Introduction
Bone sarcomas are a heterogenous group of different
types of malignant bone tumor characterized by various degrees of
mesenchymal differentiation (1).
Chondrosarcoma is a rare but aggressive malignant tumor of the
cells that produce the cartilage matrix; it is the third most
common bone tumor, accounting for >30% of primary bone cancers
in the USA, and is resistant to chemotherapy and radiation;
therefore, novel therapeutic approaches are urgently required
(1–4).
Cancer stem cells (CSCs) have the ability to
self-renew and differentiate, which are typical characteristics of
any stem cell (5). It has been
reported that a tumor contains different stem cell-like populations
due to decreased cell differentiation within a tumor, and this
heterogenous population includes CSCs (5). DNA damage may render tissue stem cells
to become CSCs during the process of differentiation and to
accumulate to form different types of tumor (6). Increasing evidence suggests that CSCs
exist as a sub-population of quiescent cells within the dominant
tumor bulk of heterogeneous tumor cells. These typically dormant
cells are resistant to standard antitumor therapies such as
chemotherapy and radiation, and they appear capable of self-renewal
and differentiation, which suggests that CSCs are responsible for
tumor repopulation following removal of the bulk tumor (7–9).
Proline-rich polypeptide-1 (PRP-1), also termed
galarmin, is synthesized by the brain neurosecretory cells and
comprises 15 amino acids (10).
PRP-1 is an mTOR kinase (mTORC1) inhibitor in chondrosarcoma that
inhibits cell proliferation (11).
Furthermore, PRP-1 has been demonstrated to upregulate several
tumor suppressors of the desmosomal protein family, such as
desmoglein, and plakoglobin (12),
as well as tumor suppressors involved in inflammatory pathways
(13) and microRNAs (miRNAs), and
to downregulate onco-miRNAs in the human chondrosarcoma JJ012 cell
line through inhibition of mTORC1 (3,14).
PRP-1 also serves an antiproliferative role via downregulation of
the embryonic stem cell marker, miRNA (miR)-302-c and its targets
nanog homeobox (NANOG), c-Myc and polycomb protein B lymphoma
Mo-MLV insertion region 1 homolog (BMI-1) (3). Pattern recognition receptors of the
adaptive immune system, including Toll-like receptor 1 (TLR1),
TLR2, TLR6 and secreted mucin 5B, have been recently identified as
binding partners for PRP-1, resulting in tumor suppressive effects
(4). PRP-1-treated cells have been
demonstrated to accumulate in the S phase, thus delaying cell cycle
progression (15). The cytostatic,
antiproliferative, immunomodulatory and tumor suppressive roles of
PRP-1 indicate its potential as a therapeutic agent against human
chondrosarcoma cells resistant to radiation and chemotherapy
(3,4,10–12,14).
The aforementioned properties were further investigated in the
present study to determine the ability of PRP-1 to target
chondrosarcoma CSCs.
Aldehyde dehydrogenase (ALDH) is an established
marker of CSCs in several neoplasms. Cells expressing high levels
of ALDH have been isolated in a number of human sarcoma cell lines,
including the human chondrosarcoma SW-1353 cell line (16). Identification of normal and
malignant stem/progenitor cells by the same marker reinforces the
concept that stem and progenitor cells are primary targets of
transformation, which further supports the CSC hypothesis.
Furthermore, the ability to identify stem/progenitor cells by ALDH1
expression permits analysis of cancer initiation and progression
from the normal to the pre-malignant and the malignant states
(2,17). A widely accepted method for
identifying CSCs is based on detecting the enzymatic activity of
ALDH1, a detoxifying enzyme responsible for the oxidation of
intracellular aldehydes (16). The
self-renewal capacity, proliferative and metastatic roles of ALDH
demonstrate its potential as a therapeutic target against human
chondrosarcoma (18).
The present study aimed to determine the underlying
molecular mechanism by which PRP-1 decreases the CSC population,
with particular focus on the mammalian Switch/sucrose non
fermentable (SWI/SNF)-complex (19), which can either activate or repress
transcription, and to identify additional biomarkers for
chondrosarcoma.
Materials and methods
Tissue culture
The human JJ012 chondrosarcoma cells were obtained
from Dr Joel Block's Laboratory (Rush University, Chicago, USA).
The cells were maintained in complete growth medium containing DMEM
+ GlutaMAX™ supplemented with F-12 + GlutaMAX™ Nutrient mixture
(Ham) (Thermo Fisher Scientific, Inc.), 10% FBS (Sigma-Aldrich;
Merck KGaA), 1% penicillin/streptomycin (Thermo Fisher Scientific,
Inc.), 25 µg/ml ascorbic acid, 100 ng/ml insulin and 100 nM
hydrocortisone (Sigma-Aldrich; Merck KGaA). The cells were
incubated at 37°C in a humidified atmosphere with 5% CO2
and periodically checked for mycoplasma. Estrogen negative
MDA-MB-231 breast cancer cells (ATCC® CRM-HTB-26) were
obtained from ATCC. The medium for this cell line was
ATCC-formulated Leibovitz's L-15 Medium (cat. no. 30-2008; ATCC)
supplemented with 10% FBS. The MDA-MB-231 breast cancer cells were
incubated at 37°C in a humidified atmosphere with 5%
CO2. Human B cell lymphoma (HT; cat. no. ACC567), Ly1
(cat. no. ACC722) and Ly3 (cat. no. ACC761) cell lines were
obtained from DSMZ. Ly10 cells were a gift from Dr Messner (Ontario
Cancer Institute, Canada), and HBL-1 cells were a gift from Dr Vega
(MD Anderson Cancer Center, Houston, TX, USA). Melanoma metastatic
cell line C81618 was provided by Dr Liu, University of Miami,
Miller school of Medicine.
Mycoplasma detection assay
JJ012 chondrosarcoma cells were seeded
1×105 cells/ml in full growth media and treated with 25
µg/ml Plasmocin™ (InvivoGen) for 14 days. Plasmocin™-containing
medium was replaced every 3–4 days. MycoAlert®
Mycoplasma Detection kit (cat. no. LT07-118; lot. no. 0000678423;
Lonza, Inc.) was used according to the manufacturer's instructions.
The microplate was placed in the Molecular devices Spectra Max
microplate luminometer (Lonza, Inc.) and programmed for Reading A
analysis. A total of 100 µl MycoAlert™ substrate was added to each
sample and incubated for 10 min, following which the luminometer
was programmed for Reading B analysis, and the Reading B/Reading A
ratio was calculated.
Cell viability
Cells were cultured under the same conditions as
previously described and stained with 0.4% trypan blue stain
(Gibco; Thermo Fisher Scientific, Inc.) at a 1:1 ratio. After a
72-h incubation at 37°C in the 5% CO2 incubator, cells
were observed using the Luna II Automated Cell Counter™ (Logos
Biosystem) according to the manufacturer's protocol to generate
cell count and viability data.
PRP-1 treatment
A subset of cultured human JJ012 chondrosarcoma
cells were treated with 10 µg/ml PRP-1 as previously described
(10) and incubated at 37°C in a
humidified atmosphere of 5% CO2 for 24 h prior to
performing the Aldefluor™ assay. Cell proliferation was measured in
cells seeded at 5×104 cells/ml using the Rapid Cell
Proliferation kit (EMD Millipore), according to the manufacturer's
instructions.
