Introduction
Renal cell carcinoma (RCC) is one of the most
commonly diagnosed human malignant neoplasms with more than 300,000
new patients diagnosed worldwide each year (1). The major type of kidney tumor (80–90%)
originates from the epithelial lining of the proximal convoluted
tubules and exhibits highly vascularized and metastatic
characteristics (2). To date, the
primary therapy for localized RCC is surgery (radical nephrectomy
and nephron-sparing surgery), while for unresectable and metastatic
RCC, the therapeutic options remain limited (3–5). RCC
is sensitive to neither traditional chemotherapy nor radiation
therapy (6). However, the existing
therapies remain ineffective against metastatic and unresectable
RCC. Therefore, exploring effective and safe strategies for the
treatment of RCCs is crucial.
Tumor necrosis factor-related apoptosis-inducing
ligand (TRAIL), a member of the tumor necrosis factor (TNF) family,
is an optimal anticancer agent (7).
The ability of TRAIL to induce apoptosis depends on the interaction
of TRAIL and its membrane receptors death receptor (DR)4 and DR5
(named as TRAIL-R1 and TRAIL-R2) (8). Upon ligand stimulation, DR4 and DR5
bind Fas-associated death domain protein (FADD) through the death
domain, which results in the formation of the death-inducing
signaling complex (DISC). Caspase 8 is then recruited to DISC where
it initiates the downstream apoptotic cascade. Activation of
caspase 8 induces apoptosis via two well-elucidated apoptotic
pathways: The extrinsic pathway (stimulating the effector caspases
3, 6, and 7) and the intrinsic-mitochondrial pathway [stimulating
Bax and Bak, and releasing mitochondrial cytochrome c and
mitochondrial-derived activator of caspase (Smac)] (9–11). As
death receptors, DR4 and DR5 are normally upregulated in tumor
cells, thus the TRAIL signaling pathway can be an optimal target
for cancer therapy (12–14). Accumulating evidence from basic and
clinical studies indicates that various cancer types are not
sensitized to TRAIL-induced apoptosis (15,16).
TRAIL-based drug development has attracted significant interest to
identify an effective combination regimen, which can overcome TRAIL
resistance in cancer cells.
In renal cancer, a cancer type highly resistant to
chemotherapy, the identification of specific agents that are able
to sensitize TRAIL-induced apoptosis of unresponsive renal
carcinoma cells holds the utmost importance for the targeted
treatment of renal cancer. In the present study, our data showed
that andrographolide (Andro), a major constituent of
Andrographis paniculate, an annual herbaceous plant in the
family Acanthaceae, remarkably improved the sensitivity of
RCC cells to TRAIL-induced growth inhibition. The combined
treatment stimulated caspase-dependent apoptosis, and enhanced DR4
expression. Our study provides proof-of-concept evidence for the
clinical application of this traditional anti-inflammatory medical
agent, andrographolide, in the treatment of renal cancers.
Materials and methods
Cell culture and treatments
The RCC cell lines 786-0, OS-RC-2, and ACHN were
purchased from the Type Culture Collection of the Chinese Academy
of Sciences (Shanghai, China). 786-0 cells were cultured in
RPMI-1640 medium (HyClone; Cytiva). OS-RC-2 and ACHN cells were
cultured in the DMEM medium (HyClone; Cytiva). All media were
supplemented with 10% fetal bovine serum (FBS) (Biological
Industries, USA) and penicillin/streptomycin solution. All cells
were cultured under standard incubator conditions (37°C, 5%
CO2).
Chemicals, reagents, and
antibodies
Andrographolide (MedChemExpress, MCE) was dissolved
in DMSO at 10 mmol/l as a stock solution, and recombinant human
TRAIL (R&D Systems, Inc.) was prepared in PBS containing 0.1%
bovine serum albumin at 20 µg/ml. Z-VAD (HY-16658) and
Necrostatin-1 (HY-15760) were purchased from MCE. Antibodies used
in this study were as follows: Phospho-HistoneH2A.X (product
#9718), PARP1 (product #9532), DR4 (product #42533), caspase 9
(product #9502), caspase 8 (product #4790), GAPDH (product #51332)
(from Cell Signaling Technology, Inc.), Bax (cat. #633601,
BioLegend), DR5 (LM11912, Novus, USA), β-actin (ab8227, Abcam),
anti-rabbit IgG (product #7054) and anti-mouse IgG (product #7056)
(from Cell Signaling Technology, Inc.).
Cell viability assay
Cell viability was assessed by measuring the
formazan production following the addition of
3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulphop-henyl)-2H-tetrazolium,
inner salt (MTS) (Promega Corp.). Approximately 5,000 cells/well
were seeded in a 96-well plate and incubated at 37°C in a 5%
CO2 incubator for 24 h. The cells were incubated in a
medium containing 20 µl MTS for 3 h at 37°C post treatment under
different conditions for 24 h. Absorbance was detected using a
BioTek ELISA reader (BioTek Instruments, Inc.) at a wavelength of
490 nm.
Cell proliferation assay
We used EdU (5-ethynyl-2′-deoxyuridine) and colony
formation assays to evaluate the effect of Andro and/or TRAIL on
cell proliferation. For the EdU Assay, 2×105 cells/well
were seeded in a 12-well plate, treated under Andro and/or TRAIL
for 24 h, and then, cell proliferation was determined using
BeyoClick EdU cell proliferation kit (Beyotime Biotech Inc.)
according to the manufacturer's instructions. Images (×400
magnification) of the cells were acquired on a confocal microscope
using OLYMPUS cellSens Standard software (Olympus).
For the colony formation assays, 200 cells/well were
seeded in a 6-well plate in 2 ml of medium, treated under different
conditions, and subjected to growth for 12 days. After 12 days of
incubation, the cells were washed once with cold phosphate-buffered
saline (PBS). Then, 4% paraformaldehyde was used to fix the cells
for 20 min. Cells were then stained with 0.1% crystal violet
solution for 15 min at 25°C, and then washed with water thrice and
air-dried for counting using an inverted microscope (×100
magnification), where cell colonies (>50 cells) were counted.
