Introduction
Esophageal cancer (EC) is the 10th most prevalent
malignancy worldwide and the seventh leading cause of
cancer-associated mortalities. According to statistics, in 2020,
there were 604,100 new cases of and 544,076 deaths from esophageal
cancer worldwide (1). The most
prevalent histological form of EC, squamous cell carcinoma (SCC),
accounts for ~85% of all cases globally (2). Despite the advances in medical
technology, the prognosis for patients with esophageal SCC (ESCC)
remains unsatisfactory (3).
Understanding the molecules involved in the development of ESCC may
lead to new insight for developing methods of improving patient
prognosis.
During the last 20 years, 22 human and 34 mouse
members of the nucleotide binding and oligomeric domain-like
receptor (NLR) family have been identified (4). The sole NLR member found in
mitochondria is NLRX1, a member of the NLRC subfamily. It is
involved in immune, inflammatory, autophagic, mitochondrial
regulatory and metabolic processes (5). In addition, NLRX1 is associated with
the initiation and progression of cancer (6–9). In an
azomethane (AOM)-induced colorectal cancer model, as well as in
estrogen receptor (ER)/progesterone receptor (PR)- breast cancer
and in human papillomavirus-induced head and neck SCC, it has been
reported that NLRX1 can enhance tumor growth. NLRX1 can also slow
down the growth of pancreatic cancer, ER/PR+ breast cancer,
hepatocellular carcinoma, histiocyte sarcoma and a dextran sodium
sulfate/AOM-induced colitis model (6,7,10–14).
This suggests that NLRX1 can stimulate or repress tumor growth via
as-yet-unidentified mechanisms. However, the association between
NLRX1 and ESCC has so far remained elusive.
The present study aimed to identify potential links
between NLRX1 and ESCC. First, bioinformatics were used to examine
NLRX1 expression, prognosis, potential biological activities,
interaction with immune infiltration and treatment sensitivity in
ESCC. Next, in vitro assays were performed to investigate
the possible biological function of NLRX1 in ESCC. The present
findings imply that NLRX1 may be a crucial tumor suppressor in
ESCC.
Materials and methods
Data and patient specimen
collection
From the Gene Expression Omnibus (GEO) database
(https://www.ncbi.nlm.nih.gov/geo/),
the ESCC gene expression datasets GSE20347, GSE23400, GSE53625,
GSE67269 and GSE161533) were downloaded (15–18).
Furthermore, information on the gene expression and survival of 95
patients with ESCC was gathered from The Cancer Genome Atlas (TCGA;
http://cancergenome.nih.gov/). R v4.2.1
software was used for analysis in the current study (19,20).
All transcriptome data underwent the necessary preprocessing before
statistical analysis, including averaging multiple expression
values of the same gene in each dataset, followed by logarithmic
transformation and normalization of the dataset using the R package
‘limma’ (21).
Furthermore, paraffin blocks of cancerous and
healthy tissues adjacent to the tumor were collected from patients
with ESCC who visited The Second Affiliated Hospital of Zhengzhou
University (Zhengzhou, China) between January 2022 and March 2023.
The inclusion criteria were as follows: All patients had ESCC, were
aged 18–75 years, all surgical specimens had been taken before any
neo-adjuvant anti-tumor treatment and all patients had
corresponding informed consent. Exclusion criteria were the
presence of other malignant tumors, psychological abnormalities or
contraindications to anti-tumor treatment.
Cell lines and culture
ECA109 and KYSE450 cells were obtained from the
American Type Culture Collection, while KYSE150 cells were
purchased from Shanghai Zhongqiao Xinzhou Biotechnology Co., Ltd.
The culture medium used contained 89% RPMI 1640 medium (cat. no.
ZQ0449; Zhongqiao Xinzhou Biotechnology Co., Ltd.), 10% fetal
bovine serum (cat. no. 04-001-1ACS; Biological Industries), 1% 100
IU/ml penicillin and 100 g/ml streptomycin (cat. no. CSP006;
Zhongqiao Xinzhou Biotechnology Co., Ltd.). Cells were cultured at
37°C and 5% CO2 saturated humidity.
Expression and clinical correlation
analysis of NLRX1
First, the ‘limma’ package was used to perform
differential analysis on the transcriptome data of the datasets.
Next, patients with ESCC in the GSE53625 and the TCGA dataset
containing clinical data were divided into two groups using the
median expression value of NLRX1: GSE53625 high: n=89, low: n=90;
and TCGA high: n=47, low: n=48. Subsequently, Kaplan-Meier survival
analysis was performed on them.
The GSE53625 dataset, which had the largest sample
size, was used for subsequent analysis using the Wilcoxon
signed-rank or χ2 tests to analyze the relationship
between NLRX1 and clinical reference data. Subsequently, a receiver
operating characteristic (ROC) curve was plotted to evaluate the
diagnostic value of NLRX1, and Cox regression analysis was used to
help judge the prognostic value of NLRX1. Finally, the ‘survival’
package was used for regression analysis. A nomogram was
constructed and a calibration curve was drawn to verify its
reliability.
Enrichment and variation analysis
First, based on the transcriptome data of the
GSE53625 dataset, the correlation between NLRX1 and the expression
of other genes was analyzed, and genes highly correlated with NLRX1
were identified with |correlation coefficient|>0.5 and P<0.05
as selection criteria. Next, Gene Ontology (GO) and Kyoto
Encyclopedia of Genes and Genomes (KEGG)-enrichment analyses were
performed on these genes, while Gene Set Enrichment Analysis (GSEA)
and Gene Set Variation Analysis (GSVA) were used to explore the
pathways enriched in the low expression group of NLRX1 in GSE53625.
This step utilized the ‘clusterProfiler’, ‘GSEABase’ and ‘GSVA’
packages.
Immune and drug correlation
analysis
Based on the data from GSE53625, patients were
divided into high and low groups according to the median expression
value of the NLRX1, and immune-related functional analysis was
performed using the ‘GSEABase’ and ‘GSVA’ packages (22,23).
Next, the ‘Corrplot’ package was used for immune checkpoint
correlation analysis, while the ‘CIBERSORT’ package was used to
analyze the degree of immune infiltration in samples and screen out
meaningful samples for differential analysis and correlation
testing. Finally, from the Genomics of Drug Sensitivity in Cancer
website (https://www.cancerrxgene.org/), drug information was
downloaded and differences in drug sensitivity between patients
with high and low expression of NLRX1 were analyzed.
Immunohistochemical staining
Immunohistochemical staining was performed as
previously described (24). In
brief, paraffin-embedded tissues were dewaxed, hydrated and
subjected to antigen retrieval and blocking of endogenous
peroxidase prior to incubation with anti-NLRX1 antibody (1:200
dilution; cat. no. ab107611; Abcam) at room temperature for 30 min.
Next, a conjugated secondary antibody was added (1:100 dilution;
cat. no. SA00001-2; Proteintech Group, Inc.) and incubated at room
temperature for 30 min, followed by staining with DAB, hematoxylin
and alkaline bluing solution. Next, a neutral resin was applied for
sealing after dehydration before observing the slides under an
optical microscope at ×200 magnification and acquiring images.
A semi-quantitative scoring technique based on the
percentage of positive cells and staining intensity was used to
grade the immunohistochemical staining results as follows: 0,
negative staining; 1, pale yellow (weak); 2, yellow (moderate); and
3, brown (strong). The positivity score was assigned as 0 for
<5% positive cells; 1 for 6–25% positive cells; 2 for 26–50%
positive cells; 3 for 51–75% positive cells; and 4 for >75%
positive cells. The two aforementioned scores were then multiplied
and 0 was considered to indicate negative expression, <5 low
expression and ≥5 high expression. Two pathologists independently
evaluated the immunohistochemical staining.
Cell transfection and plasmid
extraction
The plasmids (pcDNA3.1-NLRX1) and small interfering
RNAs (siRNAs) used in the present study were produced by TsingKe
Biological Technology and plasmid extraction was carried out in
line with the instructions provided by the manufacturer of FastPure
EndoFree Plasmid Mini Kit (cat. no. DC203-01; Vazyme Biotech Co.,
Ltd.). Approximately 4×105 KYSE450 cells or
3×105 EAC109 cells were seeded into 6-well plates and
transfection was performed the following day when the cell density
reached 70–80%. Transfection was carried out according to the
manufacturer's instructions of Lipo8000 transfection reagent (cat.
no. C0533; Beyotime Institute of Biotechnology) and subsequent
experiments were conducted within 48 h. The siRNA sequences were as
follows: siNLRX1 forward, 5′-GGACUACUACAACGAUGAUTT-3′ and reverse,
5′-AUCAUCGUUGUAGUAGUCCTT-3′; and negative control (NC) forward,
5′-UUCUCCGAACGUGUCACGUTT-3′ and reverse,
5′-ACGUGACACGUUCGGAGAATT-3′.