Aldefluor™ assay and
fluorescence-activated cell sorting (FACS)
The Aldefluor™ assay (StemCell Technologies, Inc.)
was performed according to the manufacturer's protocol, in order to
detect cells with ALDH activity. Briefly, the JJ012 cells were
harvested and resuspended in Aldefluor™ assay buffer at a density
of 1×106/ml. The cells were incubated with 25 µl DMSO
with 25 µl 2N HCl for 15 min, followed by addition of 360 µl assay
buffer in order to activate the Aldefluor™ reagent, and incubated
for 45 min at 37°C. N,N-Diethylaminobenzaldehyde (DEAB), a specific
ALDH inhibitor, was used as the negative control. Following
incubation, all tubes were centrifuged for 5 min at 250 × g at 4°C,
the supernatant was discarded and the cells were resuspended in
Aldefluor™ assay buffer. The cells were then transferred to 5 ml
Falcon polystyrene round bottom tubes with cell strainer caps.
After labeling, the samples with and without PRP-1 treatment were
sorted using a FACSAria II flow cytometer (BD Biosciences) with
FACSDiva software (version 6.1.3; BD Biosciences) into
ALDHlow and ALDHhigh cells. Data analysis was
performed using FlowJo software (version 10; FlowJo, LLC).
Gel electrophoresis and western
blotting
Chondrosarcoma JJ012 cells were cultured to 100%
confluency. Cells were collected using trypsin, seeded into petri
dishes at a density of 1×106 cells/ml and incubated for
24 h at 37°C in a 5% CO2 incubator. After 24 h, cells
were washed with ice-cold PBS, and protease inhibitor was added to
the CelLytic M cell lysis reagent (Sigma-Aldrich; Merck KGaA) at a
1:100 ratio, and kept in a 5% CO2 incubator for 10 min.
Following collection of cells with a rubber scraper and cell
membrane lysis with an 18-gauge needle, the cells were centrifuged
at 15,000 × g at 4°C for 10 min. The supernatant was collected, and
the protein content was measured using a NanoDrop®
spectrophotometer. The supernatant was frozen at −80°C until
loading into gels (20 µg/lane). Polyacrylamide gel electrophoresis
and western blotting reagents were supplied by Lonza Group, Ltd.,
and the experiments were performed according to the manufacturer's
protocols. The reagents used were as follows: Pager Gold Precast
Gels (10% Tris-Glycine; Lonza Group, Ltd.); ProSieve Quad Color
Protein marker (4.6–300 kD; Lonza Group, Ltd.); 20X reducing agent
for ProSieve ProTrack Dual Color Loading buffer (Lonza Group,
Ltd.); ProTrack Loading buffer (Lonza Group, Ltd.); ProSieve
ProTrack Dual Color Loading buffer EX running buffer (Lonza Group,
Ltd.); ProSieve EX Western Blot Transfer buffer (Lonza Group, Ltd.)
and Immobilon®-P Polyvinylidene difluoride membranes
(Sigma-Aldrich; Merck KGaA).
For the blocking step, Western Blocker solution
(Sigma-Aldrich; Merck KGaA) was used. Primary antibodies were
diluted in blocking buffer at the ratio of 1:1,000, and the
membranes were incubated at 4°C overnight with gentle agitation.
The secondary antibodies horseradish peroxidase-conjugated goat
anti-rabbit IgG (cat. no. A0545; Sigma-Aldrich; Merck KGaA) and
anti-mouse IgG (cat. no. A4416; Sigma-Aldrich; Merck KGaA) were
diluted to 1:5,000, and the membranes were incubated for 2 h at
room temperature with gentle agitation. Western blot visualization
was performed using ECL reagents (GE Healthcare Life Sciences) for
1 h at room temperature.
Antibodies for western blotting
The primary antibodies used for western blot
analysis were as follows: CD133 (cat. no. 19898; Abcam), CD144
(VE-cadherin) monoclonal (16B1) PE-Cyanine7 (cat. no. 25-1449-4;
Invitrogen; Thermo Fisher Scientific, Inc.), c-Kit (Ab 81) (cat.
no. 13508; Santa Cruz Biotechnology, Inc.), rabbit LGR5 (GPR49)
(cat. no. 9205024; MyBioSource, Inc.), SOX2 (cat. no. 97959;
Abcam), human ALDH1A1 (cat. no. MAB5869; R&D Systems, Inc.),
polycomb complex protein oncogene BMI-1 (BMI-1; cat. no. 38295;
Abcam), c-Myc (9E10) (cat. no. 40; Santa Cruz Biotechnology, Inc.),
CD44 (cat. no. 6124; Abcam), FOXO3 (cat. no. MAB6165; R&D
Systems), FOXC1 (cat. no. MAB 6329; R&D Systems), NANOG clone
7F7.1 (cat. no. MABD24; Sigma-Aldrich; Merck KGaA), anti CD34 +
mesenchymal stem cell monoclonal antbody (STRO-1; cat. no. 39-8401;
Thermo Fisher Scientific, Inc.), CD117 c-Kit (Ab 81) (cat. no.
13508; Santa Cruz Biotechnology, Inc.), BRG1-associated factor 170
(BAF170) or SMARCC2 (cat. no. 9401447; MyBioSource, Inc.),
BRG1-associated factor 155 (BAF155) or SMARCC1 (cat. no. 8245507;
MyBioSource, Inc.), protein Brahma homolog 1 (BRG1; cat. no. 4081;
Abcam), Brahma protein (BRM) or SMARCA 2 (cat. no. 610390; BD
Biosciences), ARF tumor suppressor, p14ARF antibody (ARF 4C6/4;
cat. no. 53392; Santa Cruz Biotechnology, Inc.), cyclin-dependent
kinase inhibitor 2A (CDKN2A) P15/P16 (p15, INK4b; p16, INK4a; cat.
no. 377412; Santa Cruz Biotechnology, Inc.), p-topoisomerase IIα
(TOP2A) (Ser1469) (cat. no. 13072; Cell Signaling Technology,
Inc.), tubulin (cat. no. T5168; Sigma Aldrich; Merck KGaA) and
topoisomerase IIα SER1469 [cat. no. 13072 (E); Cell Signaling
Technology, Inc.].
Densitometry analysis of western
blots
Quantitative analysis and densitometry were obtained
using integrated density analysis on ImageJ software 1.52e
(National Institutes of Health) to calculate relative optical
density (OD) of the previously mentioned proteins compared with the
housekeeping protein tubulin; the bulk untreated JJ012 was used as
the control.
Apoptosis assay and cell cycle
analysis
Staining was performed using the Annexin V Apoptosis
Detection kit according to the manufacturer's protocol. Modified
Annexin V/propidium iodide (PI) apoptosis assay (cat. no.
88-8007-72; eBioscience; Thermo Fisher Scientific, Inc.) was used
for apoptosis experiments with 1×106 cellss/ml. The
LRS-II Analyzer was used with DiVa-8 software (BD Biosciences). The
antibodies used for flow cytometric analysis were as follows:
APC-conjugated human CD133 (cat. no. FAB11331A-025; R&D
Systems, Inc.), Alexa Fluor® 647 mouse anti-human BMI-1
clone P51-311 (cat. no. 562637; BD Biosciences), CD144
(VE-cadherin) (16B1) phycoerythrin (PE)-cyanine7 (cat. no.
