Introduction
Glutathione S-transferases (GSTs) are phase-II
detoxification enzymes that catalyze the conjugation of glutathione
to the electrophilic centers of multiple types of endogenous and
exogenous electrophiles, including genotoxins, carcinogens and
anticancer agents (1,2). Based on their biochemical properties,
the cytosolic GSTs are divided into the following subclasses: alpha
(α), mu (μ), omega (ω), pi (π), sigma (σ), theta (θ) and zeta (ζ).
GST pi-1 (GSTP-1) is encoded by a single gene mapped to chromosome
11q13 (3,4) and is frequently overexpressed in many
types of tumors, including colorectal cancers (5,6). In
addition to its enzymatic functions, GSTP-1 is also thought to
possess non-enzymatic functions. For example, GSTP-1 acts as a
regulator of cell signaling in apoptosis through its interactions
with other proteins, such as c-Jun NH2-terminal kinase (7,8).
These enzymatic and non-enzymatic functions of GSTP-1 are
correlated with drug resistance, failure of therapy and poor
survival in patients with tumors expressing high levels of GSTP-1
(8,9).
Currently, the role of GSTP-1 in carcinogenesis is
controversial. Loss of GSTP-1 function is associated with multiple
types of cancers, corroborating the notion that GSTP-1 may be a
tumor suppressor. For example, mice lacking GSTP-1 exhibit
increased skin tumorigenesis (10)
and it has been proposed that hypermethylated GSTP-1 represents a
potential epigenetic biomarker for prostate cancer (11). By contrast, overexpression of
GSTP-1 is associated with the development of other types of cancer
as well as with the acquisition of drug resistance in some
neoplasms (12,13), suggesting that GSTP-1 can also act
as an oncogene. In particular, GSTP-1 is overexpressed in all
stages of colorectal cancer, from aberrant crypt foci to advanced
carcinomas (5,6). Colorectal cancer is the third most
commonly diagnosed cancer in the world, and an increased incidence
of colon cancer has been reported in recent years in Asian
countries including Korea (14).
Colorectal cancer develops through multiple steps that involve
sequential acquisition of genetic alterations in key tumor
suppressors and oncogenes (15).
To attempt to resolve the controversy regarding the role of GSTP-1,
we investigated the epigenetic alteration of GSTP-1 in human
colorectal cancers and explored the underlying mechanisms in
detail.
The two major epigenetic controls involved in
transcriptional silencing are covalent modification of histones and
DNA CpG-island methylation (16).
Post-translational modifications of histones include acetylation,
methylation, phosphorylation and ubiquitination. Each of these
modifications recruits regulatory factors that influence chromatin
structure and transcription. Reversible histone acetylation
regulates nucleosome-nucleosome and nucleosome-DNA interactions,
which in turn influence the access of transcriptional factors to
DNA (17). The balance of
acetylation is regulated by two types of enzymes with opposing
activities, acetyltransferases and deacetylases (18). DNA methylation is responsible for
transcriptional regulation and has been implicated in
transcriptional silencing (19).
Hypermethylation of a promoter region leads to the formation of a
repressor complex composed of methyl-CpG-binding proteins (MBDs),
DNA methyltransferases (DNMTs), and histone deacetylases (HDACs)
(20,21). This DNA-protein complex induces
histone deacetylation and catalyzes the methylation of lysine 9 and
27 in histone H3, compacting the structural arrangement of
nucleosomes and thereby generating heterochromatin (22,23).
By contrast, the absence of methylated CpG sites,
DNMTs, MBDs and HDACs from gene promoters is a hallmark of
transcriptionally active euchromatin. Abnormal gene-specific
demethylation can lead to overexpression of genes that contribute
to disease (24). Additionally,
modifications of lysine 4 in histone H3, such as trimethylation
(H3K4me3), are also associated with transcriptionally permissive
chromatin (25,26). Analysis of the human GSTP-1
proximal promoter region revealed the presence of putative binding
sites for SP-1, AP-1 and NF-κB, indicating that these transcription
factors play important roles in the regulation of GSTP-1 expression
(27–29).
