct MiR-27b-3p-ZEB1 axis. Front Oncol. 12:8228392022. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Chen X, Sun H, Zhao Y, Zhang J, Xiong G,
Cui Y and Lei C: CircRNA circ_0004370 promotes cell proliferation,
migration, and invasion and inhibits cell apoptosis of esophageal
cancer via miR-1301-3p/COL1A1 axis. Open Med (Wars). 16:104–116.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Xu P, Wang L, Liu Q, Gao P, Hu F, Xie X,
Jiang L, Bi R, Ding F, Yang Q and Xiao H: The abnormal expression
of circ-ARAP2 promotes ESCC progression through regulating
miR-761/FOXM1 axis-mediated stemness and the
endothelial-mesenchymal transition. J Transl Med. 20:3182022.
View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Song B, Liu X, Dong H and Roy R:
miR-140-3P induces chemotherapy resistance in esophageal carcinoma
by targeting the NFYA-MDR1 axis. Appl Biochem Biotechnol.
195:973–991. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Zhao F, Tian H, Wang Y, Zhang J, Liu F and
Fu L: LINC01004-SPI1 axis-activated SIGLEC9 in tumor-associated
macrophages induces radioresistance and the formation of
immunosuppressive tumor microenvironment in esophageal squamous
cell carcinoma. Cancer Immunol Immunother. 72:1835–1851. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Wu J, Liu Y, Huang X, Cheng Y, Qian Z, Ni
X, Chen S, Lin M and Luo J: LncRNA DGCR5 silencing enhances the
radio-sensitivity of human esophageal squamous cell carcinoma via
negatively regulating the Warburg effect. Radiat Res. 199:264–272.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Zuo J, Zhao M, Fan Z, Liu B, Wang Y, Li Y,
Lv P, Xing L, Zhang X and Shen H: MicroRNA-153-3p regulates cell
proliferation and cisplatin resistance via Nrf-2 in esophageal
squamous cell carcinoma. Thorac Cancer. 11:738–747. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Qiao Y, Zhang C, Li A, Wang D, Luo Z, Ping
Y, Zhou B, Liu S, Li H, Yue D, et al: IL6 derived from
cancer-associated fibroblasts promotes chemoresistance via CXCR7 in
esophageal squamous cell carcinoma. Oncogene. 37:873–883. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Che Y, Wang J, Li Y, Lu Z, Huang J, Sun S,
Mao S, Lei Y, Zang R, Sun N and He L: Cisplatin-activated PAI-1
secretion in the cancer-associated fibroblasts with paracrine
effects promoting esophageal squamous cell carcinoma progression
and causing chemoresistance. Cell Death Dis. 9:7592018. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Zhuang J, Shen L, Li M, Sun J, Hao J, Li
J, Zhu Z, Ge S, Zhang D, Guo H, et al: Cancer-associated
fibroblast-derived miR-146a-5p generates a niche that promotes
bladder cancer stemness and chemoresistance. Cancer Res.
83:1611–1627. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Kang SH, Oh SY, Lee KY, Lee HJ, Kim MS,
Kwon TG, Kim JW, Lee ST, Choi SY and Hong SH: Differential effect
of cancer-associated fibroblast-derived extracellular vesicles on
cisplatin resistance in oral squamous cell carcinoma via
miR-876-3p. Theranostics. 14:460–479. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Zhao Q, Huang L, Qin G, Qiao Y, Ren F,
Shen C, Wang S, Liu S, Lian J, Wang D, et al: Cancer-associated
fibroblasts induce monocytic myeloid-derived suppressor cell
generation via IL-6/exosomal miR-21-activated STAT3 signaling to
promote cisplatin resistance in esophageal squamous cell carcinoma.
Cancer Lett. 518:35–48. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Guo D, Jin J, Liu J, Dong X, Li D and He
Y: MicroRNA-29b regulates the radiosensitivity of esophageal
squamous cell carcinoma by regulating the BTG2-mediated cell cycle.