25-1449-4; Invitrogen; Thermo Fisher Scientific), BV605 mouse
anti-human CD117 clone 104D2 (cat. no. 562687; BD Biosciences),
PE-CF594 rat anti-human LGR5 (N-terminal) clone 8F2 (cat. no.
563470; BD Biosciences), PE mouse anti-SOX2 clone 245610 (cat. no.
560291; BD Biosciences), BV421 mouse anti-human CD49b (cat. no.
564119; BD Biosciences), CD10 (SN5c) PerCP-eFluor 710 (cat. no.
46-0108-41; Invitrogen; Thermo Fisher Scientific, Inc.), PE-CF594
mouse anti-human CD221 clone 1H7 (cat. no. 562535; BD Biosciences)
and Alexa Fluor® 647 mouse anti-human CD271 clone
C40-1457 (cat. no. 560326; BD Biosciences).
Aldefluor™ assay and
fluorescence-activated cell sorting (FACS)
To measure cells with ALDH activity, the Aldefluor™
assay (cat. no. 01700; StemCell Technologies, Inc.) was performed
according to manufacturer's protocol. Briefly, cells were harvested
and resuspended in Aldefluor™ assay buffer at a density of
1×106 cells/ml. To activate the Aldefluor™ reagent,
first 25 µl DMSO was added and incubated for 15 min with 25 µl 2N
HCl, and then 360 µl assay buffer was added to the vial. The cells
were incubated with the activated Aldefluor™ reagent for 45 min at
37°C. Diethylaminobenzaldehyde (DEAB), a specific ALDH inhibitor,
was added as a negative control. Following incubation, all tubes
were centrifuged for 5 min at 250 × g, at 4°C and the supernatant
was removed; the cells were then resuspended in Aldefluor™ assay
buffer, transferred and strained into a Falcon 5 ml polystyrene
round bottom tube with a cell strainer cap (cat. no. 352235; Thermo
Fisher Scientific, Inc.). After labeling, the samples were sorted
using a Special Order Research Product (SORP) FACSAria II (BD
Biosciences) using BD FACSDiva software (version 6.1.3; BD
Biosciences) into ALDHlow and ALDHhigh cells
with and without PRP-1 treatment. Data analysis was performed using
FlowJo software version 10 (FlowJo LLC).
Bromodomain-containing protein 4
(BRD4) inhibitor screening
The cells were seeded at 4,000 cells/well in
384-well plates. BRD4 inhibitor screening was performed by BPS
Bioscience Inc., using the BPS Bioscience TR-FRET Assay kit BRD4
according to the manufacturer's instructions.
Statistical analysis
All statistical analyses were performed using
GraphPad Prism (version 8.0.2; GraphPad Software, Inc.).
Statistical analyses were performed using individual unpaired
t-tests for flow cytometry experiments (repeated 10 times).
Statistical analyses of relative ODs were completed using one-way
ANOVA, followed by Tukey's or Dunnett's post hoc multiple
comparison test, and the data were expressed as 95% confidence
intervals of the mean difference. All western blots were performed
in triplicate. P<0.05 was considered to indicate a statistically
significant difference. Data are presented as the mean ± standard
error of the mean in all graphs with *P<0.05, **P<0.01 and
***P<0.001.
Results
The inhibitory action of PRP-1 is
disease-specific
It has previously been reported that PRP-1 displays
disease-specific inhibition of tumor growth; however, in
chondrosarcoma, PRP-1 inhibits the stemness signature cluster of
miR-302-367 (3). For example, in
glioblastoma, miR-302-367 is strongly induced during serum-mediated
stemness suppression, which prevents PRP-1 from exerting inhibitory
effects on tumor cell proliferation (3). Thus, the present study assessed
different types of malignancies, such as lymphoma and melanoma.
PRP-1 failed to inhibit the growth of two major
subtypes of diffuse large B-cell lymphoma: Activated B-cell subtype
lymphoma 3, 10 (Ly3, Ly10), human B line (HBL1) and germinal center
subtype (Ly1 and HT cell lines). The results also demonstrated that
proliferation of the metastatic melanoma cell line C81618 was not
inhibited by PRP-1. The effect of PRP-1 on cell proliferation was
assessed in vitro (data not shown).
ALDH1A1 is be the only biomarker for
CSC population in human chondrosarcoma JJ012 cell line
In order to identify additional biomarkers for
chondrosarcoma CSCs in the JJ012 cell line, two different
approaches were employed, the traditional side population (SP)
approach and the enzymatic ALDH1A1 approach. The SP analysis
results demonstrated that no SPs for JJ012 cells were detected
(data not shown) despite previous reports of successful
identification of SP in sarcomas (20). Subsequently, known biomarkers of
different types of cancer were assessed using the human
chondrosarcoma JJ012 cell line. A number of well-known markers and
the Aldefluor assay were employed in order to determine ALDH
expression in human chondrosarcoma JJ012 cells, with and without
PRP-1 treatment, and the cells were divided into ALDHlow
and ALDHhigh populations, respectively.
Human chondrosarcoma JJ012 cells were stained with
Aldefluor™ in the presence and absence of the ALDH inhibitor DEAB.
The DEAB (+) group was used to determine ALDHlow and
ALDHhigh populations (Fig.
1A). JJ012 fractions treated with PRP-1 were labeled as
ALDHlow-PRP-1 (21).
These gates were used in the analysis of surface staining of
well-established CSC markers for sarcoma. Similar to the control
MDA-MB-231 breast cancer cells, JJ012 cells expressed CD133, a
known CSC marker (22–25). However, no difference was observed
in CD133 expression between the ALDHhigh and
ALDHlow groups (Fig.
1B). The cells were stained with other surface antibodies that
also demonstrated no difference in the expression levels of BMI-1,
SOX2, CD144, CD117, CD221, LGR5, CD271, CD10 and CD49b between the
ALDHhigh and ALDHlow cells (Fig. 1C). Thus, none of the aforementioned
markers could serve as a CSC marker.