In this study, we tested the hypothesis that
overexpression of GSTP-1 in human colorectal cancers is partially
due to the lower degree of DNA methylation and histone modification
in tumor cells relative to normal colon cells and tissues, and that
transcription factors are involved in this regulation.
Materials and methods
Materials
Primary mouse monoclonal anti-GSTP-1, anti-DNMT1,
anti-MBD2 antibodies and primary rabbit polyclonal anti-histone H3
(tri-methyl K4) and anti-histone H3 (tri-methyl K9) antibodies were
purchased from Abcam, Inc. (Cambridge, MA, USA). Primary rabbit
anti-acetyl-histone H3 (K9) and anti-c-Jun antibodies were
purchased from Cell Signaling Technology, Inc. (Beverly, MA, USA).
Primary mouse monoclonal anti-HDAC1 antibody was purchased from
Santa Cruz Biotechnology (Santa Cruz, CA, USA).
[3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium] bromide
(MTT) and primary rabbit anti-β-actin antibody were purchased from
Sigma-Aldrich Chemical Co. (St. Louis, MO, USA). All other
chemicals and reagents were of analytical grade.
Patient tissues
The biospecimens and data used in this study were
provided by the Biobank of Jeju National University Hospital, a
member of Korea Biobank Network (Jeju, Korea) (A03-2). This study
was approved by the institutional review board for ethics of Jeju
National University Hospital (IRB 2011-38), and informed written
consent was obtained from all the patients.
Cell culture
The human colon cancer cell lines Caco-2, HCT-116,
HT-29, SNU-407 and SNU-1033 were obtained from the Korean Cell Line
Bank (Seoul, Korea), and the normal human colon cell line FHC was
purchased from the American Type Culture Collection (Rockville, MD,
USA). Cells were maintained at 37°C in an incubator with a
humidified atmosphere containing 5% CO. HCT-116, HT-29, SNU-407 and
SNU-1033 cells were cultured in RPMI-1640 medium containing 10%
heat-inactivated fetal bovine serum (FBS), streptomycin (100 μg/ml)
and penicillin (100 U/ml). Caco-2 cells were cultured in MEM medium
containing 10% heat-inactivated FBS, streptomycin (100 μg/ml) and
penicillin (100 U/ml). FHC cells were cultured in a 1:1 mixture of
Ham’s F12 and DMEM containing HEPES (25 mM), cholera toxin (10
ng/ml; Calbiochem-Novabiochem Corp., La Jolla, CA, USA), insulin (5
μg/ml), transferrin (5 μg/ml), hydrocortisone (100 ng/ml) and 10%
FBS.
Proteomic analysis
Two samples were pooled from 10 colon cancer and
normal tissue, respectively. The extracted proteins from tissues
were digested using modified gel-assisted digestion protocol
(30). The gel pieces containing
proteins were reduced and alkylated. Proteolytic digestion was
conducted with 100 ng/μl trypsin and incubated at 37°C overnight.
The peptides were extracted from the gel. The nanoscale LC
separation of tryptic peptides was performed by using a nanoAcquity
system (Waters Corp., Milford, MA, USA), equipped with a Symmetry
C18 5 μm, 5 mm × 300 μm precolumn and an BEH C18 1.7 μm, 25 cm × 75
μm analytical reversed phase column (Waters Corp.). The samples
were initially transferred, with an aqueous 0.1% formic acid
solution, to the precolumn at a flow rate of 10 μl/min for 3 min.