Strahlenther Onkol. 197:829–835. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Wu Q, Zhang H, Yang D, Min Q, Wang Y,
Zhang W and Zhan Q: The m6A-induced lncRNA CASC8 promotes
proliferation and chemoresistance via upregulation of hnRNPL in
esophageal squamous cell carcinoma. Int J Biol Sci. 18:4824–4836.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Jia Y, Tian C, Wang H, Yu F, Lv W, Duan Y,
Cheng Z, Wang X, Wang Y, Liu T, et al: Long non-coding RNA
NORAD/miR-224-3p/MTDH axis contributes to CDDP resistance of
esophageal squamous cell carcinoma by promoting nuclear
accumulation of β-catenin. Mol Cancer. 20:1622021. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Tong Y, Yang L, Yu C, Zhu W, Zhou X, Xiong
Y, Wang W, Ji F, He D and Cao X: Tumor-secreted exosomal lncRNA
POU3F3 promotes cisplatin resistance in ESCC by inducing fibroblast
differentiation into CAFs. Mol Ther Oncolytics. 18:1–13. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Wang P, Yang Z, Ye T, Shao F, Li J, Sun N
and He J: lncTUG1/miR-144-3p affect the radiosensitivity of
esophageal squamous cell carcinoma by competitively regulating
c-MET. J Exp Clin Cancer Res. 39:72020. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Liu Z, Lu X, Wen L, You C, Jin X and Liu
J: Hsa_circ_0014879 regulates the radiosensitivity of esophageal
squamous cell carcinoma through miR-519-3p/CDC25A axis. Anticancer
Drugs. 33:e349–e361. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Cheng J, Zhang R, Yan M and Li Y: Circular
RNA hsa_circ_0000277 promotes tumor progression and DDP resistance
in esophageal squamous cell carcinoma. BMC Cancer. 22:2382022.
View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Zhou S, Guo Z, Lv X and Zhang X: CircGOT1
promotes cell proliferation, mobility, and glycolysis-mediated
cisplatin resistance via inhibiting its host gene GOT1 in
esophageal squamous cell cancer. Cell Cycle. 21:247–260. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Liu Z, Gu S, Wu K, Li L, Dong C, Wang W
and Zhou Y: CircRNA-DOPEY2 enhances the chemosensitivity of
esophageal cancer cells by inhibiting CPEB4-mediated Mcl-1
translation. J Exp Clin Cancer Res. 40:3612021. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Zhu H, Du F and Cao C: Restoration of
circPSMC3 sensitizes gefitinib-resistant esophageal squamous cell
carcinoma cells to gefitinib by regulating miR-10a-5p/PTEN axis.
Cell Biol Int. 45:107–116. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Tang Q, Zhao S, Zhou N, He J, Zu L, Liu T,
Song Z, Chen J, Peng L and Xu S: PD-1/PD-L1 immune checkpoint
inhibitors in neoadjuvant therapy for solid tumors (review). Int J
Oncol. 62:492023. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Yi M, Zheng X, Niu M, Zhu S, Ge H and Wu
K: Combination strategies with PD-1/PD-L1 blockade: Current
advances and future directions. Mol Cancer. 21:282022. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Jiang W, Pan S, Chen X, Wang ZW and Zhu X:
The role of lncRNAs and circRNAs in the PD-1/PD-L1 pathway in
cancer immunotherapy. Mol Cancer. 20:1162021. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Fang Z, Jiang C and Li S: The potential
regulatory roles of circular RNAs in tumor immunology and
immunotherapy. Front Immunol. 11:6175832021. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Gao C, Xu YJ, Qi L, Bao YF, Zhang L and
Zheng L: CircRNA VIM silence synergizes with sevoflurane to inhibit
immune escape and multiple oncogenic activities of esophageal
cancer by simultaneously regulating miR-124/PD-L1 axis. Cell Biol
Toxicol. 38:825–845. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Jin W, Wang L, Cheng S and Lv H:
Prognostic value of microRNA-378 in esophageal cancer and its
regulatory effect on tumor progression. Exp Ther Med. 22:7042021.