Western blot (WB) analysis
RIPA lysis buffer (cat. no. R0010; Solarbio Group,
Inc.) was used to lyse cells, and protein was quantified using the
BCA method. Total protein (20 µg of protein was added to each lane)
was separated by 7.5% SDS-PAGE prepared according to the
instructions of the Epizyme PAGE Gel Express Preparation Kit (cat.
no. PG111; Epizyme, Inc.; Ipsen Biopharmaceuticals, Inc.). The
electrophoresis and PVDF membrane transfer times were adjusted
according to the molecular weight of the proteins. A fastblocking
solution (cat. no. G2052; Wuhan Servicebio Technology Co., Ltd.)
was used to block the membrane at room temperature for 10 min,
while an antibody diluent (cat. no. BMU103-CN; Abbkine Scientific
Co., Ltd.) was used to dilute the primary antibodies to an
appropriate concentration prior to incubation at 4°C overnight.
Next, the membrane was incubated with a secondary antibody at room
temperature for 2 h. The expression of proteins was then visualized
by enhanced chemiluminescence agents (cat. no. BMU102-CN; Abbkine
Scientific Co., Ltd.). Anti-NLRX1 (1:1,000 dilution; cat. no.
ab107611; Abcam), anti-PI3K/AKT signaling pathway panel (1:1,000;
cat. no. ab283852; Abcam) and anti-GAPDH (1:10,000; cat. no.
10494-1-AP; Proteintech Group, Inc.) were used as primary
antibodies. The phosphorylation sites of phosphorylated (p)-AKT
were S472, S473 and S474, and anti-GAPDH was used as a loading
control for normalization. HRP-conjugated Affinipure goat
anti-rabbit IgG (H+L) (1:2,000; cat. no. SA00001-2; Proteintech
Group, Inc.) was used as the secondary antibody.
Cell proliferation assay
For the cell-proliferation experiment, cells
transfected on a 6-well culture plate were collected and 2,000
cells per well were added to a fresh 96-well culture plate. When
the cells had adhered to the wall after 6 h of incubation, 10 µl
Cell Counting Kit-8 (CCK-8) reagent (cat. no. BMU106-CN; Abbkine
Scientific Co., Ltd.) was added. After 2 h, the optical density
(OD) values at 450 nm were measured by a microplate reader (Thermo
Fisher Scientific, Inc.) and considered as time=0 h. Next, the OD
values at 450 nm were measured at 24, 48 and 72 h.
Wound-healing assay
At the back of a 6-well culture plate, three
horizontal lines were drawn in advance with a black pen, while
three vertical lines were drawn on the 6-well culture plate with a
pipette tip at 48 h post-transfection (cell confluence rate reached
>90%) (25). Cells were cultured
in medium containing 2% serum (26)
and the migration of cells at the intersection of the horizontal
and vertical lines was recorded at 0, 24, 48 and 72 h. Results were
calculated as Mobility (%)=(A0-AN)/A0, where A0 represents the
initial wound width and AN represents the remaining wound width at
the metering point.
Transwell assays
In the Transwell migration assay, cells were
collected and resuspended in serum-free medium and 40,000 cells/200
µl were added to each upper chamber of a Transwell plate (8.0-µm
pore size; Corning, Inc.), while 600 µl complete medium was added
to each lower chamber. After 36 h, the cells were fixed at room
temperature for 30 min with 4% paraformaldehyde and then stained at
room temperature for 10 min with crystal violet. After staining,
the cells at the top of the pore filter were gently wiped off with
a wet cotton swab. The number of migrated cells was observed under
a microscope. For the invasion assay, the culture medium was
replaced with serum-free culture medium for starvation treatment 1
day in advance. Next, Matrigel® (cat. no. 356234; BD
Biosciences) was diluted with serum-free medium and 100 µl of this
solution was added to each well. After incubation at 37°C for 1 h,
any uncured diluent was aspirated and the next steps were the same
as for the migration assay.
Apoptosis analysis
Approximately 1×105 KYSE450 cells or
7.5×104 ECA109 cells into were inoculated in each well
of a 24-well plate. According to the instructions of the
manufacturer of the apoptosis kit (cat. no. KTA0002; Abbkine
Scientific Co., Ltd.), a mixture was prepared at a ratio of
apoptosis inducer to complete culture medium of 1:3,000. After 48 h
of transfection, 1 ml mixture was added to KYSE450 or ECA109 cells
cultured in a 24-well culture plate. After 24 h of induction,
Annexin V/PI staining reagent was added and the fluorescence
emission of the sample on the slide was observed using a
fluorescence microscope. Green fluorescence (Annexin V) represents
cell membrane staining and red fluorescence [propidium iodide (PI)]
represents cell nuclear staining. Cells exhibiting only green
fluorescence represented early apoptotic cells, while cells
exhibiting only red fluorescence represented necrotic cells and
cells exhibiting both red and green fluorescence represented late
apoptotic cells. Furthermore, the cells treated with the
above-mentioned kit were prepared into a suspension and analyzed by
flow cytometry (BeamCyte Flow Cytometer; Bidake Biotechnology Co.,
Ltd.), with 10,000 effective cells recorded each time to detect the
apoptosis rate, and the software used was CytoSYS v1.0 (Bidake
Biotechnology Co., Ltd.).
Statistical analysis
SPSS 26.0 (IBM Corp.) was used for statistical
analysis. Each experiment was performed as 3 repeats. Continuous
variables were presented as the mean ± standard error of the mean.
To determine the significance of the difference between the two
groups, an unpaired Student's t-test, Fisher's exact test or
χ2 test was employed as appropriate. If the variances
within the groups were not homogenous, nonparametric Mann-Whitney U
or Wilcoxon signed-rank tests were used. P<0.05 was considered
to indicate a statistically significant difference.
Results
Expression of NLRX1 at the mRNA and
protein levels
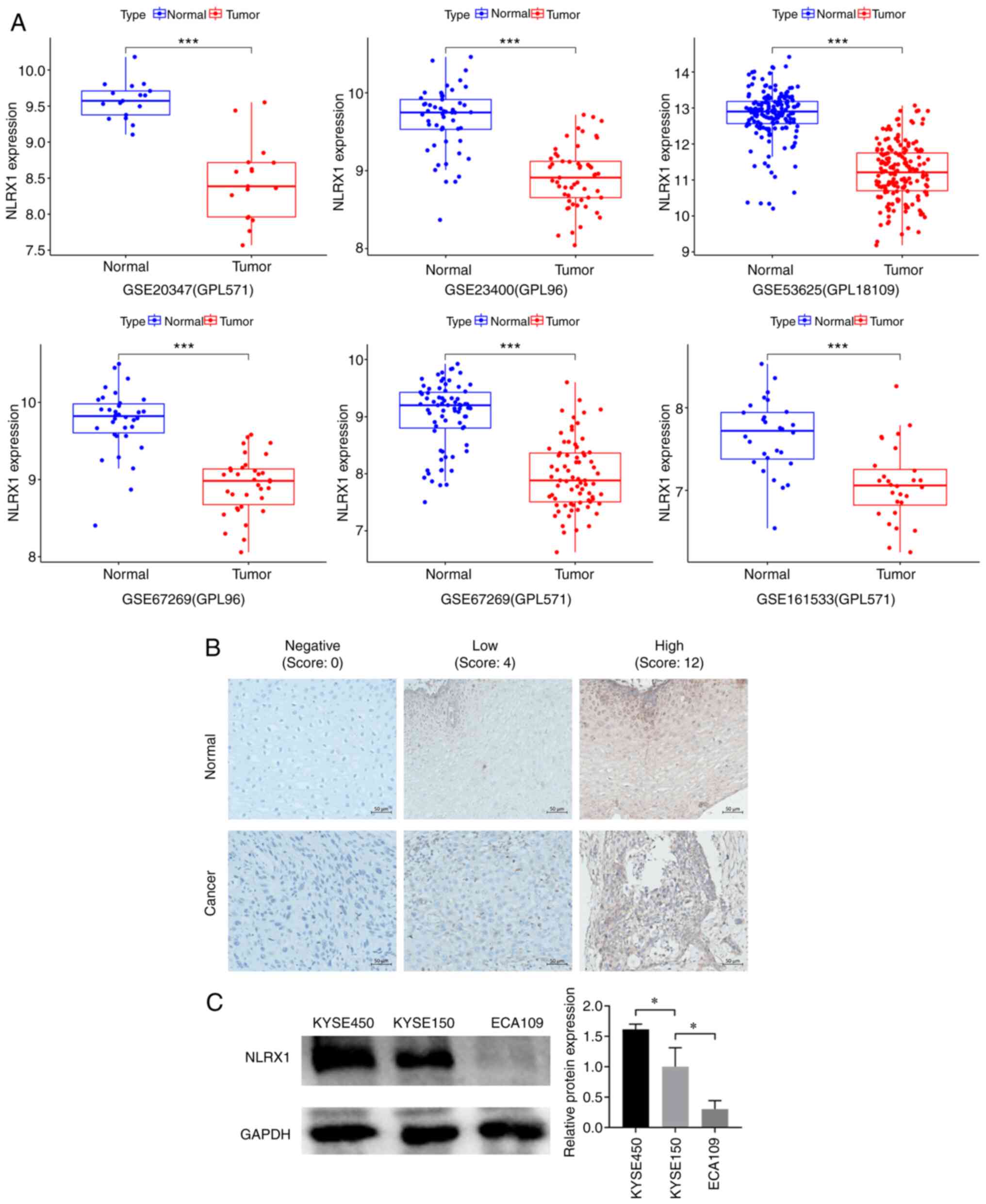
Data mining revealed that, in contrast to normal
adjacent tissue, NLRX1 was expressed at lower mRNA levels in ESCC
tissue (Fig. 1A). To determine
whether there were changes in the expression of NLRX1 at the
protein level in 36 pairs of cancerous and adjacent normal tissues
[24 males and 12 females; 11 patients aged <65 years and 25
patients aged ≥65 years; median age, 68 years (range, 18–75 years),
immunohistochemistry was employed (Fig.