All experiments were repeated thrice.
Cell migration assay
For cell migration, 2×105 cells/well were
seeded in a 6-well plate and incubated in an incubator. When the
cells reached 90% confluence, straight scratches were made by using
a sterile 200-µl pipette tip and the cells were then washed thrice
with PBS. Then the cells were incubated in an incubator with
serum-free medium containing Andro or/and TRAIL or DMSO for 24 h.
An inverted microscope (×100 magnification) was used to monitor
cell migration at 0, 6, 12, 18 and 24 h post scratching. Images of
cells were acquired on a confocal microscope using OLYMPUS cellSens
Standard software. Data were analyzed with Image J software
(version 1.8.0, National Institutes of Health, Bethesda, MD,
USA).
Flow cytometric analysis
For cell cycle analysis, cells treated under
different conditions for 24 h were detached from the 6-well culture
plates, washed twice with ice-cold PBS, and pelleted by
centrifugation at 1,000 × g. The cells were then suspended in 75%
ethanol overnight at −20°C. Following an overnight suspension,
cells were centrifuged at 1,000 × g for 5 min and washed twice with
ice-cold PBS. The cell pellets were resuspended in buffer
containing PI (propidium iodide) and RNase for 1 h in the dark at
37°C, and the cell cycle distribution was examined by flow
cytometry (BD Bioscience) after filtration.
Apoptotic cells were identified and quantified by
using the Annexin V-FITC apoptosis detection kit (KeyGENBioTECH).
After treatment under different conditions for 24 h, the cells were
digested and collected with trypsin solution without EDTA, which
were then washed twice with PBS and then centrifuged at 1,200 × g
for 5 min to collect the cells. In the next step, the cells were
re-suspended in binding buffer and incubated with Annexin V-FITC
and PI for 15 min in the dark at 37°C. A fluorescence-activated
cell sorting (FACS) flow cytometer (BD Bioscience) was used to
analyze cell apoptosis.
Cell senescence assay
Senescent cells were identified and quantified by
Senescence β-Galactosidase staining kit (Cell Signaling Technology,
Inc.). Following treatment under different conditions for 24 h, the
cells were washed with PBS, fixed by the fixative solution for 15
min at 25°C, and determined using Senescence β-Galactosidase
(pH=6.0) staining for 24 h. An inverted microscope (×400
magnification) was used to monitor senescent cells and to count
them. Images of the cells were acquired on a confocal microscope
using OLYMPUS cellSens Standard software.
Immunoblot assay
Whole-cell extracts, which were treated under
different conditions for the corresponding times, were separated by
12% sodium dodecyl sulfate polyacrylamide gel electrophoresis and
electrophoretically transferred to nitrocellulose membranes (EMD
Millipore), and 5-bromo-4-chloro-3-indolyl phosphate and nitro blue
tetrazolium (EMD Millipore) were used to visualize the protein
bands. Images of the western blotting were acquired on a scanner
(Epson Perfection V330 Photo) using Scan-n-Stitch Deluxe software
(version 1.1.9, Arcsoft).
siRNAs for the construction of
knockdown cells
Synthetic siRNA [negative-control siRNA, DR4 siRNA,
and DR5 siRNA] which can specifically knock down the
TNFRSF10A (DR4) gene and TNFRSF10B (DR5) gene, were
obtained from GenePharma (Shanghai, China). The cellular delivery
of siRNA was performed using Lipofectamine 3000 (Thermo Fisher
Scientific, Inc.), optimized using various siRNA concentrations,
and evaluated by immunoblot assay. The siRNA sequences are listed
in Table SI.
Data collection and bioinformatics
analysis
We downloaded fragments per kilobase million (FPKM)
values of RNA-sequencing profiles of RCC patients including 414 RCC
tissues and 19 normal tissues from The Cancer Genome Atlas (TCGA)
databse's official website (https://portal.gdc.cancer.gov/). RNA expression
datasets were processed using the R software version 3.6.6
(https://www.r-project.org/).
Statistical analysis
Differences among test groups were analyzed by
GraphPad Prism software (version 8.0; GraphPad Software Inc.). Data
are expressed as the mean ± standard deviations (SD). An unpaired
two-tailed Student's t test was performed to detect statistical
difference between two individual experimental groups. For multiple
comparisons, statistical analyses were performed using one-way
analysis of variance (ANOVA) and two-way ANOVA with Dunnett and
Tukey post-test. P<0.05 was considered to indicate a
statistically significant difference.