View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Kim S, Kim GH, Park SJ, Kwon CH, I H, Lee
MW and Lee BE: Exosomal MicroRNA analyses in esophageal squamous
cell carcinoma cell lines. J Clin Med. 11:44262022. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Wen J, Wang G, Xie X, Lin G, Yang H, Luo
K, Liu Q, Ling Y, Xie X, Lin P, et al: Prognostic value of a
four-miRNA signature in patients with lymph node positive
locoregional esophageal squamous cell carcinoma undergoing complete
surgical resection. Ann Surg. 273:523–531. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Okuda Y, Shimura T, Iwasaki H, Fukusada S,
Nishigaki R, Kitagawa M, Katano T, Okamoto Y, Yamada T, Horike SI
and Kataoka H: Urinary microRNA biomarkers for detecting the
presence of esophageal cancer. Sci Rep. 11:85082021. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Zhang J, Ling X, Fang C and Ma J:
Identification and validation of an eight-lncRNA signature that
predicts prognosis in patients with esophageal squamous cell
carcinoma. Cell Mol Biol Lett. 27:392022. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Cao S, Wang X, Liu X, Li J, Duan L, Gao Z,
Lun S, Zhu Y, Yang H, Zhang H and Zhou F: Integrative analysis of
angiogenesis-related long non-coding RNA and identification of a
six-DEARlncRNA signature associated with prognosis and therapeutic
response in esophageal squamous cell carcinoma. Cancers (Basel).
14:41952022. View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Liu Y, Wang L, Liu H, Li C and He J: The
prognostic significance of metabolic syndrome and a related
Six-lncRNA signature in esophageal squamous cell carcinoma. Front
Oncol. 10:612020. View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Xie K, Zheng C, Gu W, Jiang Z, Luo C, Luo
J, Diao Y, Wang G, Cong Z, Yao X, et al: A RASSF8-AS1 based
exosomal lncRNAs panel used for diagnostic and prognostic
biomarkers for esophageal squamous cell carcinoma. Thorac Cancer.
13:3341–3352. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
151
|
Huang E, Fu J, Yu Q, Xie P, Yang Z, Ji H,
Wang L, Luo G, Zhang Y and Li K: CircRNA hsa_circ_0004771 promotes
esophageal squamous cell cancer progression via miR-339-5p/CDC25A
axis. Epigenomics. 12:587–603. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Wang W, Zhu D, Zhao Z, Sun M, Wang F, Li
W, Zhang J and Jiang G: RNA sequencing reveals the expression
profiles of circRNA and identifies a four-circRNA signature acts as
a prognostic marker in esophageal squamous cell carcinoma. Cancer
Cell Int. 21:1512021. View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Hu X, Wu D, He X, Zhao H, He Z, Lin J,
Wang K, Wang W, Pan Z, Lin H and Wang M: circGSK3β promotes
metastasis in esophageal squamous cell carcinoma by augmenting
β-catenin signaling. Mol Cancer. 18:1602019. View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Bu J, Gu L, Liu X, Nan X, Zhang X, Meng L,
Zheng Y, Liu F, Li J, Li Z, et al: The circRNA circADAMTS6 promotes
progression of ESCC and correlates with prognosis. Sci Rep.
12:137572022. View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Wang Q, Liu H, Liu Z, Yang L, Zhou J, Cao
X and Sun H: Circ-SLC7A5, a potential prognostic circulating
biomarker for detection of ESCC. Cancer Genet. 240:33–39. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Zhao Q, Zhu X, Ke JM, Su XY, Yi J, Wu DL,
Lin J and Deng ZQ: Circular RNA BMI1 serves as a potential target
for diagnosis and treatment in esophageal cancer. Technol Cancer
Res Treat. 20:153303382110330752021. View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Tang Y, Gao Z and Liu R: Identification
and function of circular RNA hsa_circ_0071106: A novel biomarker
for differentiation degree of esophageal squamous cell carcinoma.