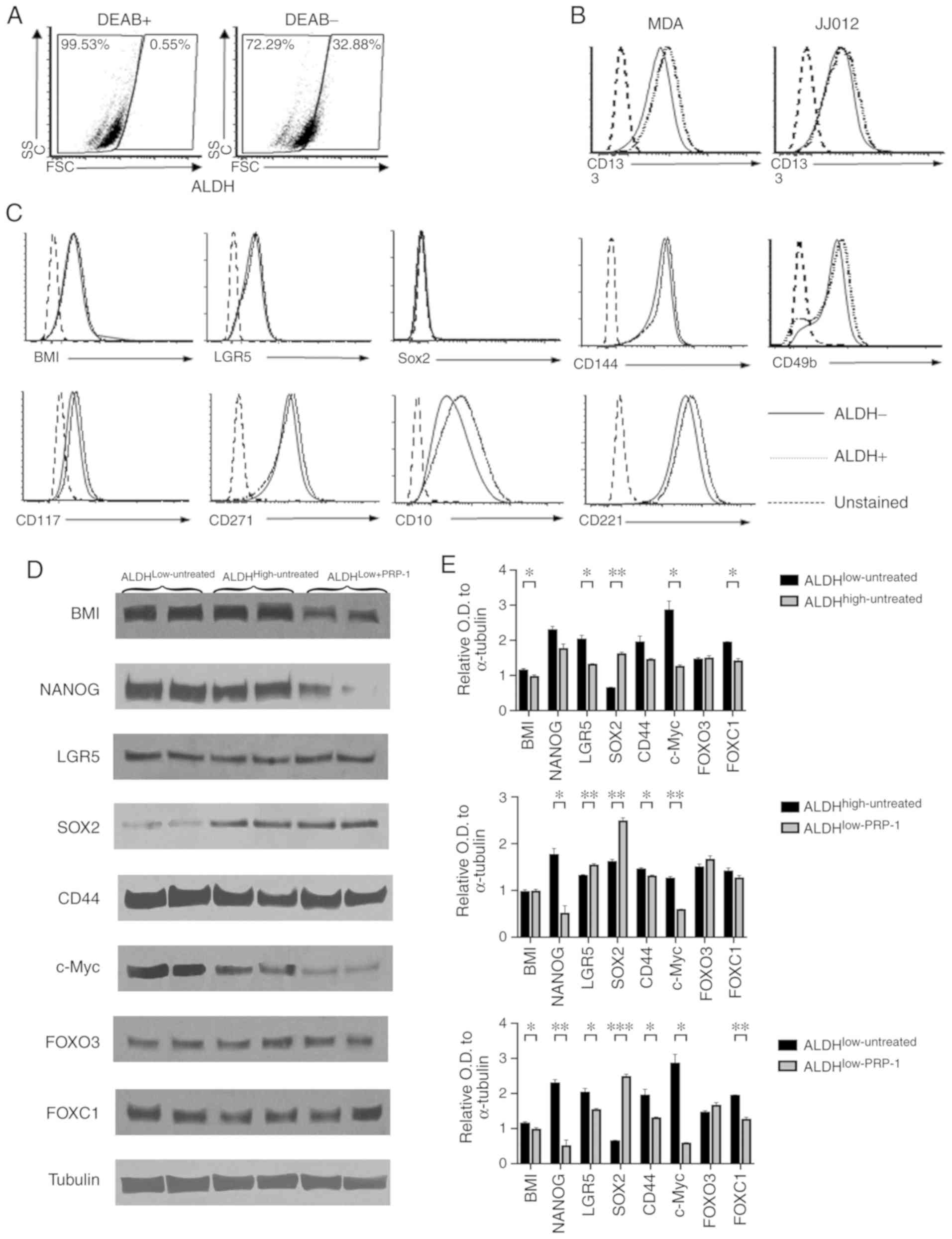 | Figure 1.Identification of additional
biomarkers for human chondrosarcoma. (A-C) Flow cytometry
experiments. (A) The DEAB (+) group was used to determine
ALDHlow and ALDHhigh populations. (B) CD133
was not different between the two populations. (C) Staining with
BMI-1, SOX2, CD144,CD117, CD221, LGR5, CD271, CD10 and CD49b on
ALDHhigh and ALDHlow cells verified that none
of them was a biomarker for JJ012 human chondrosarcoma cells. (D)
Western blot analysis of LGR5, SOX2, CD44, FOX03, FOXC1, BMI-1,
c-Myc and NANOG. Only BMI-1, c-Myc and NANOG in
ALDHlow+PRP-1 exhibited decreases in protein expression
expression compared with that in ALDHlow and
ALDHhigh untreated cells. (E) Histograms of the relative
OD in ALDH fractions. *P<0.05, **P<0.01 and ***P<0.001.
ALDH, aldehyde dehydrogenase; OD, optical density; BMI-1, B
lymphoma Mo-MLV insertion region 1 homolog; NANOG, nanog homeobox;
LGR5, leucine-rich repeat-containing G protein-coupled receptor 5;
FOXO3, forkhead box O3; FOXC1, forkhead box C1; DEAB,
N,N-diethylaminobenzaldehyde. |
Western blot analysis demonstrated decreased
expression levels of BMI, c-Myc and NANOG in the
ALDHlow-PRP-1 fraction following treatment with PRP-1
compared with untreated ALDHlow and ALDHhigh
fractions (Fig. 1D). However, no
differences in the expression levels of CD44, FOX03 and FOXC1 were
demonstrated between the fractions. Of note, compared with that in
the ALDHlow-untreated group, the expression of SOX2
stemness-associated factor in cancer was upregulated in
ALDHhigh-untreated and ALDHlow-PRP-1, and was
downregulated in the ALDHlow -untreated fraction
(Fig. 1E). Individual one-way ANOVA
of the protein markers demonstrated significant differences between
ALDHlow-untreated, ALDHhigh-untreated and
ALDHlow-PRP-1 cells in the expression levels of BMI-1,
NANOG, LGR5, SOX2, c-Myc and FOXC1 (Table SI). No differences of means between
the groups were detected for FOXO3. ALDHlow-PRP-1 cells
exhibited a significantly lower BMI-1 expression level compared
with ALDHhigh-untreated cells. Furthermore, NANOG
expression was significantly lower in ALDHlow-PRP-1
cells compared with ALDHlow-untreated and
ALDHhigh-untreated cells. ALDHhigh-untreated
cells and ALDHlow-PRP-1 cells had significantly lower
expression levels of LGR5 compared with
ALDHlow-untreated cells (Fig. 1E).
In a JJ012 bulk untreated subpopulation, 4.2%
untreated cells were ALDHlow and 42.9%
ALDHhigh (Fig. 2A),
whereas a PRP-1-treated subpopulation demonstrated a notable change
in ALDHlow-PRP-1 (82.6%) and ALDHhigh-PRP-1
(0.6%) cells (Fig. 2B). Fig. 2A demonstrates one of the 10
representative experiments of the untreated JJ012 cells, whereas
Fig. 2B shows one of the 10
representative experiments of the PRP-1 treated JJ012 cells;
Fig. 2C is the average of the 10
experiments with the SEM represented. The individual results are
presented to demonstrate how the gating was performed by the flow
cytometry software. The mean value for ALDHlow-untreated
cells was 45.03% vs. 87.53% for ALDHlow-PRP-1 treated
cells. The mean value for ALDHhigh-untreated was 46.72%
vs. 8.27% for ALDHhigh-PRP-1 treated cells (Fig. 2C; Table
SII). Subpopulations were collected using flow cytometry and
consisted of the following; ALDHhigh cells sorted from
untreated JJ012 (ALDHhigh-untreated), ALDHlow
cells sorted from untreated JJ012 (ALDHlow-untreated)
and ALDHlow cells sorted from PRP-1-treated JJ012
(ALDHlow-PRP-1). ALDHhigh cells from
PRP-1-treated JJ012 (ALDHhigh-PRP-1) were difficult to
collect as a result of the exceedingly low levels of these CSC
populations following treatment with PRP-1; however, this was
eventually accomplished following multiple experiments and
compilation of the sorting results (Table SII).
The cell viability assay demonstrated an increase in
non-viable cells in the PRP-1 treated cells compared with the
untreated cells (Fig. 2D; Tables SIII and SIV). One-way ANOVA analysis demonstrated
significant differences amongst the controls and subpopulations
following 72 h PRP-1 treatment between the JJ012 control and PRP-1
treated JJ012 (Tables SIII and
SIV; Fig. 1E).