Results
Andro sensitizes TRAIL-induced
survival and proliferation inhibition in renal cancer cells
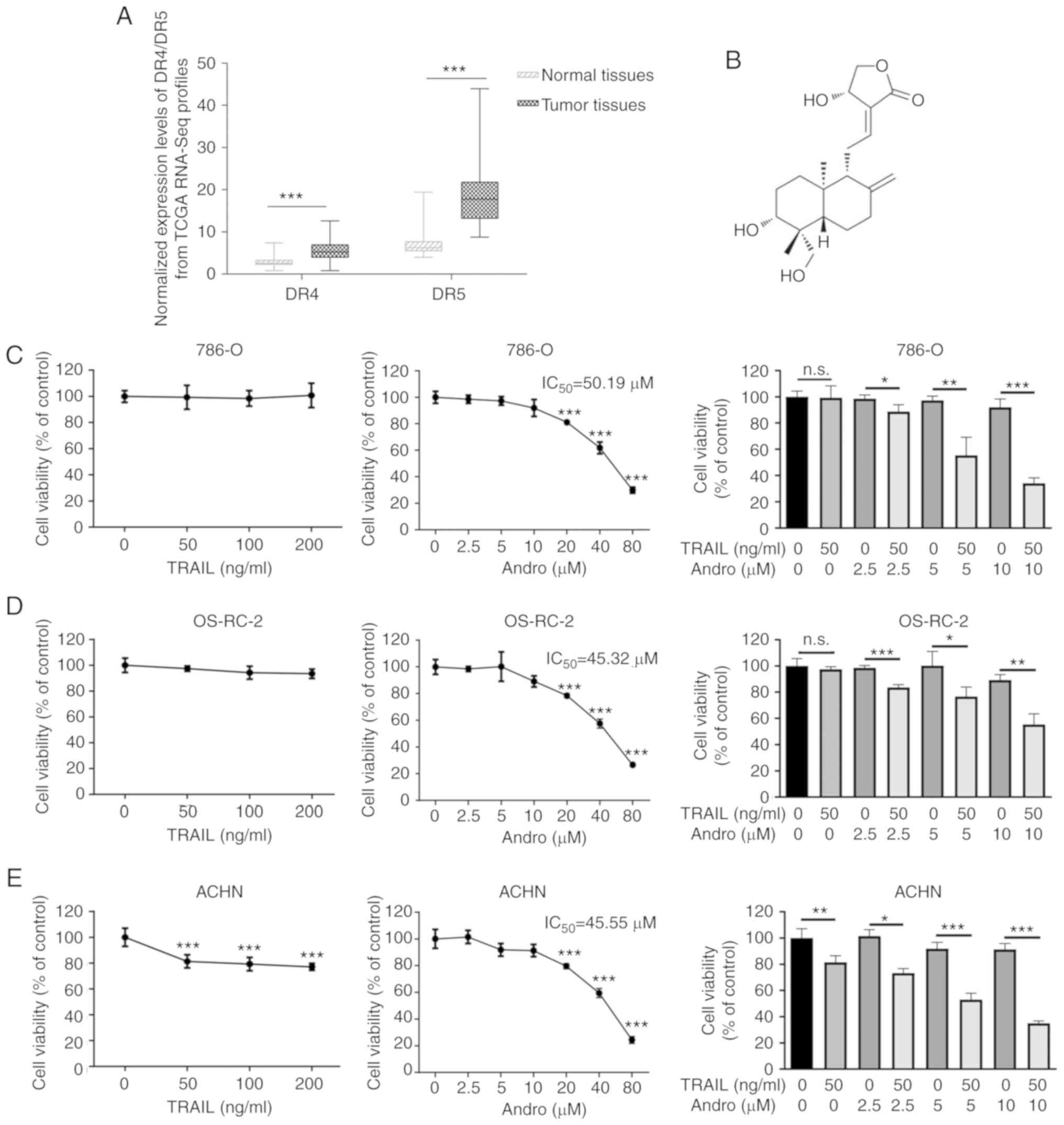
As DR4 and DR5 are canonical TRAIL receptors
involved in its antitumor effects, we analyzed mRNA expression data
of RCC patients from the TCGA database. We found that the mean DR5
mRNA expression in renal cancer tissues exceeded that in normal
tissues, whereas a mild was found in DR4 between tumor and normal
tissues (Fig. 1A). These data
hinted that TRAIL signaling could be a potential target for renal
cancer therapy. However, our experiments indicated that renal
cancer 786-0, OS-RC-2, and ACHN cells were resistant to the
TRAIL-mediated suppression even at an extremely high concentration
(200 ng/ml), while our previous study demonstrated that the 50%
inhibitory concentration IC50 value of TRAIL in bladder
cancer T24 cells was 38.35 ng/ml (Fig.
1C-E) (17). As noted,
andrographolide (Andro), a diterpene lactone
(C20H30O) (Fig.
1B), represents a potential agonist for TRAIL therapy. The
IC50 of Andro was 50.19 µM in 786-0 cells, 45.32 µM in
OS-RC-2 cells, and 45.55 µM in ACHN cells (Fig. 1C-E). Interestingly, cell viability
of the RCC cell lines treated with the combination of Andro and
TRAIL for 24 h was significantly decreased as compared with that of
the cells treated with TRAIL or Andro alone (Fig. 1C-E).
Next, we evaluated the ability of Andro to sensitize
TRAIL-mediated proliferation inhibition in RCC cells. As shown in
Fig. 2B, TRAIL (50 ng/ml) or Andro
(5 µM or 10 µM) alone mildly inhibited the growth rate of renal
cancer cells. In contrast, Andro significantly sensitized 786-0
cells to TRAIL-mediated proliferation inhibition at a concentration
of 5 µM. In agreement with this result, the morphological changes
in treated RCC cells further supported that the combined treatment
with TRAIL and Andro inhibited the survival and proliferation of
786-0 (Fig. 2A), OS-RC-2 (Fig. S1A) and ACHN cells (Fig. S2A) more potently than single-drug
treatment. Furthermore, EdU cell proliferation assay showed that
786-0 cells treated with the combination of TRAIL and Andro
proliferated much more slower than the cells exposed to single-drug
treatment (Fig. 2C).
 | Figure 2.TRAIL combined with Andro inhibits
the cell proliferation, colony formation, and migration of 786-0
cells. (A) Images (×200 magnification) show 786-0 cell morphology
after treatment with various concentrations of TRAIL and/or Andro
for 24 h. (B) Cell proliferation of 786-0 cells after treated with
various concentrations of TRAIL and/or Andro for 24, 48 and 72 h
(two-way ANOVA, Tukey). (C) Images (×200 magnification) show cells
that were treated with TRAIL (50 ng/ml) and/or Andro (5 µM) for 24
h, and then proliferation was determined by BeyoClick EdU cell
proliferation kit. Hoechst staining shows the entire nucleus, and
EdU shows the nucleus which was proliferating. The histogram
(right) shows the ratio of proliferation (one-way ANOVA, Tukey).
(D) Effects of TRAIL (50 ng/ml) and Andro (0.5 µM) on the
clonogenic formation of 786-0 cells. The histogram indicates the
percentage of each group's clony number compared to the control
group (one-way ANOVA, Tukey). (E) Images (×100 magnification) show
the effects of TRAIL (50 ng/ml) and Andro (5 µM) on the migratory
ability of 786-0 cells. The histogram indicates the percentage of
migration compared to the initial width (one-way ANOVA, Tukey).