Pathol Res Pract. 233:1538752022. View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Chen B, Dragomir MP, Yang C, Li Q, Horst D
and Calin GA: Targeting non-coding RNAs to overcome cancer therapy
resistance. Signal Transduct Target Ther. 7:1212022. View Article : Google Scholar : PubMed/NCBI
|
|
159
|
Veneziano D, Di Bella S, Nigita G, Laganà
A, Ferro A and Croce CM: Noncoding RNA: Current deep sequencing
data analysis approaches and challenges. Hum Mutat. 37:1283–1298.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Anastasiadou E, Faggioni A, Trivedi P and
Slack F: The nefarious nexus of noncoding RNAs in cancer. Int J Mol
Sci. 19:20722018. View Article : Google Scholar : PubMed/NCBI
|
|
161
|
Nemeth K, Bayraktar R, Ferracin M and
Calin GA: Non-coding RNAs in disease: From mechanisms to
therapeutics. Nat Rev Genet. 25:211–232. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
162
|
Sugihara H, Ishimoto T, Miyake K, Izumi D,
Baba Y, Yoshida N, Watanabe M and Baba H: Noncoding RNA expression
aberration is associated with cancer progression and is a potential
biomarker in esophageal squamous cell carcinoma. Int J Mol Sci.
16:27824–27834. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
163
|
Li R, Chai L, Lei L, Guo R and Wen X:
MiR-671-5p sponging activity of circMMP1 promotes esophageal
squamous cancer progression. Thorac Cancer. 14:2924–2933. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
164
|
Nan Y, Luo Q, Wu X, Chang W, Zhao P, Liu S
and Liu Z: HCP5 prevents ubiquitination-mediated UTP3 degradation
to inhibit apoptosis by activating c-Myc transcriptional activity.
Mol Ther. 31:552–568. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
165
|
Qiao G, Zhang W and Dong K: Regulation of
ferroptosis by noncoding RNAs: A novel promise treatment in
esophageal squamous cell carcinoma. Mol Cell Biochem.
477:2193–2202. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
166
|
Song W and Zou SB: Prognostic role of
lncRNA HOTAIR in esophageal squamous cell carcinoma. Clin Chim
Acta. 463:169–173. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
167
|
Sang L, Yang L, Ge Q, Xie S, Zhou T and
Lin A: Subcellular distribution, localization, and function of
noncoding RNAs. Wiley Interdiscip Rev RNA. 13:e17292022. View Article : Google Scholar : PubMed/NCBI
|
|
168
|
Shi X, Liu X, Pan S, Ke Y, Li Y, Guo W,
Wang Y, Ruan Q, Zhang X and Ma H: A novel autophagy-related long
non-coding RNA signature to predict prognosis and therapeutic
response in esophageal squamous cell carcinoma. Int J Gen Med.
14:8325–8339. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
169
|
Zhu T, Ma Z, Wang H, Wei D, Wang B, Zhang
C, Fu L, Li Z and Yu G: Immune-related long non-coding RNA
signature and clinical nomogram to evaluate survival of patients
suffering esophageal squamous cell carcinoma. Front Cell Dev Biol.
9:6419602021. View Article : Google Scholar : PubMed/NCBI
|
|
170
|
Liao G, Tang J and Bai J: Early
development of esophageal squamous cell cancer: Stem cells,
cellular origins and early clone evolution. Cancer Lett.
555:2160472023. View Article : Google Scholar : PubMed/NCBI
|
|
171
|
Hu XY, Wang R, Jin J, Liu XJ, Cui AL, Sun
LQ, Li YP, Li Y, Wang YC, Zhen YS, et al: An EGFR-targeting
antibody-drug conjugate LR004-VC-MMAE: potential in esophageal
squamous cell carcinoma and other malignancies. Mol Oncol.
13:246–263. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
172
|
Mimura K, Kono K, Maruyama T, Watanabe M,
Izawa S, Shiba S, Mizukami Y, Kawaguchi Y, Inoue M, Kono T, et al:
Lapatinib inhibits receptor phosphorylation and cell growth and
enhances antibody-dependent cellular cytotoxicity of EGFR- and
HER2-overexpressing esophageal cancer cell lines. Int J Cancer.
129:2408–2416. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
173
|
Meng L, Wu H, Wu J, Ding Pa, He J, Sang M
and Liu L: Mechanisms of immune checkpoint inhibitors: Insights
into the regulation of circular RNAS involved in cancer hallmarks.