Following subpopulation collection and cell lysis,
western blot analysis demonstrated a significant decrease of
ALDH1A1 protein expression between subpopulations with and without
PRP-1 treatment (Fig. 2F).
Densitometry analysis demonstrated a statistically significant
difference in ALDH1A1 protein expression between
ALDHhigh-untreated cells vs. ALDHlow-PRP-1
cells. When comparing the mean relative OD of ALDH between
ALDHhigh-untreated (mean OD, 3.590±0.040; n=2) and
ALDHlow-PRP-1 (mean OD, 1.224±0.009; n=2) cells,
Student's t-test demonstrated a significant 2.93-fold decrease in
ALDH expression in ALDHlow-PRP-1 cells [t(2)=57.23;
P=0.0003; Fig. 2G; Table SV].
Epigenetic mechanism of CSC regulation
by PRP-1 in chondrosarcoma JJ012 cell line
The epigenetic effect of PRP-1 on CSCs was assessed.
In order to further elucidate the molecular mechanism by which
PRP-1 decreases CSC-population, the effect of PRP-1 on important
epigenetic readers, such as the BD and BET proteins (26). TOP2A is an important enzyme in the
regulation of DNA structure and cell proliferation (27–31).
However, the results of the present study failed to demonstrate
downregulation of TOP2A activity following treatment with PRP-1
(data not shown).
Comparison of BAF170 expression between
ALDHhigh-untreated and ALDHlow-PRP-1 cells
demonstrated a significant decrease in the mean relative OD in
ALDHlow-PRP-1 cells (Fig.
3). In addition, BRG1 expression was significantly decreased in
ALDHlow-PRP-1 cells compared with that in
ALDHhigh-untreated cells. ALDHlow-PRP-1 cells
demonstrated a significant decrease in BRM expression compared with
ALDHhigh-untreated cells (Table SVI). No differences were observed
between fractions with or without PRP-1 treatment for BAF155 or
STRO-1 mesenchymal stem cell factor (Fig. 3A). The downstream targets for BMI-1,
such as p14 ARF and p15/p16 did not exhibit any changes at the
protein level (Fig. 3B), which
suggested that the oncogenic function of BMI-1 in chondrosarcoma
was independent from these downstream targets.
 | Figure 3.(A) Western blot analysis of
Switch/sucrose non-fermenting complex key components in
ALDHhigh-untreated and ALDHlow-PRP-1
fractions. PRP-1 induced decreases in the protein expression of BAF
155, BRG 1 and BAF170 in ALDHlow-PRP-1 fractions
compared with that in ALDHhigh-untreated fractions.
Mesenchymal stem cell factor STRO-1 protein levels were not
different between the fractions and were not affected by PRP-1.
BMI-1 oncogene targets p14ARF, and p15/p16 displayed the same
protein levels, and PRP-1 did not cause any changes. (B) Histograms
of the relative OD in the western blots. *P<0.05, **P<0.01
and ***P<0.001. ALDH, aldehyde dehydrogenase; OD, optical
density; PRP-1, proline-rich polypeptide-1; BRG1, protein Brahma
homolog 1; BAF 170, BRM BRG1-associated factor 170; BAF 155,
BRG1-associated factor155; BMR, bromodomain protein; p14ARF, ARF
tumor suppressor; p15/p16, cyclin-ependent kinase inhibitor 2A. |
Discussion
The aforementioned conventional and mesenchymal stem
cell markers CD144, CD117, CD221, LGR5, CD271, CD10, BMI, FOXC1 and
c-Myc are notable in bone sarcoma stem cells; however, there is no
definitive marker for bone chondrosarcoma stem cells other than
ALDH1A1 (16). The present study
aimed to identify novel specific markers for chondrosarcoma stem
cells. The results demonstrated that none of the known markers for
other types of sarcoma were identified as chondrosarcoma
biomarkers. Thus, the search for potential epigenetic players that
decrease the CSC population following PRP-1 treatment remains
critical. A previous study has reported that PRP-1 attenuates
miR-302 (part of miR-302-367 cluster) expression and targets the
embryonic stem cell markers Nanog, polycomb protein BMI-1 and c-Myc
(2). A previous study on PRP-1 in
the human chondrosarcoma JJ012 cell line investigated the effect of
peptides on JJ012 subpopulations, and the outcome indicated that
PRP-1 treatment notably decreased ALDHhigh cells from
bulk JJ012 population (25).
In the present study, low activity of embryonic stem
cell markers was observed in chondrosarcoma JJ012 cells treated
with PRP-1, which were identified as
ALDHlow-PRP-1-treated following sorting.
The oncogene BMI-1 is a member of the
polycomb-group family of proteins, and is frequently overexpressed
in different types of tumor to promote carcinogenesis and drive
stem cell-like properties (32–43).
BMI-1 knockdown using RNA interference was demonstrated to
significantly impair osteosarcoma cell viability, in vitro
colony formation and in vivo tumorigenesis, as well as to
sensitize osteosarcoma cells to cisplatin-induced apoptosis
(41). BMI-1 targets Ink4a/ARF,
which encodes vital cell cycle inhibitors such as the human product
of CDKN2A gene p16 (Ink4a) (17),
which helps regulate stem cells. BMI-1 represses the
Ink4a/ARF locus and the onset of senescence in human
embryonic fibroblasts (43).
However, the inhibitory effect of PRP-1 on BMI-1 was demonstrated
to be independent of Ink4a/ARF at the protein levels in the
present study, as no difference in protein expression was observed
with or without PRP-1 treatment.
As PRP-1 has been demonstrated to inhibit Nanog
(1), the present study assessed its
upstream regulator, BRD4, which is a member of the bromodomain (BD)
and extraterminal domain (BET), an important hallmark of cancer and
epigenetic regulation and marker for stem cell self-renewal
(44). BRD4 binds to acetylated
histones at enhancers and promoters via its BDs in order to
regulate transcriptional elongation and gene expression programs
that have pivotal roles in inflammation and cancer development
(44–47). BRD4 JQ1, a BET inhibitor, suppresses
c-Myc and inhibits cell proliferation, and induces cell senescence
and apoptosis in human chondrosarcoma cells (48). The cytostatic effect of PRP-1 has
been demonstrated to be mediated by c-Myc inhibition in human
chondrosarcoma cells (11). Over
the past decade, the BD, BRD4, BET proteins have emerged as an
important class of epigenetic readers (27). However, PRP-1 did not act an
inhibitor of BRD4 in the present study.
TOP2A is an important enzyme in the regulation of
DNA structure and cell proliferation (28–31).
High TOP2A expression has been reported in advanced leiomyosarcoma,
high mitotic index and advanced stage tumors (31). TOP2A is highly expressed in soft
tissue sarcomas and is an independent predictor of poor prognosis
(30). However, the results of the
present study failed to demonstrate downregulation of TOP2A
activity following treatment with PRP-1.
Gene transcription is dynamically regulated in
stemness and cell proliferation (34,35).