1B). The results showed that NLRX1 protein expression was lower
in cancer tissues (19 cases of low expression) than in adjacent
normal tissues (10 cases of low expression), with a statistically
significant difference (χ2=4.677, P=0.031, Table I).
 | Table I.Expression of NLRX1 in esophageal
squamous cell carcinoma and normal tissues. |
Table I.
Expression of NLRX1 in esophageal
squamous cell carcinoma and normal tissues.
| Group | Cancer (n=36) | Normal (n=36) | χ2 | P-value |
|---|
| NLRX1 |
|
| 4.677 | 0.031 |
|
Low | 19 | 10 |
|
|
|
High | 17 | 26 |
|
|
Among the KYSE450, KYSE150 and ECA109 cell lines,
NLRX1 expression was higher in KYSE450 cells, while the expression
level of NLRX1 in ECA109 cells was lower (P<0.05 vs. KYSE150;
Fig. 1C). Therefore, in subsequent
experiments, NLRX1 expression was knocked down in KYSE450 cells and
overexpressed in ECA109 cells.
Clinical and prognostic value of NLRX1
in ESCC
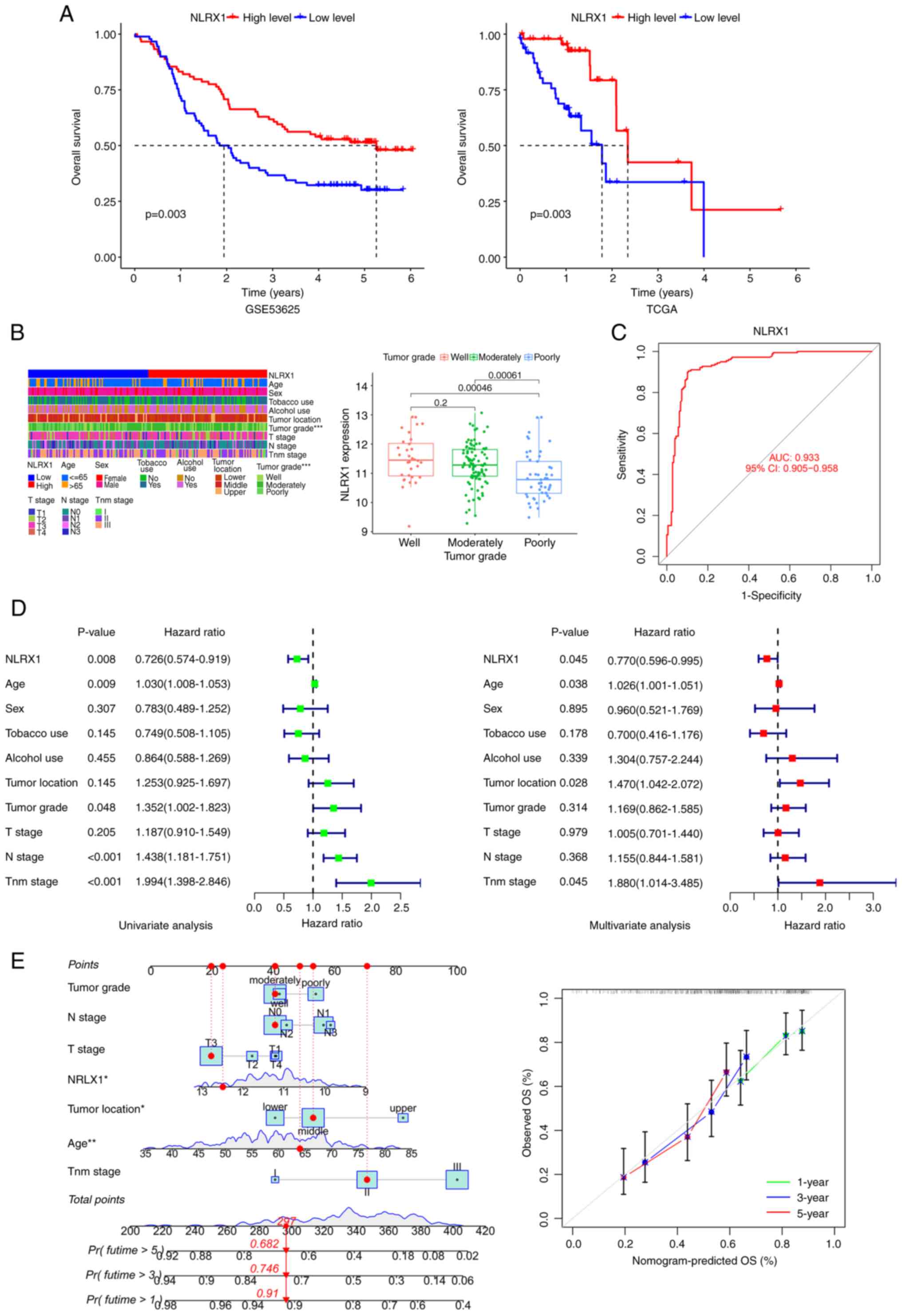
Patients with low NLRX1 levels had shorter survival
times in the GSE53625 and TCGA datasets (Fig. 2A). An association between NLRX1 and
the pathological parameter of tumor grade was found in patients
with ESCC (Fig. 2B). In the ROC
curve, the area under the curve of NLRX1 expression was 0.933,
indicating that NLRX1 has a good diagnostic value (Fig. 2C). Univariate and multivariate Cox
regression analyses showed that NLRX1 was an independent prognostic
factor for OS with a hazard ratio of 0.726 [95% confidence interval
(CI), 0.574–0.919] and 0.770 (95% CI, 0.596–0.995), respectively
(Fig. 2D). Next, a nomogram was
created based on the NLRX1 expression and clinical data contained
in the GSE53625 dataset as parameters to predict the prognosis of
patients with ESCC. The calibration curves of OS showed good
consistency between the predicted OS and the observed OS,
indicating that the predicted results of the nomogram are in good
agreement with the actual results (Fig.
2E).