Cell Death Dis. 15:32024. View Article : Google Scholar : PubMed/NCBI
|
|
174
|
Ding L, Lu S and Li Y: Regulation of
PD-1/PD-L1 pathway in cancer by noncoding RNAs. Pathol Oncol Res.
26:651–663. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
175
|
Peng Y, Xu Y, Zhang X, Deng S, Yuan Y, Luo
X, Hossain MT, Zhu X, Du K, Hu F, et al: A novel protein
AXIN1-295aa encoded by circAXIN1 activates the Wnt/β-catenin
signaling pathway to promote gastric cancer progression. Mol
Cancer. 20:1582021. View Article : Google Scholar : PubMed/NCBI
|
|
176
|
Zheng X, Chen L, Zhou Y, Wang Q, Zheng Z,
Xu B, Wu C, Zhou Q, Hu W, Wu C and Jiang J: A novel protein encoded
by a circular RNA circPPP1R12A promotes tumor pathogenesis and
metastasis of colon cancer via Hippo-YAP signaling. Mol Cancer.
18:472019. View Article : Google Scholar : PubMed/NCBI
|
|
177
|
Imazeki H and Kato K: Development of
chemotherapeutics for unresectable advanced esophageal cancer.
Expert Rev Anticancer Ther. 20:1083–1092. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
178
|
Fukuda T, Baba H, Okumura T, Kanda M,
Akashi T, Tanaka H, Miwa T, Watanabe T, Hirano K, Sekine S, et al:
miR-877-3p as a potential tumour suppressor of oesophageal squamous
cell carcinoma. Anticancer Res. 43:35–43. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
179
|
Yu J, Chen S, Niu Y, Liu M, Zhang J, Yang
Z, Gao P, Wang W, Han X and Sun G: Functional significance and
therapeutic potential of miRNA-20b-5p in esophageal squamous cell
carcinoma. Mol Ther Nucleic Acids. 21:315–331. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
180
|
Xing Y, Zha WJ, Li XM, Li H, Gao F, Ye T,
Du WQ and Liu YC: Circular RNA circ-Foxo3 inhibits esophageal
squamous cell cancer progression via the miR-23a/PTEN axis. J Cell
Biochem. 121:2595–2605. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
181
|
Jin Y, Meng Q, Zhang B, Xie C, Chen X,
Tian B, Wang J, Shih TC, Zhang Y, Cao J, et al: Cancer-associated
fibroblasts-derived exosomal miR-3656 promotes the development and
progression of esophageal squamous cell carcinoma via the
ACAP2/PI3K-AKT signaling pathway. Int J Biol Sci. 17:3689–3701.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
182
|
Shou Y, Wang X, Liang Y, Liu X and Chen K:
Exosomes-derived miR-154-5p attenuates esophageal squamous cell
carcinoma progression and angiogenesis by targeting kinesin family
member 14. Bioengineered. 13:4610–4620. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
183
|
Yang J, Zhang Q, Zhao P, Qiao T, Cao Z,
Gao F, Liu M and Wu S: DNA methyltransferase 3 beta regulates
promoter methylation of microRNA-149 to augment esophageal squamous
cell carcinoma development through the ring finger protein
2/Wnt/β-catenin axis. Bioengineered. 13:4010–4027. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
184
|
Yan J, Shi L, Lin S and Li Y:
MicroRNA-624-mediated ARRDC3/YAP/HIF1alpha axis enhances esophageal
squamous cell carcinoma cell resistance to cisplatin and
paclitaxel. Bioengineered. 12:5334–5347. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
185
|
Suyal G, Pandey P, Saraya A and Sharma R:
Tumour suppressor role of microRNA-335-5p in esophageal squamous
cell carcinoma by targeting TTK (Mps1). Exp Mol Pathol.