Data are shown as mean ± SD; ***P<0.001; n=3). Andro,
andrographolide; TRAIL, tumor necrosis factor-related
apoptosis-inducing ligand. |
Andro promotes TRAIL-dependent
inhibition of the clone formation and migration of renal cancer
cells
Subsequently, we conducted clonogenic assays to
determine the long-term anti-proliferative effects of Andro and
TRAIL in invasive renal cancer cells. Our data indicated that the
colony formation in case of cells treated with the combination of
Andro and TRAIL was significantly (75%) inhibited compared to that
in cells treated with only Andro or TRAIL in 786-0 (Fig. 2D), OS-RC-2 (Fig. S1B) and ACHN cells (Fig. S2B) cells.
To determine whether Andro increases the ability of
TRAIL to suppress RCC migration, we applied wound healing
measurements as functional readings. The results indicated that
TRAIL or Andro alone modestly (<25%) decreased the migration of
RCC 786-0 (Fig. 2E), OS-RC-2
(Fig. S1C) and ACHN cells
(Fig. S2C) cells. However, there
was an approximately 95% decrease in RCC migration induced by the
combined treatment of Andro and TRAIL. These findings demonstrated
that Andro effectively enhanced the suppression of the growth and
migration of renal cancer cells mediated by TRAIL.
Andro enhances TRAIL-induced G2 cell
cycle arrest and senescence in renal cancer cells
To understand the mechanism through which the
combined treatment of Andro and TRAIL inhibited cell proliferation,
we investigated the effects of the indicated drugs on the cell
cycle distribution of RCC cells, and demonstrated that TRAIL (50
ng/ml) or Andro (5 µM) alone did not have a significant effect on
cell cycle distribution. Yet, in the case of 786-0 cells treated
with the same amounts of TRAIL and Andro significant cell cycle
arrest at the G2 phase was triggered (Fig. 3A and B).
Moreover, RCC cells treated with a combination of
Andro and TRAIL appeared larger, and flat morphological changes
with time were exacerbated, which indicated that cell senescence
was exacerbated. This was confirmed by the β-galactosidase staining
assay (Fig. 3C and D).
Combination of Andro and TRAIL
triggers apoptosis in renal cancer cells
In RCC 786-0 cells treated with the combination of
Andro and TRAIL, we observed apoptotic features, such as cell
contraction, rounding, and floating. We then evaluated the roles of
Andro in apoptosis progression using Annexin V-fluorescein
isothiocyanate (FITC)/propidium iodide (PI)-labeled flow cytometry.
Compared with groups that were solely treated with TRAIL
(4.07±0.29%) or Andro (6.33±0.24%), the groups treated with their
combination for 24 h exhibited 39.26±1.17% apoptosis (Fig. 4A). Immunoblot assays were used to
analyze changes in protein content in 786-0 cells treated with
TRAIL and/or Andro. The results indicated that the combined