Identification of relevant genes for
NLRX1 and enrichment analysis
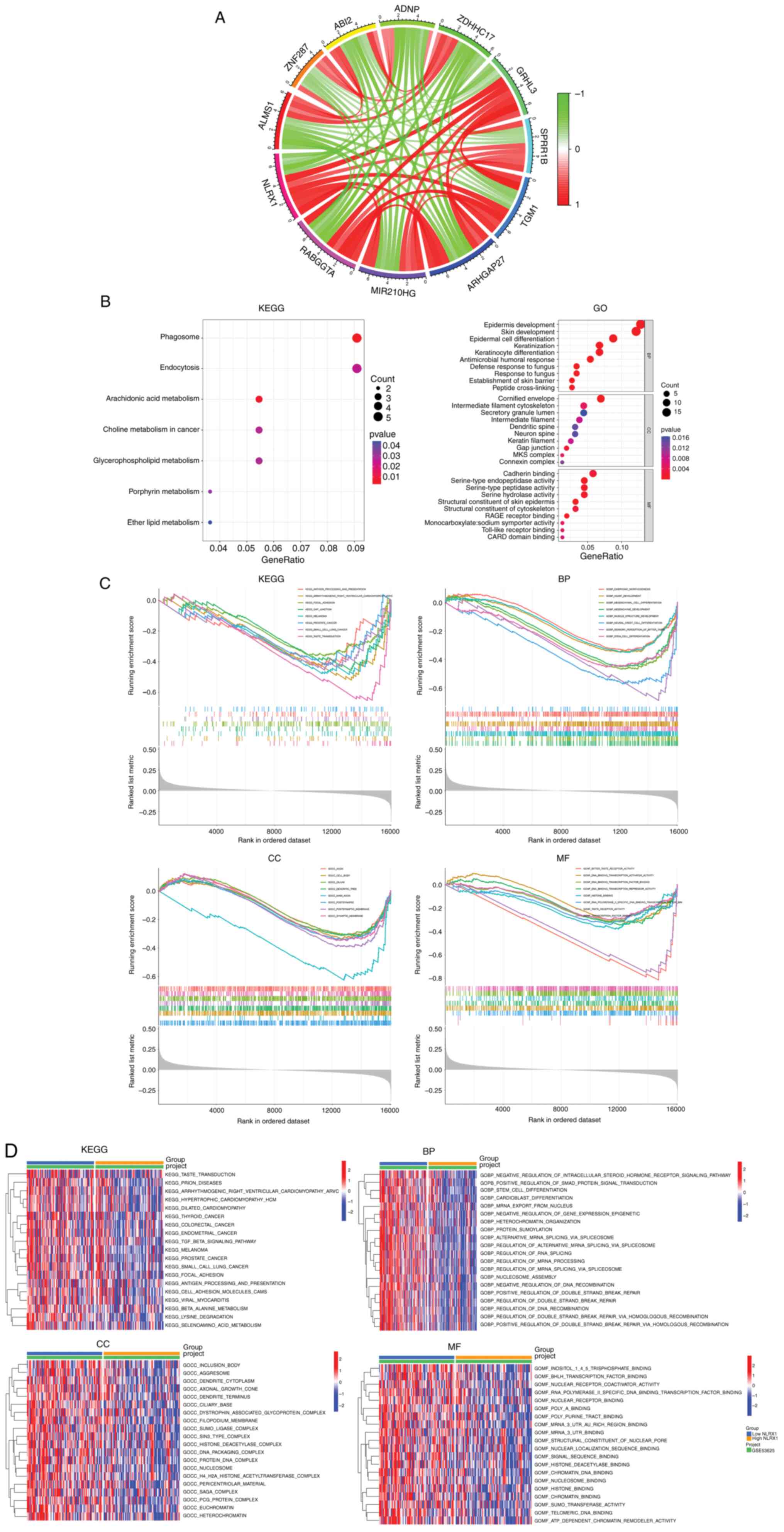
Based on co-expression analysis, 181 genes were
found to be highly correlated with NLRX1 (Table SI), and Circos plots were used to
show the 6 genes that were most positively correlated with NLRX1
and the 5 genes that were most negatively correlated with NLRX1
(Fig. 3A). In the KEGG analysis,
NLRX1-related genes were mainly enriched in ‘phagosome’,
‘arachidonic acid metabolism’ and ‘endocytosis’, while in the GO
analysis, they were mainly enriched in ‘skin development’,
‘cornified envelope’ and ‘structural constituent of skin epidermis’
(Fig. 3B).
Next, the tumor samples of GSE53625 were divided
into two groups according to the median expression value of NLRX1,
and GSEA and GSVA were performed. The results showed that
pathways/functional terms including ‘taste transduction’,
‘arrhythmogenic right ventricular cardiomyopathy’, ‘melanoma’,
‘focal adhesion’, ‘small cell lung cancer’, ‘antigen processing and
presentation’, ‘prostate cancer’, ‘stem cell differentiation’,
‘polymerase II specific DNA binding transcription factor binding’
and ‘histone binding’ were enriched in the NLRX1 low expression
group (n=90) (P<0.05; Fig. 3C and
D).
Immunological and drug sensitivity
analysis
To investigate whether NLRX1 is immune-related in
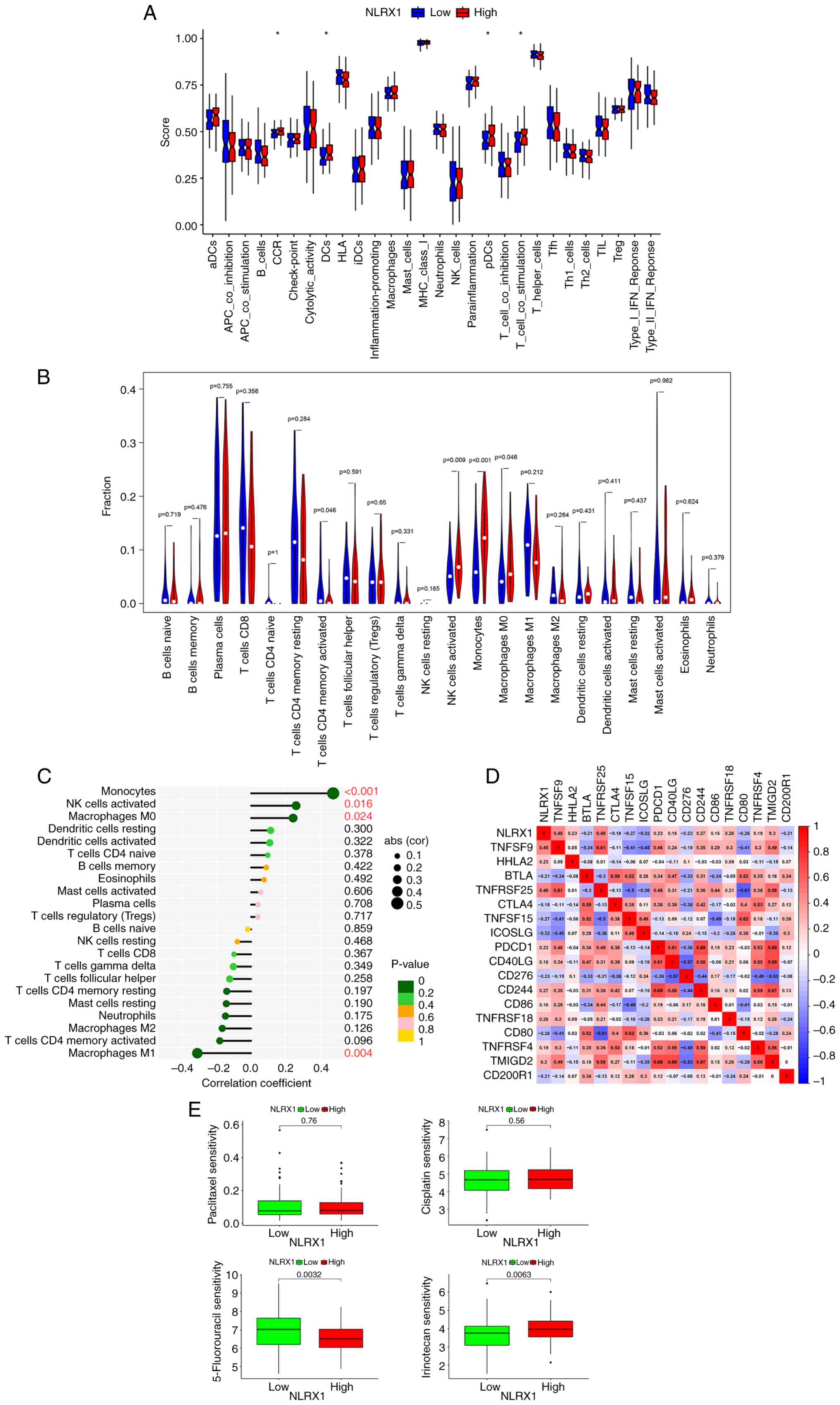
ESCC, an immune-related functional analysis was first performed.
The findings demonstrated that in 179 ESCC samples, the group with
low NLRX1 expression (n=90) had lower scores for CC chemokine
receptors (CCR), dendritic cells (DCs), plasma-like dendritic cells
(pDCs) and T-cell co-stimulation (Fig.
4A, P<0.05; Table SII
contains definitions of the terms). Afterward, 85 meaningful
samples (P<0.05) were analyzed using the ‘CIBERSORT’ package for
immune infiltration analysis. According to the median expression
value of NLRX1, they were divided into two groups. In the
differential analysis and correlation testing, the degree of
natural killer (NK) cells activated, monocytes and macrophages M0
infiltration decreased in the low NLRX1 group (n=43) and was highly
positively correlated with NLRX1 expression (Fig. 4B and C). Immunological checkpoint
analysis showed that TNFRSF25, TNFSF9, TMIGD2, CD244, TNFRSF18,
HHLA2, PDCD1, TNFRSF4, CD40LG and CD86 were positively correlated
with NLRX1, while ICOSLG, TNFSF15, CD80, CD276, BTLA, CD200R1 and
CTLA4 were negatively correlated with NLRX1 (Fig. 4D, P<0.05). Drug sensitivity
analysis revealed that the group with low NLRX1 expression was more
resistant to 5-fluorouracil and more sensitive to irinotecan
(Fig. 4E).