124:1047382022. View Article : Google Scholar : PubMed/NCBI
|
|
186
|
Zhang L, Yu S, Yin X, Tu M, Cai L, Zhang
Y, Yu L, Zhang S, Pan X and Huang Y: MiR-942-5p inhibits tumor
migration and invasion through targeting CST1 in esophageal
squamous cell carcinoma. PLoS One. 18:e02770062023. View Article : Google Scholar : PubMed/NCBI
|
|
187
|
Wang J, Li J, Cheng D, Zhang K, Liu W, Xue
Q, Du R, Zhou L, Yeung YT, Bai R, et al: miR-132-3p promotes heat
stimulation-induced esophageal squamous cell carcinoma
tumorigenesis by targeting KCNK2. Mol Carcinog. 62:583–597. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
188
|
Xu J, Wang J, Liu L, Chen L, Hu S and Liu
F: MicroRNA-196b is related to the overall survival of patients
with esophageal squamous cell carcinoma and facilitates tumor
progression by regulating SOCS2 (Suppressor Of Cytokine Signaling
2). Bioengineered. 12:7737–7746. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
189
|
Xue J, Xiao P, Yu X and Zhang X: A
positive feedback loop between AlkB homolog 5 and miR-193a-3p
promotes growth and metastasis in esophageal squamous cell
carcinoma. Hum Cell. 34:502–514. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
190
|
Li P, Liu X, Xing W, Qiu H, Li R, Liu S
and Sun H: Exosome-derived miR-200a promotes esophageal cancer cell
proliferation and migration via the mediating Keap1 expression. Mol
Cell Biochem. 477:1295–1308. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
191
|
Luo XJ, He MM, Liu J, Zheng JB, Wu QN,
Chen YX, Meng Q, Luo KJ, Chen DL, Xu RH, et al: LncRNA TMPO-AS1
promotes esophageal squamous cell carcinoma progression by forming
biomolecular condensates with FUS and p300 to regulate TMPO
transcription. Exp Mol Med. 54:834–847. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
192
|
Zhao Y, Zhang Q, Liu H, Wang N, Zhang X
and Yang S: lncRNA PART1, manipulated by transcriptional factor
FOXP2, suppresses proliferation and invasion in ESCC by regulating
the miR-18a-5p/SOX6 signaling axis. Oncol Rep. 45:1118–1132. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
193
|
Tang J, Xu H, Liu Q, Zheng J, Pan C, Li Z,
Wen W, Wang J, Zhu Q, Wang Z and Chen L: LncRNA LOC146880 promotes
esophageal squamous cell carcinoma progression via
miR-328-5p/FSCN1/MAPK axis. Aging (Albany NY). 13:14198–14218.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
194
|
Niu Y, Guo Y, Li Y, Shen S, Liang J, Guo W
and Dong Z: LncRNA GATA2-AS1 suppresses esophageal squamous cell
carcinoma progression via the mir-940/PTPN12 axis. Exp Cell Res.
416:1131302022. View Article : Google Scholar : PubMed/NCBI
|
|
195
|
Chen L, Lu J, Xu T, Yan Z, Guo Y, Dong Z
and Guo W: KTN1-AS1, a SOX2-mediated lncRNA, activates
epithelial-mesenchymal transition process in esophageal squamous
cell carcinoma. Sci Rep. 12:201862022. View Article : Google Scholar : PubMed/NCBI
|
|
196
|
Dong Z, Yang L, Lu J, Guo Y, Shen S, Liang
J and Guo W: Downregulation of LINC00886 facilitates
epithelial-mesenchymal transition through SIRT7/ELF3/miR-144
pathway in esophageal squamous cell carcinoma. Clin Exp Metastasis.
39:661–677. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
197
|
Pan C, Chen G, Zhao X, Xu X and Liu J:
lncRNA BBOX1-AS1 silencing inhibits esophageal squamous cell cancer
progression by promoting ferroptosis via miR-513a-3p/SLC7A11 axis.