The mobile phase A was composed of water with 0.1% formic acid, and
the mobile phase B was composed of 0.1% formic acid in
acetonitrile. The peptides were separated with a gradient of 3–40%
mobile phase B over 180 min at a flow rate of 300 nl/min, followed
by a 25 min rinse with 90% of mobile phase B. The analysis of
tryptic peptides was performed using a Q-Tof Premier mass
spectrometer (Waters Corp.). Tryptic peptide was detected within
the retention time range of approximately 0.12 min and at mass
precision of approximately 3.1 ppm from three independent runs.
Accurate mass LC-MS data were collected in an alternating, low
energy and elevated energy mode of acquisition (31). Continuum LC-MS data were processed
for generating peaklist and searched, using ProteinLynx
GlobalServer v2.3 IdentityE (Waters Corp.). All MS/MS data were
searched against the IPI human protein database (version 3.36;
containing 69,012 sequences and 29,002,682 residues) using the
Proteinlynx Global Server v2.3 IdentityE software. All database
searches allowed for fixed modification in regard to the cysteine
residue and variable modification in regard to the methionine. The
relative quantitation was performed using the precursor intensity
measurements available in the clustered exact mass and retention
time. The redundant quantitative measurements provided by the
multiple tryptic peptides from each protein were used to determine
an average relative fold-change. A 95% confidence interval was
determined for each average fold-change from the standard deviation
of the observed quantitative measurements and the total number of
observed tryptic peptides.
Western blot analysis
Cells were lysed on ice for 30 min in 100 μl lysis
buffer [120 mM NaCl, 40 mM Tris (pH 8.0), 0.1% NP-40] and
centrifuged at 13,000 × g for 15 min. Supernatants were collected
and the protein concentrations were determined. Aliquots containing
40 μg of protein were boiled for 5 min and electrophoresed on 10%
SDS-polyacrylamide gels. Proteins were transferred onto
nitrocellulose membranes, which were subsequently incubated with
anti-GSTP-1 antibody overnight at 4°C. The membranes were further
incubated with secondary horseradish peroxidase-conjugated
anti-immunoglobulin-G (Pierce, Rockford, IL, USA). Protein bands
were detected using an enhanced chemiluminescence western blotting
detection kit (Amersham, Little Chalfont, Buckinghamshire, UK).
Immunohistochemistry for detection of
GSTP-1
For detection of GSTP1 in tissue specimens, tissue
specimens were fixed in 10% buffered formalin and embedded in
paraffin. The same paraffin-embedded tissues as those used for the
original hematoxylin and eosin stained sections were used for
immunohistochemistry. Tissue blocks were cut into 3 μm thick slices
and mounted on SuperFrost® Plus coated slides. Sections
were then deparaffinized in xylene and rehydrated through a graded
ethanol series. A standard immunohistochemical technique was
performed using a Ventana BenchMark XT immunostainer with the
anti-GSTP-1 antibody at a dilution of 1:100. Antigen retrieval on
the immunostainer was carried out for 30 min. The anti-GSTP-1
antibody was incubated at 37°C for 60 min, and
3,3′-diaminobenzidine was used as a chromogen; slides were
counterstained with hematoxylin prior to mounting. All staining
procedures were performed according to the manufacturer’s
recommendations. Intramucosal mononuclear cells in the tissue
samples, which stain strongly, served as internal positive
controls. Negative controls for non-specific binding were obtained
by omitting the primary antibody. The immunohistochemical slides
were evaluated and interpreted by a pathologist. For detection of
GSTP-1 in cells, cells plated on coverslips were fixed with 4%
paraformaldehyde for 30 min and permeabilized with PBS containing
0.1% Triton X-100 for 2.5 min. Cells were then treated with
blocking solution (PBS containing 3% bovine serum albumin) for 1 h,
and then incubated for 2 h with anti-GSTP-1 antibody diluted in
blocking solution. Immunoreacted primary anti-GSTP-1 antibody was
detected by incubating samples for 1 h with a FITC-conjugated
secondary antibody at a dilution of 1:200 (Jackson Immuno Research
Laboratories, West Grove, PA, USA). After washing with PBS, stained
cells were mounted onto microscope slides in mounting medium.