Downregulation of NLRX1 expression
promotes the growth and development of KYSE450 cells
In order to further explore the biological function
of NLRX1 in ESCC, in vitro experiments were conducted. As
confirmed by WB, the expression of NLRX1 was knocked down in
KYSE450 cells (Fig. 5A). The
results of the CCK8 assay showed that knockdown of NLRX1
significantly promoted the growth of KYSE450 cells (Fig. 5B, P<0.05). Furthermore, the
scratch wound-healing and Transwell assays showed that silencing
NLRX1 expression significantly promoted the migration and invasion
of KYSE450 cells (Fig. 5C and D,
P<0.05). The fluorescence microscopy and flow cytometry results
of the apoptosis experiment also showed that knockdown of NLRX1
significantly reduced the apoptosis of KYSE450 cells (Fig. 5E, P<0.05).
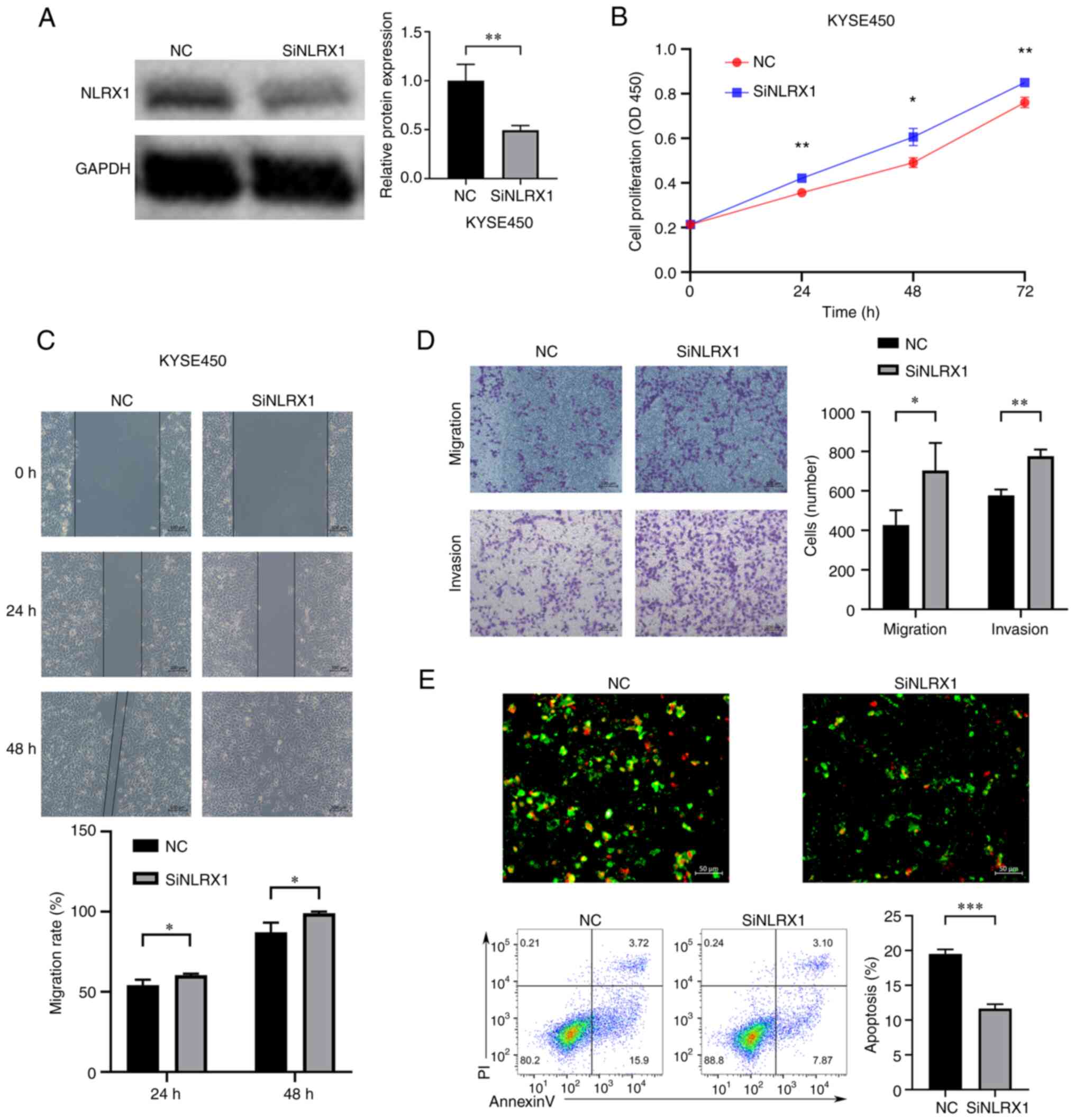 | Figure 5.Downregulation of NLRX1 expression
promotes the growth and development of KYSE450 cells. (A) WB
results demonstrate knockdown efficiency. (B) Determination of the
effect of NLRX1 on the proliferation of KYSE450 cells through Cell
Counting Kit-8 assays. (C) Evaluation of the effect of NLRX1 on
KYSE450 cell migration using the scratch wound-healing assay (scale
bar, 100 µm). (D) Evaluation of the impact of NLRX1 on KYSE450 cell
migration and invasion by the Transwell assay (scale bar, 100 µm).
(E) Evaluation of the effect of NLRX1 on KYSE450-cell apoptosis
through fluorescence microscopy (scale bar, 50 µm; green
fluorescence represents cell membrane staining and red fluorescence
represents cell nuclear staining. Cells exhibiting only green
fluorescence represent early apoptotic cells, while cells
exhibiting only red fluorescence represented necrotic cells and
cells exhibiting both red and green fluorescence represent late
apoptotic cells) and flow cytometry. *P<0.05, **P<0.01,
***P<0.001. NLRX1, nucleotide binding and oligomeric domain-like
receptor X1; NC, negative control; siNLRX1, small interfering RNA
targeting NLRX1; OD450, optical density at 450 nm; P-AKT,
phosphorylated AKT; PI, propidium iodide; WB, western blot. |
Upregulation of NLRX1 expression
inhibits the growth and development of ECA109 cells
In ECA109 cells, NLRX1 was overexpressed (Fig. 6A). According to our findings,
upregulation of NLRX1 expression in ECA109 cells significantly
inhibited cell growth, migration and invasion, and significantly
increased apoptosis (Fig. 6B-E,
P<0.05).
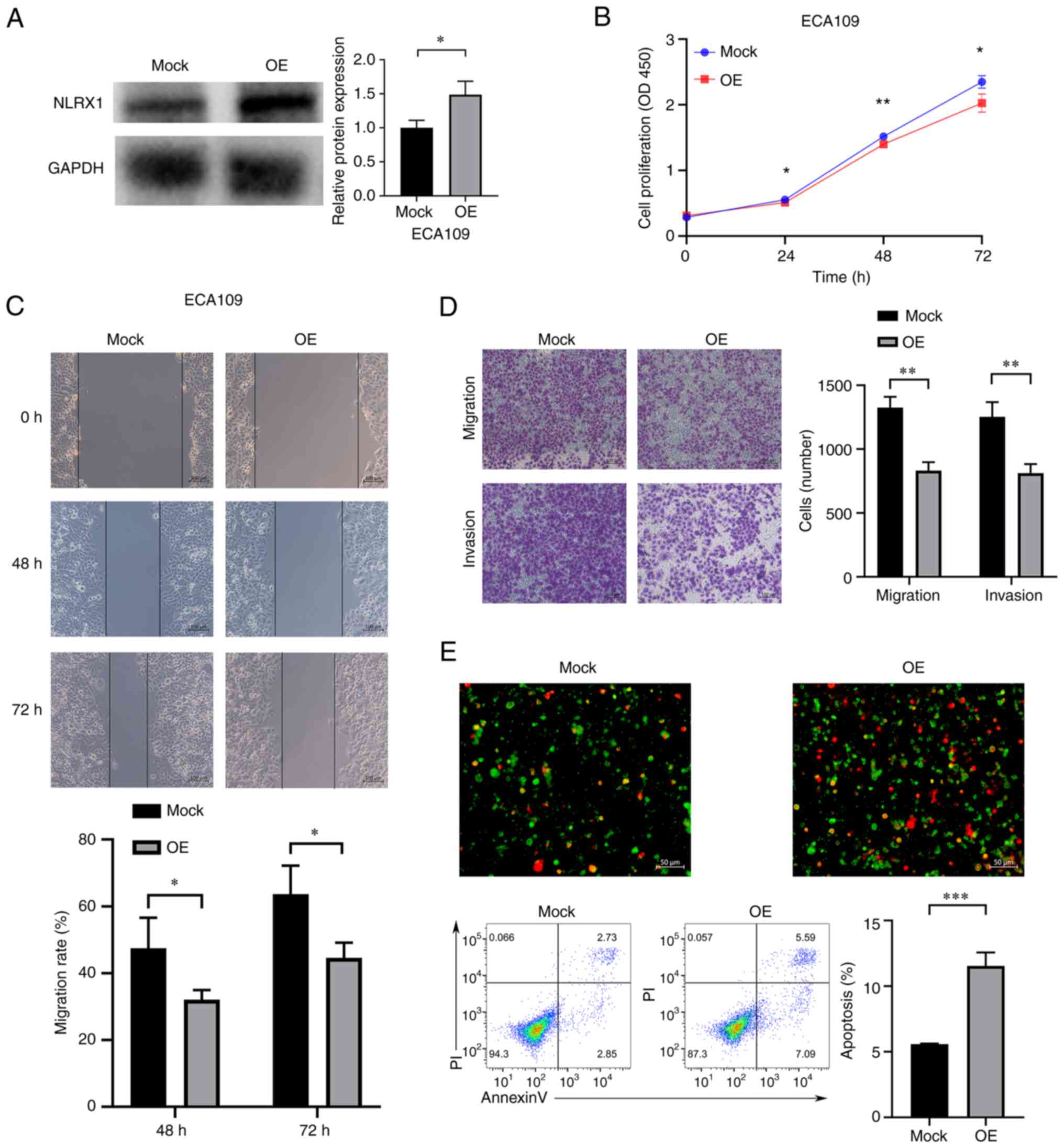 | Figure 6.Upregulation of NLRX1 expression
inhibits the growth and development of EC109 cells. (A) WB results
demonstrate overexpression efficiency. (B) Determination of the
effect of NLRX1 on the proliferation of ECA109 cells through Cell
Counting Kit-8 assays. (C) Evaluation of the effect of NLRX1 on
ECA109 cell migration using the scratch wound-healing assay (scale
bar, 100 µm). (D) Evaluation of the impact of NLRX1 on ECA109 cell
migration and invasion by the Transwell assay (scale bar, 100 µm).