treatment enhanced levels of cleaved-poly(ADP ribose) polymerase 1
(PARP1; 89 kDa), cleaved caspase 8 (17 kDa), and cleaved caspase 9
(35 kDa). It also decreased full-length caspase 8 (60 kDa) and
caspase 9 (45 kDa) expression while increasing levels of apoptosis
regulator Bax (21 kDa), indicating caspase 8 activation and
initiation of the apoptotic signal (Fig. 4B). We also noted that the levels of
the phosphorylated form of H2AX (γ-H2AX) were increased in Andro
and TRAIL combined treated groups (Fig.
4B). We also showed that the combination therapy also potently
induced apoptosis in other RCC cell lines (OSR-C and ACHN cells);
supporting that Andro enhanced TRAIL-induced apoptosis independent
of RCC cell type (Fig. S3).
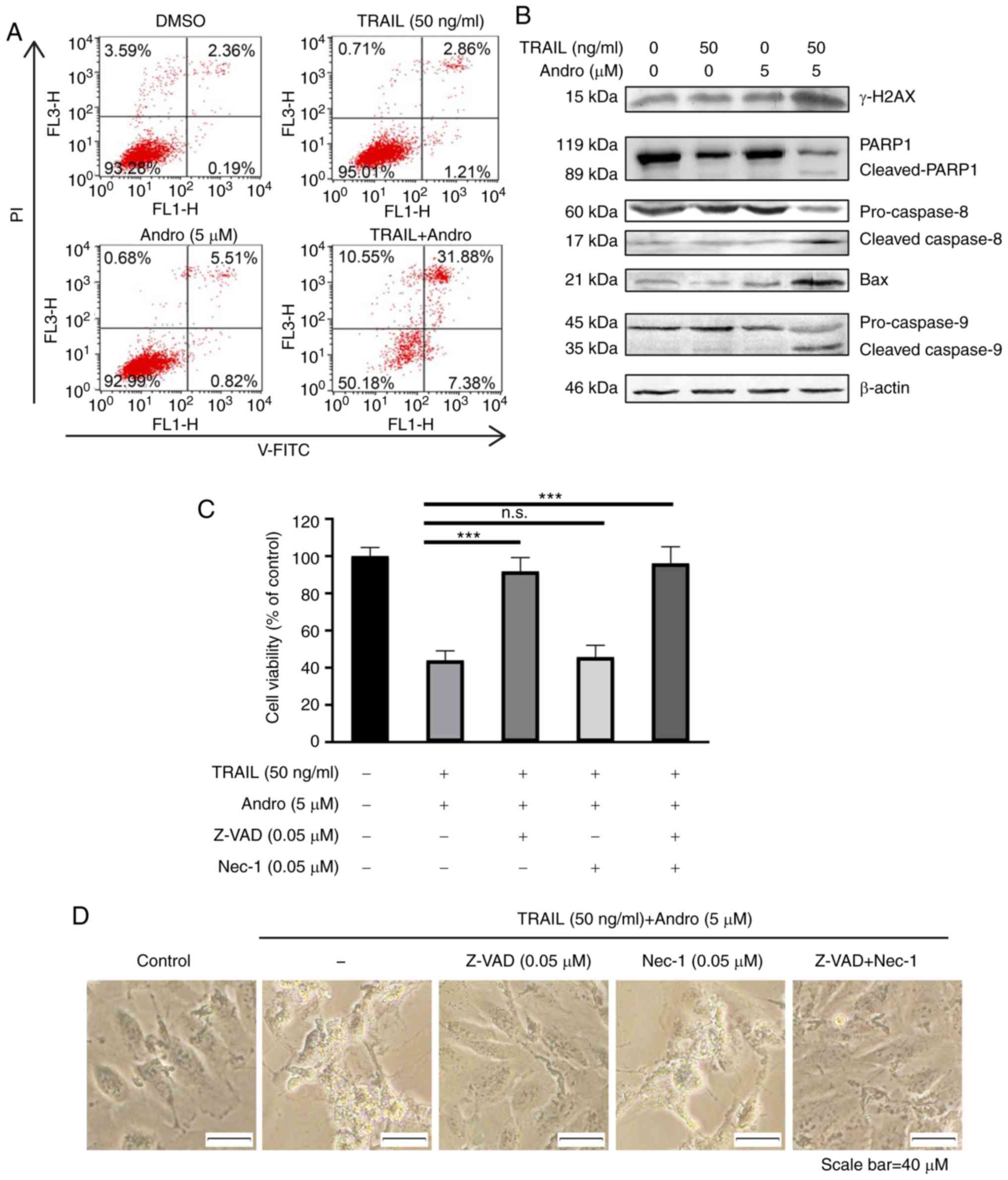 | Figure 4.Combined treatment of Andro and TRAIL
induces caspase-dependent apoptosis in 786-0 cells. (A) Cell
apoptosis was determined by Annexin V-FITC after DMSO, TRAIL (50
ng/ml), and/or Andro (5 µM) treatment for 24 h. (B) Indicated
protein levels in 786-0 cells treated with TRAIL (50 ng/ml) and/or
Andro (5 µM) for 24 h as detected by immunoblotting. (C) Cells were
treated with DMSO, TRAIL (50 ng/ml), and Andro (5 µM), pan-caspase
inhibitor Z-VAD (0.05 µM), and cell-necrosis inhibitor
necrostatin-1 (Nec-1) (0.05 µM). Then cell viability was determined
by MTS assay (one-way ANOVA, Tukey). (D) Images (magnification,
×400) show the apoptotic cells following treatment under different
conditions. Data are shown as mean ± SD; n.s. (not significant),
P>0.05, ***P<0.001, n=3). Andro, andrographolide; TRAIL,
tumor necrosis factor-related apoptosis-inducing ligand; PI,
propidium iodide; PARP1, poly(ADP ribose) polymerase 1; Bax, Bcl-2
associated X, apoptosis regulator, DR, death receptor. |
Additionally, we found that RCC apoptosis induced by
combined treatment was initiated by caspase-specific activation
that did not involve cell necrosis. The antitumor effects of the
Andro and TRAIL combined treatment was almost blocked by a
pan-caspase inhibitor Z-VAD (0.05 µM, 91.72±4.21%), but not by
cell-necrosis inhibitor necrostatin-1 (0.05 µM, 45.67±3.29%). This
further confirms that Andro enhanced TRAIL-mediated
caspase-dependent apoptotic cell death in RCC cells (Fig. 4C). Cell morphology was also
consistent with the MTS assay results (Fig. 4D).
Andro sensitizes TRAIL-induced
apoptosis via upregulation of DR4
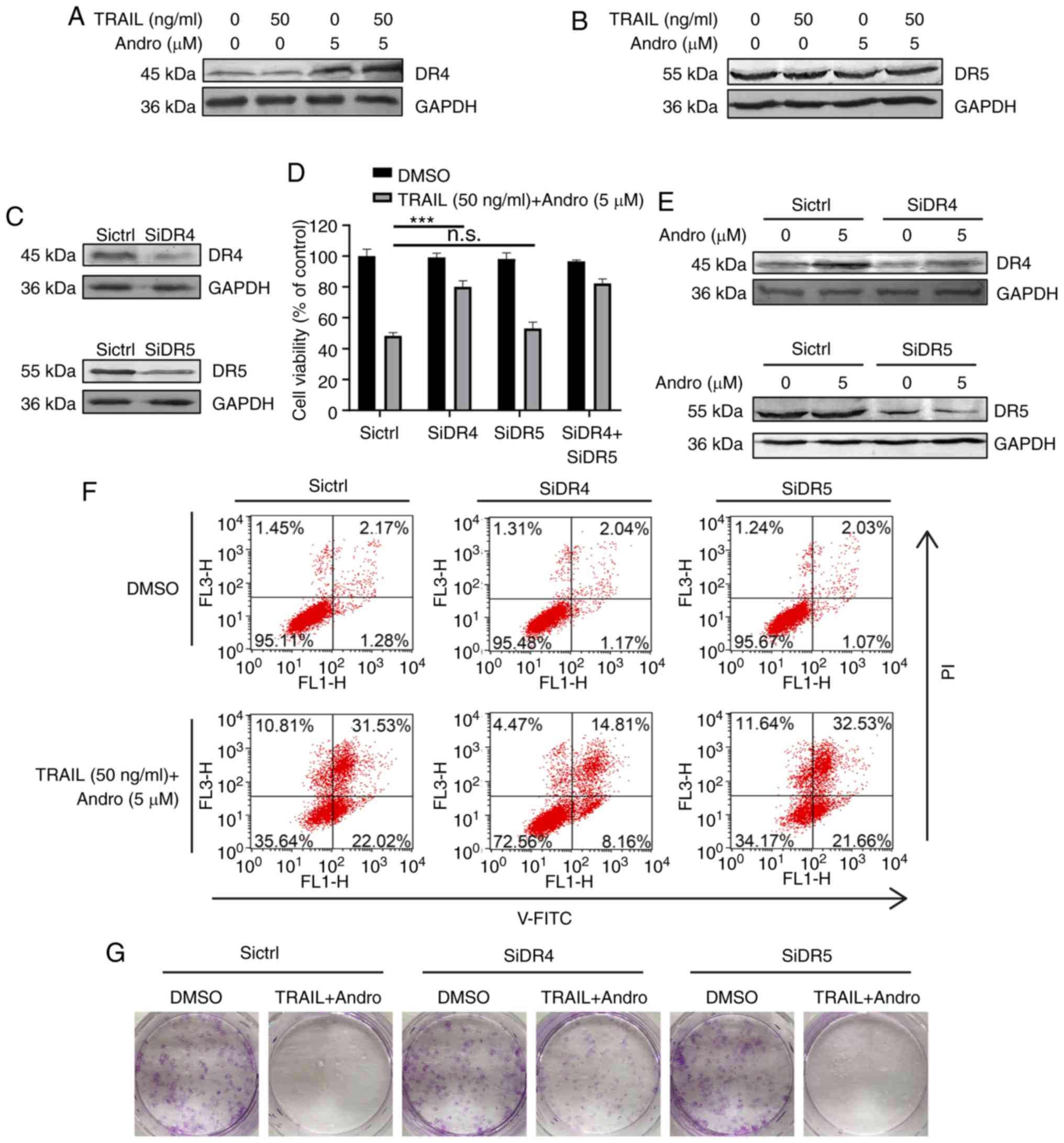
Our immunoblot assays demonstrated that Andro
treatment selectively upregulated protein levels of DR4 (Fig. 5A), but not of DR5 (Fig. 5B). To determine whether one or both
receptors are responsible for the pro-apoptotic effect of TRAIL in
RCC cells, we used small RNA interference to block endogenous
DR4/DR5 translation according to their knockdown efficiency
determined by immunoblotting (Fig.