The chromatin remodeling complexes can affect whether a gene is
activated or repressed, therefore the effect of PRP-1 on SWI/SNF
remodeling complexes was assessed, which depends on ATP as its
energy source, and its subunits BAF170 (SMARCC2), BAF155 (SMARCC1)
and BRG1 (SMARCA4) (49–52). The mammalian SWI/SNF-like
ATP-dependent chromatin remodeling complex, also termed
BAF/BRG/Brahma associated (esBAF) can either activate or repress
transcription (51). These factors
are recruited by transcription factors to the promoters of target
genes, where they can disrupt histone-DNA contacts and allow
transcription factors to access their sequence-specific DNA
(52). Depending on whether a
transcriptional activator or repressor recruits SWI/SNF,
transcription can be upregulated or downregulated accordingly
(52). The energy from ATP
hydrolysis is harnessed to disrupt histone-DNA contacts and move
nucleosomes away from the transcription start site or towards it
(52). Specifically, alterations to
mammalian SWI/SNF (mSWI/SNF or BAF) ATP-dependent chromatin
remodeling complexes and polycomb repressive complexes cause
disease-specific changes in the chromatin architecture and gene
expression across a number of sarcoma subtypes such as Ewing's,
synovial sarcoma and chondrosarcoma (48). SWI/SNF is essential for self-renewal
of stem cells and is critical for cell differentiation (49). A previous study on chondrosarcoma
reported that poorly differentiated types of tumor were
demonstrated to exhibit similarities with mesenchymal stem cells at
pre-chondrogenic stages, whereas more differentiated types of tumor
shared similarities with fully differentiated chondrocytes
(53). This suggested that
chondrosarcoma progression may be parallel to the deregulated
chondrocyte differentiation process of MSCs (53). Future studies will investigate
whether the inhibitory effect of PRP-1 on CSC leads to the revival
of the differentiation program.
Well-established interactions between esBAF and
polycomb complexes have been documented, which can act
antagonistically () or synergistically cooperate with polycomb
group proteins to support pluripotency (54,55).
BRG1 and BRM have the ability to act as either oncogenes or tumor
suppressors (54). The results of
the present study demonstrated that inhibition of BMI-1 with PRP-1
was accompanied by inhibition of both subunits of the esBAF complex
(BRG1 and BRM), which possess oncogenic properties in
chondrosarcoma. Depletion of BAF170 (SMARCC2) instead of BAF155
(SMARCC1) resulted in the loss of stem cell properties in hESCs,
and this pattern was different from that observed in rodent stem
cells (i.e., mESCs and mEpiSCs) (55–58).
Deletion of BRG1 (SMARCA4) in embryonic stem cells results
in the loss of both self-renewal capacity and pluripotency
(57). In the present study, BAF
155 was not affected by PRP-1 treatment, whereas BAF170 protein
expression, BRM and BRG1 were all demonstrated to be inhibited
following treatment compared with untreated cells Genome-wide
studies have indicated that BRG1 and BRM have cooperative and
antagonistic interactions in transcription (36). The results of the present study
demonstrated that downregulation of BAF 170, BRG and BRM by PRP-1
may act in favor of synergistic regulation of BRG and BRM in
chondrosarcoma, and that they may possess oncogenic properties in
this microenvironment.
Cytostatic drugs can lead to decreased cell
viability (12–14). Autophagy is a tightly regulated
catabolic process of cellular self-digestion by which cellular
components are targeted to lysosomes for their degradation
(59). The present study failed to
detect any apoptotic action of PRP-1 (data not shown). In cell
cultures, certain drugs that activate autophagy have been
associated with programmed cell death, but not apoptosis. Autophagy
has been demonstrated to be essential for cell death, especially in
cell lines that are resistant to apoptosis-driven chemotherapy,
which may benefit from directly destroying tumor cells or
sensitizing them to chemotherapy (59,60).
Thus, it may be assumed that drastic elimination of chondrosarcoma
stem cells by PRP-1 may be mediated by targeting BAF chromatin
remodeling complexes and by induction of autophagy. A potential
future study will aim to investigate whether this inhibition serves
a pivotal role in the process of cancer stem cell decrease mediated
by the peptide action.
In conclusion, the results of the present study
demonstrated that ALDH1A1 is the only current biomarker in
chondrosarcoma CSCs. PRP-1 decreased CSCs across all experiments.
As a possible mechanismthe results of the present study indicated
that PRP-1 may target chromatin remodeling complexes. Our future
efforts will be directed towards understanding the interconnection
between CSC maintenance, self-renewal and BAF complexes.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Patricia Lissette
Guevara, M.A from the Flow Cytometry Core Facility, Miller School
of Medicine for her assistance in data acquisition, and analysis.
The authors would also like to thank Dr Joel Block's Laboratory
(Rush University, Chicago, USA) for providing the human JJ012
chondrosarcoma cells; Dr Messner (Ontario Cancer Institute, Canada)
for Ly10 cells; and Dr Vega (MD Anderson Cancer Center, Houston,
TX, USA) for HBL-1 cells. PRP-1 was provided by professor A.
Galoyans laboratory H, Buniatian Institute of Biochemistry of
National Academy of Sciences of Armenia. The authors would also
like to thank Dr Claude Henry Volmar, Director of the Research
Laboratory for the Center of Therapeutic Innovation, University of
Miami, Miller School of Medicine for assisting with BRD4
experiments.
Funding
The present study was supported in part by the
Ratcliffe Foundation to Miami Center of Orthopaedic Research and
Education and the Department of Orthopedics, University of Miami,
Miller School of Medicine.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article
Authors' contributions
AM conducted most of the experiments and was
involved in manuscript writing. AH and AS performed western blot
experiments, statistical and image analysis. CG participated in
data analysis and certain experiments. SS performed
fluorescence-activated cell sorting and flow cytometry data
analysis. MB performed lymphoma cell line experiments with PRP-1
and participated in the critical revision of this manuscript. LZJ
tested the PRP-1 action in melanoma cell line and contributed to
the acquisition, analysis and interpretation of the data. SAC and
JP provided substantial contributions to the conception and design
and participated in critical revision of the manuscript. JB
performed western blotting experiments. KG designed the study,
wrote parts of the manuscript and was involved in the critical
revision of it. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Dorfman HD and Czerniak B: Bone cancers.
Cancer. 75 (1 Suppl):S203–S210. 1995. View Article : Google Scholar
|
|
2
|
Fujiwara T and Ozaki T: Overcoming
therapeutic resistance of bone sarcomas: Overview of the molecular
mechanisms and therapeutic targets for bone sarcoma stem cells.
Stem Cells Int. 2016:26030922016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Galoian K, Qureshi A, D'Ippolito G,
Schiller PC, Molinari M, Johnstone AL, Brothers SP, Paz AC and
Temple HT: Epigenetic regulation of embryonic stem cell marker
miR302C in human chondrosarcoma as determinant of antiproliferative
activity of proline-rich polypeptide 1. Int J Oncol. 47:465–472.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Galoian K, Abrahamyan S, Chailyan G,
Qureshi A, Patel P, Metser G, Moran A, Sahakyan I, Tumasyan N, Lee
A, et al: Toll like receptors TLR1/2, TLR6 and MUC5B as binding
interaction partners with cytostatic proline rich polypeptide 1 in
human chondrosarcoma. Int J Oncol. 52:139–154. 2018.PubMed/NCBI
|
|
5
|
Vermeulen L, Todaro M, de Sousa Mello F,
Sprick MR, Kemper K, Perez Alea M, Richel DJ, Stassi G and Medema
JP: Single-cell cloning of colon cancer stem cells reveals a
multi-lineage differentiation capacity. Proc Natl Acad Sci USA.