(E) Evaluation of the effect of NLRX1 on ECA109-cell apoptosis
through fluorescence microscopy (scale bar, 50 µm; green
fluorescence represents cell membrane staining and red fluorescence
represents cell nuclear staining. Cells exhibiting only green
fluorescence represent early apoptotic cells, while cells
exhibiting only red fluorescence represented necrotic cells and
cells exhibiting both red and green fluorescence represent late
apoptotic cells) and flow cytometry. *P<0.05, **P<0.01,
***P<0.001. NLRX1, nucleotide binding and oligomeric domain-like
receptor X1; OE, overexpression; OD450, optical density at 450 nm;
PI, propidium iodide; WB, western blot. |
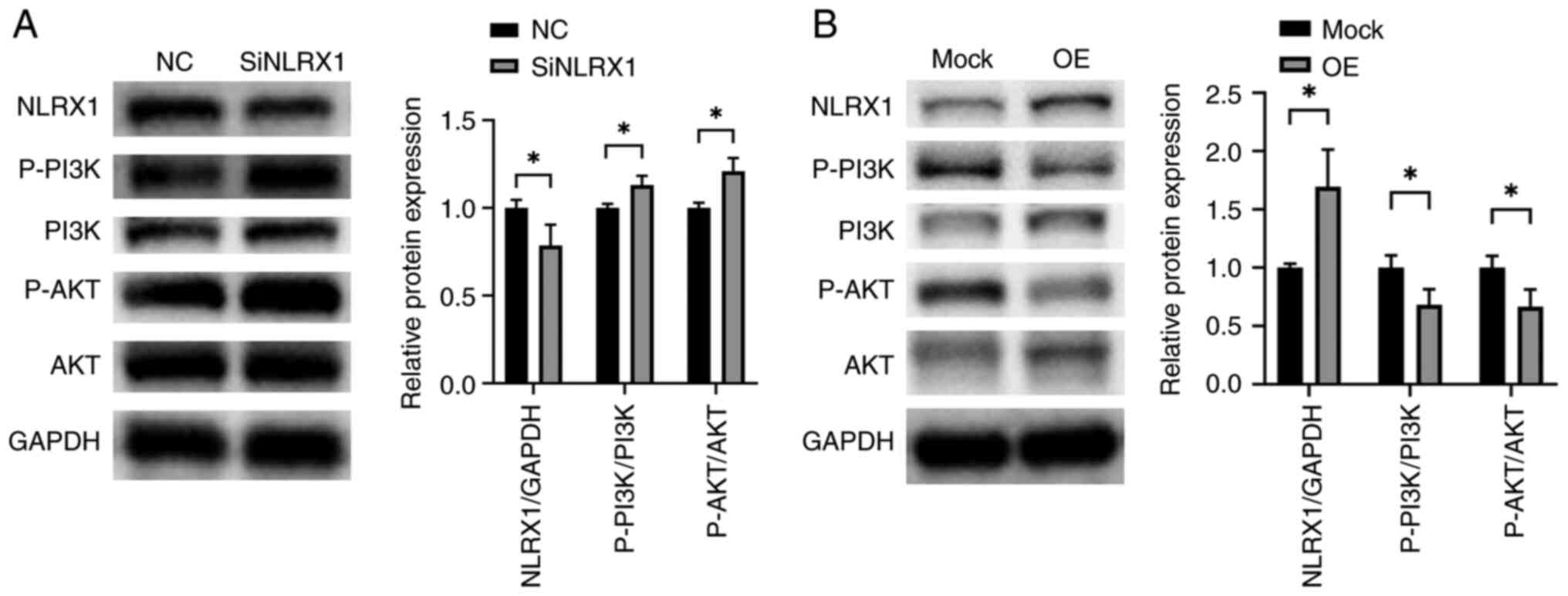
NLRX1 regulates the PI3K/AKT signaling
pathway in ESCC
To further explore the mechanism by which NLRX1
regulates the growth and development of ESCC, we experimentally
analyzed the effects of NLRX1 on the PI3K/AKT pathway. According to
the WB data, silencing NLRX1 in KYSE450 significantly activated the
PI3K/AKT pathway (Fig. 7A,
P<0.05). By contrast, upregulation of NLRX1 in ECA109
significantly inhibited the activation of the PI3K/AKT pathway
(Fig. 7B, P<0.05).
Discussion
In the present study, it was confirmed that NLRX1
functions as a tumor suppressor in ESCC using bioinformatics
analysis and experiments, and its potential biological activities
and regulatory mechanisms were investigated. These findings suggest
that NLRX1 is crucial for the emergence and progression of
ESCC.
Through data mining, it was found that NLRX1 is less
expressed in ESCC at the mRNA level, and subsequently, the findings
were validated at the protein level using immunohistochemistry
experiments. The data from GSE53625 and TCGA both indicated that
patients with low NLRX1 expression have poor prognosis. The
prognostic significance of NLRX1 has also been confirmed in other
cancer types. For instance, reduced expression of NLRX1 is
associated with poor prognosis in cholangiocarcinoma and liver
cancer (13,27,28).
The clinical correlation analysis of GSE53625 with a larger data
volume showed an association with tumor grading, ROC analysis
demonstrated good diagnostic value, and Cox analysis also indicated
that it is an independent prognostic factor for ESCC. These data
indicate that NLRX1 has significant diagnostic and prognostic value
in ESCC.
According to previous studies, NLRX1 is closely
linked to both physiological and pathological immune, inflammatory,
autophagic, mitochondrial and metabolic activities (5). To determine the function of NLRX1 in
ESCC, a correlation analysis and enrichment analysis were
conducted. Among the 11 genes highly correlated with NLRX1, the
positively correlated GRHL3 was previously shown to inhibit tumor
growth in ESCC (29,30), further strengthening our evidence of
NLRX1′s tumor suppressive function in ESCC. KEGG analysis suggested
that genes highly correlated with NLRX1 are mainly enriched in the
phagosome, endocytosis, choline metabolism in cancer, porphyrin
metabolism and lipid metabolism, which are crucial in the onset and
progression of cancer (31–34). GO analysis indicated that
NLRX1-related genes are associated with functions such as
antifungal immunity. The phagocytosis performed by phagosomes is
the core mechanism for inflammation and defense against infection
factors, and there are multiple interaction points between
phagosomes and endocytic pathways (35). In the present study, it was
speculated that one possible pathway by which NLRX1 affects immune
function is by regulating phagocytic activity, which requires
further research to confirm. Previous literature has reported that
NLRX1 can regulate lipid metabolism, including pomegranate acid and
docosahexaenoic acid (5), which is
consistent with the results of the present study. The pathway
analysis results of the GSEA and GSVA showed that multiple
cancer-related functional pathways, including melanoma, prostate
cancer and small cell lung cancer, were enriched in the low
expression group of NLRX1, indicating that NLRX1 deficiency has a
certain role in the malignant development of ESCC. In recent years,
there has been an increase in interest in the role of the tumor
immunological microenvironment in the incidence and progression of
malignancies. The innate immune response to viral infection is
reportedly weakened by NLRX1 through several signaling pathways
(5). However, it is unclear whether
NLRX1 contributes significantly to the immune system in ESCC.