Microscopic images were collected on a confocal microscope using
the Laser Scanning Microscope 5 PASCAL software (Carl Zeiss, Jena,
Germany).
Assessment of GST activity
GST enzyme activity in colon tissues or cells was
measured using the GST activity assay kit (Abcam, Inc.). The GST
substrate 1-chloro-2,4-dinitrobenzene (CDNB) was used to assess
GST-related enzyme activity. Briefly, colon tissues or cells were
homogenized in 0.5 ml GST assay buffer, and homogenates were
centrifuged at 10,000 × g for 15 min at 4°C. The supernatants were
collected and the protein concentrations were determined. Samples
were prepared with GST assay buffer in a total volume of 50 μl,
including a negative control (50 μl GST assay buffer alone) and a
positive control (10 μl of GST positive control diluted 1:50 and 40
μl GST assay buffer). To each well containing sample or control, 5
μl of glutathione was added, followed by 50 μl of substrate mixture
(45 μl of GST assay buffer and 5 μl of CDNB). After addition of the
substrate mixture, plates were shaken carefully to start the
reaction, and the absorbance at 340 nm was read once every minute
to obtain at least 5 time points. The change in absorbance (ΔA) per
minute was determined by plotting the absorbance values as a
function of time to obtain the slope of the linear portion of the
curve. The reaction rate was then determined using the CDNB
extinction coefficient at 340 nm, 0.0053 μM−1. GST
activity is expressed as nmol/min/ml.
RNA isolation and reverse
transcription-polymerase chain reaction (RT-PCR)
Total RNA was isolated from cells using the
easy-BLUE™ total RNA extraction kit (iNtRON Biotechnology, Inc.,
Korea), and cDNA was amplified using 1 μl of the reverse
transcription reaction buffer, primers, dNTPs and 0.5 U of Taq DNA
polymerase in a final volume of 25 μl. The PCR conditions were:
initial denaturation for 5 min at 94°C; 35 cycles of 94°C for 30
sec, 60°C for 30 sec, and 72°C for 30 sec; and a final elongation
step at 72°C for 7 min. The following primers were used to amplify
GSTP1 cDNA: forward, 5′-ATGCCGCCCTACACCGTG-3′; reverse,
5′-CCAGGTGACGCAGGATGG-3′. The amplified products were resolved by
electrophoresis on a 1% agarose gel, stained with RedSafe™ nucleic
acid staining solution (iNtRON Biotechnology, Inc.) and
photographed under UV light using the Image Quant™ TL analysis
software (Amersham Bioscience, Sweden).
Methylation-specific (MS)-PCR
DNA bisulfate modification was carried out using the
Methylamp™ DNA modification kit (Epigentek, Pittsburgh, PA, USA),
according to the manufacturer’s instructions. To analyze GSTP-1 DNA
methylation, MS-PCR was performed using an EpiTect MSP kit (Qiagen,
Valencia, CA, USA). Primers to perform MS-PCR on the GSTP-1
promoter were designed using the Methyl Primer Express®
software v1.0 (Applied Biosystems). The methylated and unmethylated
GSTP1 primer sets used for PCR were as follows: unmethylated GSTP-1
forward, 5′-GGAGTTTTGTTGTTGTAGTTTTT-3′, and reverse,
5′-CCACTAACAACACTAAAAACATC-3′; methylated GSTP-1 forward,
5′-GTTTCGTCGTCGTAGTTTTC-3′, and reverse,
5′-CCGCTAACGACACTAAAAAC-3′. The amplified products were resolved by
electrophoresis on a 3% agarose gel, stained with RedSafe nucleic
acid staining solution and photographed under UV light using Image
Quant™ TL analysis software.