5E). The results demonstrated that cell viability was slightly
restored in the DR5-knockdown cells (53.10±2.71%) and restored to a
higher degree in the DR4-knockdown cells (80.07±3.71%) following
combination treatment with TRAIL and Andro (Fig. 5D). Cell apoptosis assays and
clonogenic assays further supported the important roles of DR4 in
the effects of the combined treatment of TRAIL and Andro (Fig. 5F and G).
Discussion
Renal cell carcinoma (RCC) is the third most
prevalent urinary tumor and claims more than 100,000 lives each
year worldwide (18). At present,
the primary treatment for RCC, either localized RCC or locally
advanced RCC, is surgery. However, for patients with metastatic
RCC, surgery does not significantly improve the prognosis or
quality of life (4). Moreover, RCC
is neither sensitive to radiotherapy nor chemotherapy and has a low
response to cytokine therapy (19).
Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) is
a promising agent for anticancer therapy due to its ability to
selectively trigger cancer cell death (20). Moreover, in contrast to other
members of the TNF superfamily, TRAIL administration in vivo
is harmless (21–23). However, the resistance of cancer
cells to TRAIL-mediated apoptosis is a major limitation to its
clinical application (24,25). In the present study, we confirmed
that human RCC cell lines were widely unresponsive to
TRAIL-mediated cytotoxicity, which was primarily due to low
expression levels of its receptor in these RCC cells. Hence, it is
necessary to assess and find novel TRAIL sensitive agents with high
efficacy and low toxicity.
In this preclinical study, we showed that
andrographolide (Andro), a major constituent of Andrographis
paniculate, an annual herbaceous plant in the family
Acanthaceae, a natural compound, restored the sensitivity of
RCC cell lines to TRAIL-mediated apoptosis. The findings provide
critical insight into a novel therapeutic strategy for RCC
patients. Andro administration enhanced TRAIL-mediated inhibition
of cell viability, proliferation, migration, and colony formation
of RCC cell lines. Moreover, our data revealed that combination
therapy also potently inhibited the proliferation of diverse RCC
cell lines, suggesting that Andro enhanced the anticancer activity
of TRAIL independent of RCC cell type.
Most cancer cells resist apoptosis (26,27).
The combined treatment with TRAIL and Andro potently triggered cell
cycle arrest, senescence, and apoptosis in RCCs, which largely
relied on its ability to specifically increase death receptor (DR)4
expression. Elevation of membrane associated DR4 expression by
Andro treatment amplified TRAIL-mediated initiation of apoptosis,
cleavage of PARP1, and caspase activation. A pan-caspase inhibitor
(Z-VAD-FMK), but not the necrosis inhibitor, Necrostatin-1, almost
fully restored cell viability in RCC cells treated with both TRAIL
and Andro, further supporting an Andro-specific increase in the
cytotoxicity of TRAIL in RCC cells through its induction of
caspase-dependent apoptosis. All of these results revealed that
Andro treatment acts synergistically with TRAIL treatment on RCC
cells.
In-depth understanding of the causes of TRAIL
resistance in renal cancer may help to better develop drugs that
are more effective. TRAIL binding to its receptors (DR4 and DR5) to
initiate DISC assembly subsequently activates the caspase cascades
and triggers apoptosis (28).
Accumulating evidence suggests that an increase in TRAIL receptors
is an effective strategy for enhancing the sensitivity of cancer
cells to TRAIL-mediated effects (29–31).
The tumor suppressor p53 is a key apoptosis regulator limiting
cancer development via its proapoptotic function (11,32).
Transcriptional activation of death receptors by p53 is essential
for its tumor-suppressing functions. Recently, our findings and
that of other authors have demonstrated that Andro activates p53
signaling which results in DR4 or DR5 upregulation in other cancer
cell types (17,33–35).
It has also been known that p53 signaling stimulates the DR5
gene through an intronic sequence-specific DNA-binding site
(36). In addition, previous
findings that DNA damage-induced p53 activation leads to DR4
upregulation further supports the essential role of p53 signaling
in the regulation of death receptors expression (37).
Interestingly, unlike our previous report that
elevation of DR5 but not DR4 expression is one of the determinant
factors for Andro-mediated sensitization of bladder cancer cells to
TRAIL, we found that the expression levels of DR4 but not DR5 are
critical for counteracting TRAIL-resistance in RCCs by Andro
(17). These data hint that TRAIL
signaling in diverse cancer types is selectively initiated by a
certain TRAIL receptor, DR4 or DR5. Future studies need to clarify
the detailed strategies of cancer cells to evade suppression by
TRAIL. Furthermore, clinical database analysis revealed that a
modest increase in mRNA expression levels of DR4 was noted in RCC
patients which was in contrast to the dramatic elevation of DR5
mRNA levels. These results imply that the low expression of DR4 is
one determinant strategy for the evasion of TRAIL proapoptosis
signaling by renal cancer cells.