105:13427–13432. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sell S: On the stem cell origin of cancer.
Am J Pathol. 176:2584–2494. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Honoki K, Fujii H, Kubo A, Kido A, Mori T,
Tanaka Y and Tsujiuchi T: Possible involvement of stem-like
populations with elevated ALDH1 in sarcomas for chemotherapeutic
drug resistance. Oncol Rep. 24:501–505. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tanei T, Morimoto K, Shimazu K, Kim SJ,
Tanji Y, Taguchi T, Tamaki Y and Noguchi S: Association of breast
cancer stem cells identified by aldehyde dehydrogenase 1 expression
with resistance to sequential paclitaxel and epirubicin-based
chemotherapy for breast cancers. Clin Cancer Res. 15:4234–4241.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Awad O, Yustein JT, Shah P, Gul N, Katuri
V, O'Neill A, Kong Y, Brown ML, Toretsky JA and Loeb DM: High ALDH
activity identifies chemotherapy-resistant Ewing's sarcoma stem
cells that retain sensitivity to EWS-FLI1 inhibition. PLoS One.
5:e139432010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Galoyan A: Neurochemistry of brain
neuroendocrine immune system: Signal molecules. Neurochem Res.
25:1343–1355. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Galoian K, Temple TH and Galoyan A:
Cytostatic effect of the hypothalamic cytokine PRP-1 is mediated by
mTOR and cMyc inhibition in high grade chondrosarcoma. Neurochem
Res. 36:812–818. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Galoian K, Qureshi A, Wideroff G and
Temple HT: Restoration of desmosomal junction protein expression
and inhibition of H3K9-specific histone demethylase activity by
cytostatic proline-rich polypeptide-1 leads to suppression of
tumorigenic potential in human chondrosarcoma cells. Mol Clin
Oncol. 3:171–178. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Galoian K, Luo S, Qureshi A, Patel P,
Price R, Morse AS, Chailyan G, Abrahamyan S and Temple HT: Effect
of cytostatic proline rich polypeptide-1 on tumor suppressors of
inflammation pathway signaling in chondrosarcoma. Mol Clin Oncol.
5:618–624. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Galoian KA, Guettouche T, Issac B, Qureshi
A and Temple HT: Regulation of onco and tumor suppressor MiRNAs by
mTORC1 inhibitor PRP-1 in human chondrosarcoma. Tumour Biol.
35:2335–2341. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Galoian KA, Temple TH and Galoyan A:
Cytostatic effect of novel mTOR inhibitor, PRP-1 (galarmin) in MDA
231 (ER-) breast carcinoma cell line. PRP-1 inhibits mesenchymal
tumors. Tumour Biol. 32:745–751. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lohberger B, Rinner B, Stuendl N, Absenger
M, Liegl- Atzwanger B, Walzer SM, Windhager R and Leithner A:
Aldehyde dehydrogenase 1, a potential marker for cancer stem cells
in human sarcoma. PLoS One. 7:e436642012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ginestier C, Hur MH, Charafe-Jauffret E,
Monville F, Dutcher J, Brown M, Jacquemier J, Viens P, Kleer CG,
Liu S, et al: ALDH1 is a marker of normal and malignant human
mammary stem cells and a predictor of poor clinical outcome. Cell
Stem Cell. 1:555–567. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nakahata K, Uehara S, Nishikawa S, Kawatsu
M, Zenitani M, Oue T and Okuyama H: Aldehyde dehydrogenase 1
(ALDH1) is a potential marker for cancer stem cells in embryonal
rhabdomyosarcoma. PLoS One. 10:e01254542015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tolstorukov MY, Sansam CG, Lu P,
Koellhoffer EC, Helming KC, Alver BH, Tillman EJ, Evans JA, Wilson
BG, Park PJ and Roberts CW: Swi/Snf chromatin remodeling/tumor
suppressor complex establishes nucleosome occupancy at target
promoters. Proc Natl Acad Sci USA. 110:10165–10170. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ueda K, Ogasawara S, Akiba J, Nakayama M,
Todoroki K, Ueda K, Sanada S, Suekane S, Noguchi M, Matsuoka K and
Yano H: Aldehyde dehydrogenase 1 identifies cells with cancer stem
cell-like properties in a human renal cell carcinoma cell line.
PLoS One. 8:e754632013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hoyt AK, Moran A, Granger C, Sedani A,
Saigh S, Brown J and Galoian KA: PRP1 significantly decreases the
ALDHhigh cancer stem cell population and regulates the aberrant
Wnt/β-catenin pathway in human chondrosarcoma JJ012 cells. Oncol
Rep. 42:103–114. 2019.PubMed/NCBI
|
|
22
|
Kim WT and Ryu CJ: Cancer stem cell
surface markers on normal stem cells. BMB Rep. 50:285–298. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wirths S, Malenke E, Kluba T, Rieger S,
Müller MR, Schleicher S, Hann von Weyhern C, Nagl F, Fend F, Vogel
W, et al: Shared cell surface marker expression in mesenchymal stem
cells and adult sarcomas. Stem Cells Transl Med. 2:53–60. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Genadry KC, Pietrobono S, Rota R and
Linardic CM: Soft tissue sarcoma cancer stem cells: An overview.
Front Oncol. 8:4752018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mak AB, Pehar M, Nixon AM, Williams RA,
Uetrecht AC, Puglielli L and Moffat J: Post-translational
regulation of CD133 by ATase1/ATase2-mediated lysine acetylation. J
Mol Biol. 426:2175–2182. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jacques C, Lamoureux F, Baud'huin M,
Rodriguez Calleja L, Quillard T, Amiaud J, Tirode F, Rédini F,
Bradner JE, Heymann D and Ory B: Targeting the epigenetic readers
in Ewing sarcoma inhibits the oncogenic transcription factor
EWS/Fli1. Oncotarget. 7:24125–24140. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang JC: Cellular roles of DNA
topoisomerases: A molecular perspective. Nat Rev Mol Cell Biol.
3:430–440. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Pulleyblank DE: Of topo and maxwell's
dream. Science. 277:648–649. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li TK and Liu LF: Tumor cell death induced
by topoisomerase-targeting drugs. Annu Rev Pharmacol Toxicol.
41:53–77. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
da Cunha IW, De Brot L, Carvalho KC, Rocha
RM, Fregnani JH, Falzoni R, Ferreira Fde O, Aguiar S Jr, Lopes A,
Muto NH, et al: Prognostication of soft tissue sarcomas based on
chromosome 17q gene and protein status: Evaluation of TOP2A,
HER-2/neu, and survivin. Ann Surg Oncol. 19:1790–1799. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Baiocchi G, Poliseli FL, De Brot L,
Mantoan H, Schiavon BN, Faloppa CC, Vassallo J, Soares FA and Cunha
IW: TOP2A copy number and TOP2A expression in uterine benign smooth
muscle tumours and leiomyosarcoma. J Clin Pathol. 69:884–889. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Raab JR, Runge JS, Spear CC and Magnuson
T: Co-regulation of transcription by BRG1 and BRM, two mutually
exclusive SWI/SNF ATPase subunits. Epigenetics Chromatin.