Eur J Pharmacol. 934:1753172022. View Article : Google Scholar : PubMed/NCBI
|
|
198
|
Zhang H, Pan E, Zhang Y, Zhao C, Liu Q, Pu
Y and Yin L: LncRNA RPL34-AS1 suppresses the proliferation,
migration and invasion of esophageal squamous cell carcinoma via
targeting miR-575/ACAA2 axis. BMC Cancer. 22:10172022. View Article : Google Scholar : PubMed/NCBI
|
|
199
|
Lu JT, Yan ZY, Xu TX, Zhao F, Liu L, Li F
and Guo W: Reciprocal regulation of LINC00941 and SOX2 promotes
progression of esophageal squamous cell carcinoma. Cell Death Dis.
14:722023. View Article : Google Scholar : PubMed/NCBI
|
|
200
|
Fu X, Chen X, Si Y, Yao Y, Jiang Z and
Chen K: Long non-coding RNA NCK1-AS1 is overexpressed in esophageal
squamous cell carcinoma and predicts survival. Bioengineered.
13:8302–8310. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
201
|
Wang C, Zhou M, Zhu P, Ju C, Sheng J, Du
D, Wan J, Yin H, Xing Y, Li H, et al: IGF2BP2-induced circRUNX1
facilitates the growth and metastasis of esophageal squamous cell
carcinoma through miR-449b-5p/FOXP3 axis. J Exp Clin Cancer Res.
41:3472022. View Article : Google Scholar : PubMed/NCBI
|
|
202
|
Song H, Tian D, Sun J, Mao X, Kong W, Xu
D, Ji Y, Qiu B, Zhan M and Wang J: circFAM120B functions as a tumor
suppressor in esophageal squamous cell carcinoma via the
miR-661/PPM1L axis and the PKR/p38 MAPK/EMT pathway. Cell Death
Dis. 13:3612022. View Article : Google Scholar : PubMed/NCBI
|
|
203
|
Guan X, Guan X, Wang Y, Lan T, Cheng T,
Cui Y and Xu H: Circ_0003340 downregulation mitigates esophageal
squamous cell carcinoma progression by targeting miR-940/PRKAA1
axis. Thorac Cancer. 13:1164–1175. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
204
|
Liang W, Wang C, Wang J and Zhang M:
Hsa_circ_0023984 regulates cell proliferation, migration, and
invasion in esophageal squamous cancer via regulating
miR-1294/PI3K/Akt/c-Myc pathway. Appl Biochem Biotechnol. 194:1–16.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
205
|
Qian CJ, Tong YY, Wang YC, Teng XS and Yao
J: Circ_0001093 promotes glutamine metabolism and cancer
progression of esophageal squamous cell carcinoma by targeting
miR-579-3p/glutaminase axis. J Bioenerg Biomembr. 54:119–134. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
206
|
Tang R, Zhou Q, Xu Q, Lu L and Zhou Y:
Circular RNA circ_0006948 promotes esophageal squamous cell
carcinoma progression by regulating microRNA-3612/LASP1 axis. Dig
Dis Sci. 67:2158–2172. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
207
|
Wang J, Li X, Duan C and Jia Y: CircFNDC3B
knockdown restrains the progression of oesophageal squamous cell
carcinoma through miR-214-3p/CDC25A axis. Clin Exp Pharmacol
Physiol. 49:1209–1220. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
208
|
Sun Z, Zhang S, Zhang N, Wang J, Wang J
and Liu J: Circ_0005231 promotes the progression of esophageal
squamous cell carcinoma via sponging miR-383-5p and regulating
KIAA0101. Thorac Cancer. 13:1751–1762. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
209
|
Wang YM, Zhao QW, Sun ZY, Lin HP, Xu X,
Cao M, Fu YJ, Zhao XJ, Ma XM and Ye Q: Circular RNA
hsa_circ_0003823 promotes the tumor progression, metastasis and
apatinib resistance of esophageal squamous cell carcinoma by
miR-607/CRISP3 axis. Int J Biol Sci. 18:5787–5808. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
210
|
Meng F, Zhang X, Wang Y, Lin J, Tang Y,
Zhang G, Qiu B, Zeng X, Liu W and He X: Hsa_circ_0021727
(circ-CD44) promotes ESCC progression by targeting miR-23b-5p to
activate the TAB1/NFκB pathway. Cell Death Dis. 14:92023.
View Article : Google Scholar : PubMed/NCBI
|