The potential application of andrographolide in
clinical cancer treatment has several advantages (38,39).
Andro is widely distributed in various plants of the genus
Andrographis and has been used for centuries in Asia
(40). Andro possesses therapeutic
effects against various conditions, such as carcinoma, arthritis,
ischemia, pyrogenesis, and oxidative stress (41–44).
Due to its short half-life, Andro can be excreted from the body at
a high rate with almost no toxic effects to normal cells (45). Considering these features, our
results indicated that Andro counteracts TRAIL resistance in RCC
cells providing proof-of-concept evidence for the clinical
investigation of combined treatment of TRAIL and the traditional
anti-inflammatory agent, andrographolide, in renal carcinoma
therapy.
Supplementary Material
Supporting Data
Acknowledgements
We thank Guanchen Liu and Jiaxin Yang (Key
Laboratory of Organ Regeneration and Transplantation of the
Ministry of Education, Institute of Translational Medicine,
Institute of Virology and AIDS Research, The First Hospital of
Jilin University, Changchun, Jilin 130061, China) for their
technical assistance.
Funding
This research was supported in part by funding from
the National Natural Science Foundation of China (81772183), the
Department of Science and Technology of Jilin Province
(20190304033YY and 20180101127JC), the Program for JLU Science and
Technology Innovative Research Team (2017TD-08) and Fundamental
Research Funds for the Central Universities.
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
WW, CW, and YD conceived and designed the
experiments. RB, YD, CT, LX, BX performed the experiments and
collected and analyzed the data. WW with the help of CW and BR
wrote the manuscript. All authors read and approved the manuscript
and agree to be accountable for all aspects of the research in
ensuring that the accuracy or integrity of any part of the work are
appropriately investigated and resolved.
Ethics approval and consent to
participate
This study was approved by the Ethics Committee of
the First Hospital of Jilin University (Changchun, Jilin,
China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Eble JN, Sauter G, Epstein JI and
Sesterhenn IA: World Health Organization Classification of Tumours.
Pathology and genetics of tumours of the urinary system and male
genital organs. IARC Press; Lyon: 2004
|
|
3
|
Van Poppel H, Da Pozzo L, Albrecht W,
Matveev V, Bono A, Borkowski A, Colombel M, Klotz L, Skinner E,
Keane T, et al: A prospective, randomised EORTC intergroup phase 3
study comparing the oncologic outcome of elective nephron-sparing
surgery and radical nephrectomy for low-stage renal cell carcinoma.
Eur Urol. 59:543–552. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shinder BM, Rhee K, Farrell D, Farber NJ,
Stein MN, Jang TL and Singer EA: Surgical management of advanced
and metastatic renal cell carcinoma: A multidisciplinary approach.
Front Oncol. 7:1072017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li P, Wong YN, Armstrong K, Haas N, Subedi
P, Davis-Cerone M and Doshi JA: Survival among patients with
advanced renal cell carcinoma in the pretargeted versus targeted
therapy eras. Cancer Med. 5:169–181. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Banumathy G and Cairns P: Signaling
pathways in renal cell carcinoma. Cancer Biol Ther. 10:658–664.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ashkenazi A and Salvesen G: Regulated cell
death: Signaling and mechanisms. Annu Rev Cell Dev Biol.
30:337–356. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
LeBlanc HN and Ashkenazi A: Apo2L/TRAIL
and its death and decoy receptors. Cell Death Differ. 10:66–75.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Luo X, Budihardjo I, Zou H, Slaughter C
and Wang X: Bid, a Bcl2 interacting protein, mediates cytochrome c
release from mitochondria in response to activation of cell surface
death receptors. Cell. 94:481–490. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Du C, Fang M, Li Y, Li L and Wang X: Smac,
a mitochondrial protein that promotes cytochrome c-dependent
caspase activation by eliminating IAP inhibition. Cell. 102:33–42.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jin Z and El-Deiry WS: Overview of cell
death signaling pathways. Cancer Biol Ther. 4:139–163. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hall MA and Cleveland JL: Clearing the
TRAIL for cancer therapy. Cancer Cell. 12:4–6. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang S and El-Deiry WS: TRAIL and
apoptosis induction by TNF-family death receptors. Oncogene.
22:8628–8633. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Huang Y and Sheikh MS: TRAIL death
receptors and cancer therapeutics. Toxicol Appl Pharmacol.
224:284–289. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ou YC, Li JR, Kuan YH, Raung SL, Wang CC,
Hung YY, Pan PH, Lu HC and Chen CJ: Luteolin sensitizes human 786-O
renal cell carcinoma cells to TRAIL-induced apoptosis. Life Sci.
100:110–117. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wei R, Zhu G, Jia N and Yang W:
Epigallocatechin-3-gallate sensitizes human 786-O renal cell
carcinoma cells to TRAIL-induced apoptosis. Cell Biochem Biophys.