Analysis of immune-related functions showed that in the low
expression group of NLRX1, CCR, DCs, pDCs and T-cell co-stimulation
scores were decreased, indicating that immune function was
suppressed in the low expression group of NLRX1. The differential
analysis and correlation test of immune infiltration showed that
NLRX1 was highly positively correlated with ‘NK cells activated’,
‘Monocyte’ and ‘Macrophages M0’. ‘NK cells activated’ have a
powerful anti-tumor effect, ‘Monocyte’ appears to have both
pro-cancer and anti-cancer effects, and ‘Macrophages M0’, also
known as immature macrophages, display a phagocytic function and
identify pathogenic agents (36–39).
NLRX1 may affect the tumor immune microenvironment by regulating
the activity of these immune cells, thereby affecting tumor
occurrence and development. The present study also found a high
correlation between NLRX1 and immune checkpoint receptors. For
instance, TNFRSF25 and TNFSF9 are significantly positively
correlated with NLRX1. TNFRSF25, also known as death receptor 3,
can promote inflammation and survival through different pathways
and also mediate caspase-dependent cell apoptosis. A study reported
that the use of taurolidine can promote apoptosis in KYSE270 ESCC
cells, which upregulates the expression of TNFRSF25 (40); therefore, it may be speculated that
NLRX1 inhibiting cell proliferation and increasing apoptosis may be
related to the regulation of immune checkpoint-related genes,
including TNFRSF25, which needs further confirmation. It is a
co-stimulatory molecule of T cells and innate lymphoid cell
(41). Considering that the low
expression group of NLRX1 had lower T-cell co-stimulation scores,
NLRX1 may affect its function by affecting the expression of
TNFRSF25. Another example is TNFRSF9, which can mediate anti-tumor
and tumor-promoting signals in immune cells. Its dual function has
also been observed in different solid tumor cells; however,
previous studies have not yet included ESCC (42). NLRX1 may also exert its antitumor
effect by affecting its expression. In conclusion, the relationship
between NLRX1 and immunity of tumor patients requires further
study. In addition, the relationship between NLRX1 and commonly
used chemotherapy drugs for ESCC treatment in clinical practice was
analyzed. The low NLRX1 group was more sensitive to irinotecan,
indicating that ESCC with low expression of NLRX1 may achieve
better efficacy when combined with irinotecan and other drugs.
Patients with ESCC with low NLRX1 expression may obtain a better
curative effect by using the treatment scheme combined with
irinotecan. In summary, the present results strongly demonstrate
the potential of NLRX1 as an anti-tumor therapeutic target.
NLRX1 has a role in promoting or inhibiting cancer
in different solid tumors. NLRX1 is expressed more strongly in
triple-negative breast cancer cells and metastatic breast cancers
than it is in ER/PR+ breast cancer cells. In HeLa human cervical
cancer cells and MCF-7 human ER/EP- breast cancer cells,
overexpression of NLRX1 increases cell death and reduces ATP
production, and overexpression of NLRX1 in MCF-7 cells reduces cell
proliferation and migration (6). In
MDA-MB-1 human ER/EP+ breast cancer cells, knockdown of NLRX1
results in decreased cell proliferation, migration, ATP production
and inhibited TNF-α-induced mitochondrial autophagy. It is thought
that the elevated expression of NLRX1 in invasive breast cancer
supports its tumorigenic potential by controlling mitochondrial
metabolic activity and turnover, preserving energy balance and
preserving organelle function via mitochondrial autophagy (7). The latest research has also confirmed
that NLRX1 inhibits the growth and development of Pan02 mouse
pancreatic cancer cells (14). The
present study on the phenotypic effects of NLRX1 on KYSE450 and
ECA109 cells is consistent with this. The present study found that
NLRX1 negatively regulates the growth and development of ESCC
cells, which is consistent with the function found in pancreatic
cancer, liver cancer and histiocytic sarcoma (11,13,14).
The expression and functional differences of NLRX1 in different
cells may depend on multiple complex factors. Although NLRX1 has
different roles in different tumors, its biological pathway of
aggregation is consistent across different models (14). Specifically, its pathways of action
in various tumors seem to be mainly attributed to NF-κB and AKT
signal transduction (10–12,43,44).
There are also literature reports indicating that it is related to
MAPK, STAT3 and IL-6 signaling (12,14).
Through WB assays, it was indicated in the present study that
PI3K/AKT signaling is crucial to NLRX1′s function in ESCC.
The present study has certain limitations. First,
the bioinformatics analysis data have not been experimentally
validated and using only one siNLRX1 subclone in all phenotype
experiments is a limitation. The impact of NLRX1 on cell
proliferation was only confirmed using the CCK-8 assay but not by
other experiments specifically designed to detect cell
proliferation. Furthermore, the deeper mechanisms by which NLRX1
regulates the occurrence and development of ESCC remain to be
explored. Finally, the clinicopathological information of the
cohort used in this study was not recorded or missing, so the
clinicopathological information of this cohort could not be
obtained for analysis, and a larger patient cohort is needed to
validate the clinical value of NLRX1.
In conclusion, the present study found that NLRX1 is
a tumor suppressor factor in ESCC. Low NLRX1 expression is a poor
prognostic factor for patients with ESCC. NLRX1 affects the
occurrence and development of ESCC through various pathways,
including immunity and the PI3K/AKT pathway. Research on NLRX1 may
provide new insights for the treatment of ESCC.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
LZ and ZL conceived and designed the study. LG, CS
and AC performed the literature search, and performed the
experiments and data extraction. MY analyzed and interpreted the
data. LZ drafted the manuscript. LZ and ZL confirm the authenticity
of all the raw data and revised the final version of the
manuscript. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
The study was conducted in accordance with the
Declaration of Helsinki and approved by the Second Affiliated
Hospital of Zhengzhou University Ethical Review Committee
(Zhengzhou, China; approval no. 2023056). All patient specimens
used were obtained with the patients' informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
NLRX1
|
nucleotide binding and oligomeric
domain-like receptor X1
|
|
ESCC
|
esophageal squamous cell carcinoma
|
|
OS
|
overall survival
|
|
ROC
|
receiver operating characteristic
|
|
WB
|
western blot
|
|
AOM
|
azomethane
|
|
GEO
|
Gene Expression Omnibus
|
|
TCGA
|
The Cancer Genome Atlas
|
|
GSVA
|
Gene Set Variation Analysis
|
|
OD
|
optical density
|
|
GO
|
Gene Ontology
|
|
GSEA
|
Gene Set Enrichment Analysis
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Morgan E, Soerjomataram I, Rumgay H,
Coleman HG, Thrift AP, Vignat J, Laversanne M, Ferlay J and Arnold
M: The global landscape of esophageal squamous cell carcinoma and
esophageal adenocarcinoma incidence and mortality in 2020 and
projections to 2040: New estimates from GLOBOCAN 2020.
Gastroenterology. 163:649–658.e642. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang Y, Zhang Y, Peng L and Zhang L:
research progress on the predicting factors and coping strategies
for postoperative recurrence of esophageal cancer. Cells.
12:1142022. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gharagozloo M, Gris KV, Mahvelati T,
Amrani A, Lukens JR and Gris D: NLR-dependent regulation of
inflammation in multiple sclerosis. Front Immunol. 8:20122017.
View Article : Google Scholar
|
|
5
|
Liu M, Liu K, Cheng D, Zheng B, Li S and
Mo Z: The regulatory role of NLRX1 in innate immunity and human
disease. Cytokine. 160:1560552022. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Singh K, Poteryakhina A, Zheltukhin A,
Bhatelia K, Prajapati P, Sripada L, Tomar D and Singh R, Singh AK,
Chumakov PM and Singh R: NLRX1 acts as tumor suppressor by
regulating TNF-α induced apoptosis and metabolism in cancer cells.
Biochim Biophys Acta. 1853:1073–1086. 2015. View Article : Google Scholar
|
|
7
|
Singh K, Roy M, Prajapati P, Lipatova A,
Sripada L, Gohel D, Singh A, Mane M, Godbole MM, Chumakov PM and
Singh R: NLRX1 regulates TNF-α-induced mitochondria-lysosomal
crosstalk to maintain the invasive and metastatic potential of
breast cancer cells. Biochim Biophys Acta Mol Basis Dis.