Chromatin immunoprecipitation (ChIP)
assay
Cells were processed using the Simple ChIP™
enzymatic chromatin IP kit (Cell Signaling Technology, Inc.),
according to the manufacturer’s instructions. Briefly, the
procedure was initiated by cross-linking the proteins to DNA by
adding 1% formaldehyde to the culture dishes and rocking on a
platform for 10 min at room temperature. The cross-linking was
stopped by the addition of glycine solution. Cells were washed
twice with ice-cold PBS, pelleted by centrifugation and resuspended
in 1 ml cell lysis buffer containing protease inhibitor. The
soluble chromatin was sheared by sonication followed by
centrifugation at 15,000 × g. Diluted supernatants were pre-cleared
and blocked with protein A/G agarose, and the sonicated
chromatin-DNA complex was precipitated overnight with the
antibodies of interest. Bound DNA was eluted by incubating the
beads in elution buffer, purified and amplified using primers
flanking the SP-1 and AP-1 binding sites within the human GSTP1
proximal promoter. The oligonucleotide containing the transcription
factor binding site in the GSTP-1 promoter was obtained from
Bioneer (Seoul, Korea). The ChIP procedure was analyzed by PCR
using human GSTP-1 promoter-specific primers as follows (32): forward, 5′-GCAGCGGTCTTAGGGAATTT-3′;
reverse, 5′-CTTTCCCTCTTTCCCAGGTC-3′. The cycle parameters were as
follows: 95°C for 5 min; 40 cycles of 95°C for 30 sec, 60°C for 30
sec and 72°C for 30 sec; and a final extension at 72°C for 7 min.
The amplified products were resolved by electrophoresis on a 3%
agarose gel, stained with RedSafe nucleic acid staining solution
and photographed under UV light using Image Quant™ TL analysis
software.
Transient transfection of small
interfering RNA (siRNA)
SNU-407 cells were seeded at a density of
1.5×105 cells/well in 24-well plates and allowed to
reach approximately 50% confluency on the day of transfection. The
siRNA constructs were mismatched siRNA Control and siRNA GSTP-1
(Santa Cruz Biotechnology). Cells were transfected with 10–50 nM
siRNA using Lipofectamine™ 2000 (Invitrogen) according to the
manufacturer’s instructions. At 24 h after transfection, cells were
treated with 5-fluorouracil (5-FU) for 48 h, and then examined by
the MTT assay.
Cell viability
The effect of GSTP-1 on the viability of human colon
cancer cells was determined using the MTT assay, which is based on
the reduction of a tetrazolium salt by mitochondrial dehydrogenase
in viable cells (33). Cells were
treated with 5-FU at a concentration of 800 μM. MTT (50 μl of a 2
mg/ml stock solution) was added to each well to obtain a total
reaction volume of 200 μl. After incubation for 4 h, the
supernatants were aspirated. Formazan crystals present in each well
were dissolved in 150 μl of dimethyl sulfoxide, and the absorbance
at 540 nm was measured on a scanning multi-well
spectrophotometer.
Statistical analysis
The results were subjected to analysis of variance
using Tukey’s test to analyze differences. P<0.05 was considered
statistically significant.