72:157–164. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Deng Y, Bi R, Guo H, Yang J, Du Y, Wang C
and Wei W: Andrographolide enhances TRAIL-induced apoptosis via
p53-mediated death receptors up-regulation and suppression of the
NF-кB pathway in bladder cancer cells. Int J Biol Sci. 15:688–700.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zeng Z, Que T, Zhang J and Hu Y: A study
exploring critical pathways in clear cell renal cell carcinoma. Exp
Ther Med. 7:121–130. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Buti S, Bersanelli M, Sikokis A, Maines F,
Facchinetti F, Bria E, Ardizzoni A, Tortora G and Massari F:
Chemotherapy in metastatic renal cell carcinoma today? A systematic
review. Anticancer Drugs. 24:535–554. 2013.PubMed/NCBI
|
|
20
|
von Karstedt S, Montinaro A and Walczak H:
Exploring the TRAILs less travelled: TRAIL in cancer biology and
therapy. Nat Rev Cancer. 17:352–366. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ashkenazi A, Pai RC, Fong S, Leung S,
Lawrence DA, Marsters SA, Blackie C, Chang L, McMurtrey AE, Hebert
A, et al: Safety and antitumor activity of recombinant soluble Apo2
ligand. J Clin Invest. 104:155–162. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Walczak H, Miller RE, Ariail K, Gliniak B,
Griffith TS, Kubin M, Chin W, Jones J, Woodward A, Le T, et al:
Tumoricidal activity of tumor necrosis factor-related
apoptosis-inducing ligand in vivo. Nat Med. 5:157–163. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Roberts NJ, Zhou S, Diaz LA Jr and
Holdhoff M: Systemic use of tumor necrosis factor alpha as an
anticancer agent. Oncotarget. 2:739–751. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ahmed SM, Wu X, Jin X, Zhang X, Togo Y,
Suzuki T, Li Y, Kanematsu A, Nojima M, Yamamoto S, et al:
Synergistic induction of apoptosis by mapatumumab and
anthracyclines in human bladder cancer cells. Oncol Rep.
33:566–572. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yuan X, Gajan A, Chu Q, Xiong H, Wu K and
Wu GS: Developing TRAIL/TRAIL death receptor-based cancer
therapies. Cancer Metastasis Rev. 37:733–748. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Johnstone RW, Ruefli AA and Lowe SW:
Apoptosis: A link between cancer genetics and chemotherapy. Cell.
108:153–164. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen Z, Sangwan V, Banerjee S, Chugh R,
Dudeja V, Vickers SM and Saluja AK: Triptolide sensitizes
pancreatic cancer cells to TRAIL-induced activation of the death
receptor pathway. Cancer Lett. 348:156–166. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hotta M, Sakatani T, Ishino K, Wada R,
Kudo M, Yokoyama Y, Yamada T, Yoshida H and Naito Z: Farnesoid X
receptor induces cell death and sensitizes to TRAIL-induced
inhibition of growth in colorectal cancer cells through the
up-regulation of death receptor 5. Biochem Biophys Res Commun.
519:824–831. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Shishodia G, Koul S, Dong Q and Koul HK:
Tetrandrine (TET) induces death receptors Apo Trail R1 (DR4) and
Apo Trail R2 (DR5) and sensitizes prostate cancer cells to
TRAIL-induced apoptosis. Mol Cancer Ther. 17:1217–1228. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yang X, Li Z, Wu Q, Chen S, Yi C and Gong
C: TRAIL and curcumin codelivery nanoparticles enhance
TRAIL-induced apoptosis through upregulation of death receptors.
Drug Deliv. 24:1526–1536. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Aubrey BJ, Kelly GL, Janic A, Herold MJ
and Strasser A: How does p53 induce apoptosis and how does this
relate to p53-mediated tumour suppression? Cell Death Differ.
25:104–113. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhou J, Lu GD, Ong CS, Ong CN and Shen HM:
Andrographolide sensitizes cancer cells to TRAIL-induced apoptosis
via p53-mediated death receptor 4 up-regulation. Mol Cancer Ther.
7:2170–2180. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wei RJ, Zhang XS and He DL:
Andrographolide sensitizes prostate cancer cells to TRAIL-induced
apoptosis. Asian J Androl. 20:200–204. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chen M, Wang X, Zha D, Cai F, Zhang W, He
Y, Huang Q, Zhuang H and Hua ZC: Apigenin potentiates TRAIL therapy
of non-small cell lung cancer via upregulating DR4/DR5 expression
in a p53-dependent manner. Sci Rep. 6:354682016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Takimoto R and El-Deiry WS: Wild-type p53
transactivates the KILLER/DR5 gene through an intronic
sequence-specific DNA-binding site. Oncogene. 19:1735–1743. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liu X, Yue P, Khuri FR and Sun SY: p53
upregulates death receptor 4 expression through an intronic p53
binding site. Cancer Res. 64:5078–5083. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Rajagopal S, Kumar RA, Deevi DS,
Satyanarayana C and Rajagopalan R: Andrographolide, a potential
cancer therapeutic agent isolated from Andrographis
paniculata. J Exp Ther Oncol. 3:147–158. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kumar RA, Sridevi K, Kumar NV, Nanduri S
and Rajagopal S: Anticancer and immunostimulatory compounds from
Andrographis paniculata. J Ethnopharmacol. 92:291–295. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Sabu KK, Padmesh P and Seeni S:
Intraspecific variation in active principle content and isozymes of
Andrographis paniculata Nees (Kalmegh): A traditional
hepatoprotective medicinal herb of India. J Med Aromat Plant Sci.
23:637–647. 2001.
|
|
41
|
Peng T, Hu M, Wu TT, Zhang C, Chen Z,
Huang S and Zhou XH: Andrographolide suppresses proliferation of
nasopharyngeal carcinoma cells via attenuating NF-κB pathway.
Biomed Res Int. 2015:7350562015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Gupta S, Mishra KP, Singh SB and Ganju L:
Inhibitory effects of andrographolide on activated macrophages and
adjuvant-induced arthritis. Inflammopharmacology. 26:447–456. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chan SJ, Wong WS, Wong PT and Bian JS:
Neuroprotective effects of andrographolide in a rat model of
permanent cerebral ischaemia. Br J Pharmacol. 161:668–679. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Li B, Jiang T, Liu H, Miao Z, Fang D,
Zheng L and Zhao J: Andrographolide protects chondrocytes from
oxidative stress injury by activation of the Keap1-Nrf2-Are
signaling pathway. J Cell Physiol. 234:561–571. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Jaruchotikamol A, Jarukamjorn K,
Sirisangtrakul W, Sakuma T, Kawasaki Y and Nemoto N: Strong
synergistic induction of CYP1A1 expression by andrographolide plus
typical CYP1A inducers in mouse hepatocytes. Toxicol Appl
Pharmacol. 224:156–162. 2007. View Article : Google Scholar : PubMed/NCBI
|