10:622017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Douglas D, Hsu JH, Hung L, Cooper A,
Abdueva D, van Doorninck J, Peng G, Shimada H, Triche TJ and Lawlor
ER: BMI-1 promotes ewing sarcoma tumorigenicity independent of
CDKN2A repression. Cancer Res. 68:6507–6515. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Pietersen AM, Horlings HM, Hauptmann M,
Langerød A, Ajouaou A, Cornelissen-Steijger P, Wessels LF, Jonkers
J, van de Vijver MJ and van Lohuizen M: EZH2 and BMI1 inversely
correlate with prognosis and TP53 mutation in breast cancer. Breast
Cancer Res. 10:R1092008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Dhawan S, Tschen SI and Bhushan A: Bmi-1
regulates the Ink4a/Arf locus to control pancreatic beta-cell
proliferation. Genes Dev. 23:906–911. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Becker M, Korn C, Sienerth AR, Voswinckel
R, Luetkenhaus K, Ceteci F and Rapp UR: Polycomb group protein Bmi1
is required for growth of RAF driven non-small-cell lung cancer.
PLoS One. 4:e42302009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Dovey JS, Zacharek SJ, Kim CF and Lees JA:
Bmi1 is critical for lung tumorigenesis and bronchioalveolar stem
cell expansion. Proc Natl Acad Sci USA. 105:11857–11862. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Bruggeman SW, Hulsman D, Tanger E, Buckle
T, Blom M, Zevenhoven J, van Tellingen O and van Lohuizen M: Bmi1
controls tumor development in an Ink4a/Arf-independent manner in a
mouse model for glioma. Cancer Cell. 12:328–341. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang E, Bhattacharyya S, Szabolcs A,
Rodriguez-Aguayo C, Jennings NB, Lopez-Berestein G, Mukherjee P,
Sood AK and Bhattacharya R: Enhancing chemotherapy response with
Bmi-1 silencing in ovarian cancer. PLoS One. 6:e179182011.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hsu JH and Lawlor ER: BMI 1 suppresses
contact inhibition and stabilizes YAP in Ewing sarcoma. Oncogene.
30:2077–2085. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wu Z, Min L, Chen D, Hao D, Duan Y, Qiu G
and Wang Y: Overexpression of BMI-1 promotes cell growth and
resistance to cisplatin treatment in osteosarcoma. PLoS One.
6:e146482011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Liu L, Andrews LG and Tollefsbol TO: Loss
of the human polycomb group protein BMI1 promotes cancer-specific
cell death. Oncogene. 25:4370–4375. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Bracken AP, Kleine-Kohlbrecher D, Dietrich
N, Pasini D, Gargiulo G, Beekman C, Theilgaard-Mönch K, Minucci S,
Porse BT, Marine JC, et al: The Polycomb group proteins bind
throughout the INK4A-ARF locus and are disassociated in senescent
cells. Genes Dev. 21:525–530. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Liu W, Stein P, Cheng X, Yang W, Shao NY,
Morrisey EE, Schultz RM and You J: BRD4 regulates Nanog expression
in mouse embryonic stem cells and preimplantation embryos. Cell
Death Differ. 21:1950–1960. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Gonzales-Cope M, Sidoli S, Bhanu NV, Won
KJ and Garcia BA: Histone H4 acetylation and the epigenetic reader
Brd4 are critical regulators of pluripotency in embryonic stem
cells. BMC Genomics. 17:952016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Rahnamoun H, Lee J, Sun Z, Lu H, Ramsey
KM, Komives EA and Lauberth SM: RNAs interact with BRD4 to promote
enhanced chromatin engagement and transcription activation. Nat
Struct Mol Biol. 25:687–697. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhang HT, Gui T, Sang Y, Yang J, Li YH,
Liang GH, Li T, He QY and Zha ZG: The BET bromodomain inhibitor JQ1
suppresses chondrosarcoma cell growth via regulation of
YAP/p21/c-Myc signaling. J Cell Biochem. 118:2182–2192. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
McBride MJ and Kadoch C: Disruption of
mammalian SWI/SNF and polycomb complexes in human sarcomas:
Mechanisms and therapeutic opportunities. J Pathol. 244:638–649.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
De-Meng Chen X-QZ, Kai Wang and Yi-Zhou
Jiang: SWI/SNF chromatin remodeling complex in regulating
mesenchymal stem cell lineage specification. J Tissue Sci
Engineering. 6:1542015.
|
|
50
|
Yang J, Ren Z, Du X, Hao M and Zhou W: The
role of mesenchymal stem/progenitor cells in sarcoma: Update and
dispute. Stem Cell Investig. 1:182014.PubMed/NCBI
|
|
51
|
Tang L, Nogales E and Ciferri C: Structure
and function of SWI/SNF chromatin remodeling complexes and
mechanistic implications for transcription. Prog Biophys Mol Biol.
102:122–128. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Wilson BG, Wang X, Shen X, McKenna ES,
Lemieux ME, Cho YJ, Koellhoffer EC, Pomeroy SL, Orkin SH and
Roberts CW: Epigenetic antagonism between polycomb and SWI/SNF
complexes during oncogenic transformation. Cancer Cell. 18:316–328.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
de Andrea CE and Hogendoorn PC: Epiphyseal
growth plate and secondary peripheral chondrosarcoma: The
neighbours matter. J Pathol. 226:219–228. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Kadoch C, Williams RT, Calarco JP, Miller
EL, Weber CM, Braun SM, Pulice JL, Chory EJ and Crabtree GR:
Dynamics of BAF-Polycomb complex opposition on heterochromatin in
normal and oncogenic states. Nat Genet. 49:213–222. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Shao Z, Raible F, Mollaaghababa R, Guyon
JR, Wu CT, Bender W and Kingston RE: Stabilization of chromatin
structure by PRC1, a Polycomb complex. Cell. 98:37–46. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Poynter ST and Kadoch C: Polycomb and
trithorax opposition in development and disease. Wiley Interdiscip
Rev Dev Biol. 5:659–688. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Zhang X, Li B, Li W, Ma L, Zheng D, Li L,
Yang W, Chu M, Chen W, Mailman RB, et al: Transcriptional
repression by the BRG1-SWI/SNF complex affects the pluripotency of
human embryonic stem cells. Stem Cell Reports. 3:460–474. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Kahali B, Yu J, Marquez SB, Thompson KW,
Liang SY, Lu L and Reisman D: The silencing of the SWI/SNF subunit
and anticancer gene BRM in Rhabdoid tumors. Oncotarget.
5:3316–3332. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Fulda S and Kogel D: Cell death by
autophagy: Emerging molecular mechanisms and implications for
cancer therapy. Oncogene. 34:5105–5113. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Kanzawa T, Germano IM, Komata T, Ito H,
Kondo Y and Kondo S: Role of autophagy in temozolomide-induced
cytotoxicity for malignant glioma cells. Cell Death Differ.
11:448–457. 2004. View Article : Google Scholar : PubMed/NCBIPubMed/NCBIPubMed/NCBIPubMed/NCBIPubMed/NCBIPubMed/NCBIPubMed/NCBIPubMed/NCBIPubMed/NCBIPubMed/NCBIPubMed/NCBIPubMed/NCBIPubMed/NCBI
|

