1865:1460–1476. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Luo X, Donnelly CR, Gong W, Heath BR, Hao
Y, Donnelly LA, Moghbeli T, Tan YS, Lin X, Bellile E, et al: HPV16
drives cancer immune escape via NLRX1-mediated degradation of
STING. J Clin Invest. 130:1635–1652. 2020. View Article : Google Scholar
|
|
9
|
Soares F, Tattoli I, Rahman MA, Robertson
SJ, Belcheva A, Liu D, Streutker C, Winer S, Winer DA, Martin A, et
al: The mitochondrial protein NLRX1 controls the balance between
extrinsic and intrinsic apoptosis. J Biol Chem. 289:19317–19330.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tattoli I, Killackey SA, Foerster EG,
Molinaro R, Maisonneuve C, Rahman MA, Winer S, Winer DA, Streutker
CJ, Philpott DJ and Girardin SE: NLRX1 acts as an
epithelial-intrinsic tumor suppressor through the modulation of
TNF-mediated proliferation. Cell Rep. 14:2576–2586. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Coutermarsh-Ott S, Simmons A, Capria V,
LeRoith T, Wilson JE, Heid B, Philipson CW, Qin Q,
Hontecillas-Magarzo R, Bassaganya-Riera J, et al: NLRX1 suppresses
tumorigenesis and attenuates histiocytic sarcoma through the
negative regulation of NF-κB signaling. Oncotarget. 7:33096–33110.
2016. View Article : Google Scholar
|
|
12
|
Koblansky AA, Truax AD, Liu R, Montgomery
SA, Ding S, Wilson JE, Brickey WJ, Mühlbauer M, McFadden RM, Hu P,
et al: The innate immune receptor NLRX1 functions as a tumor
suppressor by reducing colon tumorigenesis and key tumor-promoting
signals. Cell Rep. 14:2562–2575. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hu B, Ding GY, Fu PY, Zhu XD, Ji Y, Shi
GM, Shen YH, Cai JB, Yang Z, Zhou J, et al: NOD-like receptor X1
functions as a tumor suppressor by inhibiting
epithelial-mesenchymal transition and inducing aging in
hepatocellular carcinoma cells. J Hematol Oncol. 11:282018.
View Article : Google Scholar
|
|
14
|
Nagai-Singer MA, Morrison HA, Woolls MK,
Leedy K, Imran KM, Tupik JD and Allen IC: NLRX1 functions as a
tumor suppressor in Pan02 pancreatic cancer cells. Front Oncol.
13:11558312023. View Article : Google Scholar
|
|
15
|
Hu N, Clifford RJ, Yang HH, Wang C,
Goldstein AM, Ding T, Taylor PR and Lee MP: Genome wide analysis of
DNA copy number neutral loss of heterozygosity (CNNLOH) and its
relation to gene expression in esophageal squamous cell carcinoma.
BMC Genomics. 11:5762010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hyland PL, Zhang H, Yang Q, Yang HH, Hu N,
Lin SW, Su H, Wang L, Wang C, Ding T, et al: Pathway, in silico and
tissue-specific expression quantitative analyses of oesophageal
squamous cell carcinoma genome-wide association studies data. Int J
Epidemiol. 45:206–220. 2016. View Article : Google Scholar
|
|
17
|
Liu J, Wang Y, Chu Y, Xu R, Zhang D and
Wang X: Identification of a TLR-induced four-lncRNA signature as a
novel prognostic biomarker in esophageal carcinoma. Front Cell Dev
Biol. 8:6492020. View Article : Google Scholar
|
|
18
|
Hu N, Wang C, Zhang T, Su H, Liu H, Yang
HH, Giffen C, Hu Y, Taylor PR and Goldstein AM: CSMD1 shows complex
patterns of somatic copy number alterations and expressions of
mRNAs and target Micro RNAs in esophageal squamous cell carcinoma.
Cancers (Basel). 14:50012022. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Team RCJMc, . R: A language and
environment for statistical computing. 12014
|
|
20
|
Sepulveda JL: Using R and bioconductor in
clinical genomics and transcriptomics. J Mol Diagn. 22:3–20. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hänzelmann S, Castelo R and Guinney J:
GSVA: Gene set variation analysis for microarray and RNA-seq data.
BMC Bioinformatics. 14:72013. View Article : Google Scholar
|
|
24
|
Zhou L, Gan L and Liu Z: Expression and
prognostic value of AIM1L in esophageal squamous cell carcinoma.
Medicine (Baltimore). 102:e346772023. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
An J, Chen X, Chen W, Liang R, Reinach PS,
Yan D and Tu L: MicroRNA expression profile and the role of miR-204
in corneal wound healing. Invest Ophthalmol Vis Sci. 56:3673–3683.
2015. View Article : Google Scholar
|
|
26
|
Zou Y, Wu F, Liu Q, Deng X, Hai R, He X
and Zhou X: Downregulation of miRNA-328 promotes the angiogenesis
of HUVECs by regulating the PIM1 and AKT/mTOR signaling pathway
under high glucose and low serum condition. Mol Med Rep.
22:895–905. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang ZJ, Huang YP, Liu ZT, Wang YX, Zhou
H, Hou KX, Tang JW, Xiong L, Wen Y and Huang SF: Identification of
immune related gene signature for predicting prognosis of
cholangiocarcinoma patients. Front Immunol. 14:10284042023.
View Article : Google Scholar
|
|
28
|
Wang X, Yang C, Liao X, Han C, Yu T, Huang
K, Yu L, Qin W, Zhu G, Su H, et al: NLRC and NLRX gene family mRNA
expression and prognostic value in hepatocellular carcinoma. Cancer
Med. 6:2660–2672. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liang G, Wang H, Shi H, Zhu M, An J, Qi Y,
Du J, Li Y and Gao S: Porphyromonas gingivalis promotes the
proliferation and migration of esophageal squamous cell carcinoma
through the miR-194/GRHL3/PTEN/Akt axis. ACS Infect Dis. 6:871–881.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Georgy SR, Rudiatmoko DR, Auden A,
Partridge D, Butt T, Srivastava S, Wong N, Swaroop D, Carpinelli
MR, Yan F, et al: Identification of a novel
GRHL3/HOPX/Wnt/β-catenin proto-oncogenic axis in squamous cell
carcinoma of the esophagus. Cell Mol Gastroenterol Hepatol.
15:1051–1069. 2023. View Article : Google Scholar
|
|
31
|
Mellman I and Yarden Y: Endocytosis and
cancer. Cold Spring Harb Perspect Biol. 5:a0169492013. View Article : Google Scholar
|
|
32
|
Sonkar K, Ayyappan V, Tressler CM, Adelaja
O, Cai R, Cheng M and Glunde K: Focus on the glycerophosphocholine
pathway in choline phospholipid metabolism of cancer. NMR Biomed.
32:e41122019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Fiorito V, Chiabrando D, Petrillo S,
Bertino F and Tolosano E: The multifaceted role of heme in cancer.
Front Oncol. 9:15402019. View Article : Google Scholar
|
|
34
|
Bian X, Liu R, Meng Y, Xing D, Xu D and Lu
Z: Lipid metabolism and cancer. J Exp Med. 218:e202016062021.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tjelle TE, Lovdal T and Berg T: Phagosome
dynamics and function. Bioessays. 22:255–263. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wu SY, Fu T, Jiang YZ and Shao ZM: Natural
killer cells in cancer biology and therapy. Mol Cancer. 19:1202020.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Olingy CE, Dinh HQ and Hedrick CC:
Monocyte heterogeneity and functions in cancer. J Leukoc Biol.
106:309–322. 2019. View Article : Google Scholar
|
|
38
|
Ugel S, Canè S, De Sanctis F and Bronte V:
Monocytes in the tumor microenvironment. Annu Rev Pathol.
16:93–122. 2021. View Article : Google Scholar
|
|
39
|
Chaintreuil P, Kerreneur E, Bourgoin M,
Savy C, Favreau C, Robert G, Jacquel A and Auberger P: The
generation, activation, and polarization of monocyte-derived
macrophages in human malignancies. Front Immunol. 14:11783372023.
View Article : Google Scholar
|
|
40
|
Daigeler A, Chromik AM, Geisler A, Bulut
D, Hilgert C, Krieg A, Klein-Hitpass L, Lehnhardt M, Uhl W and
Mittelkötter U: Synergistic apoptotic effects of taurolidine and
TRAIL on squamous carcinoma cells of the esophagus. Int J Oncol.
32:1205–1220. 2008. View Article : Google Scholar
|
|
41
|
Bittner S and Ehrenschwender M:
Multifaceted death receptor 3 signaling-promoting survival and
triggering death. FEBS Lett. 591:2543–2555. 2017. View Article : Google Scholar
|
|
42
|
Wu J and Wang Y: Role of TNFSF9
bidirectional signal transduction in antitumor immunotherapy. Eur J
Pharmacol. 928:1750972022. View Article : Google Scholar
|
|
43
|
Fan Z, Pan J, Wang H and Zhang Y: NOD-like
receptor X1, tumor necrosis factor receptor-associated factor 6 and
NF-κB are associated with clinicopathological characteristics in
gastric cancer. Exp Ther Med. 21:2082021. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Castaño-Rodríguez N, Kaakoush NO, Goh KL,
Fock KM and Mitchell HM: The NOD-like receptor signalling pathway
in Helicobacter pylori infection and related gastric cancer: A
case-control study and gene expression analyses. PLoS One.
9:e988992014. View Article : Google Scholar
|