Results
GSTP-1 protein expression and enzyme
activity in normal and colon carcinoma tissues
To compare the fold-changes of expressed proteins
from normal and colon carcinoma tissues, shotgun proteomic approach
based on label-free quantification was applied. A total of 403
non-redundant proteins were identified from the analysis of normal
and colon carcinoma tissues. Among them, 146 proteins were
identified in common. The numbers of unique proteins were 135 and
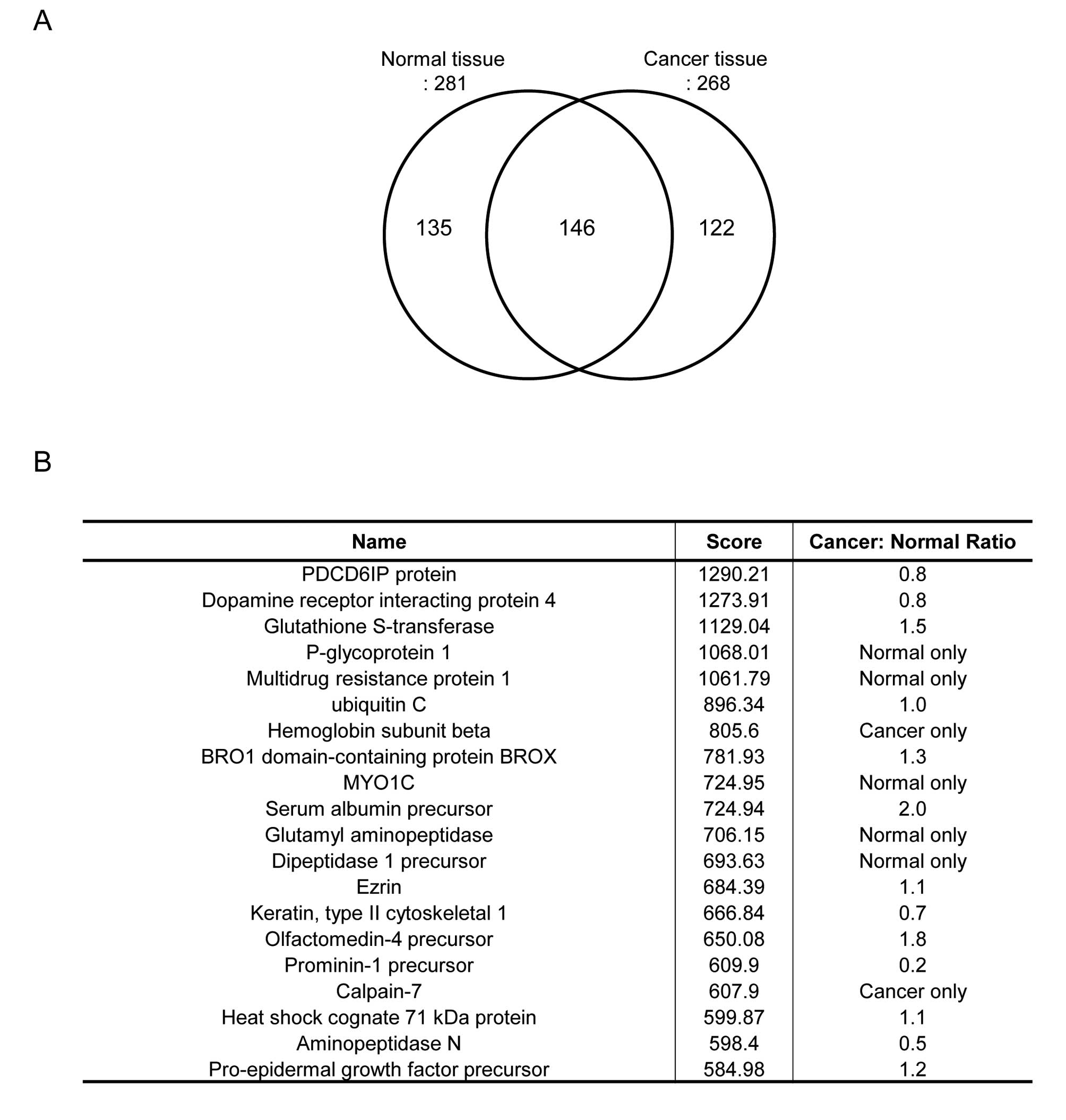
122 proteins in normal and cancer tissue, respectively (Fig. 1A). To show disease-specific
proteins, we listed up some proteins based on score of protein
recovery quantification which is proportional to accuracy of
quantification (Fig. 1B). Among
them, we focused on glutathione S-transferase which was
overexpressed in the cancer group. To confirm this proteomic
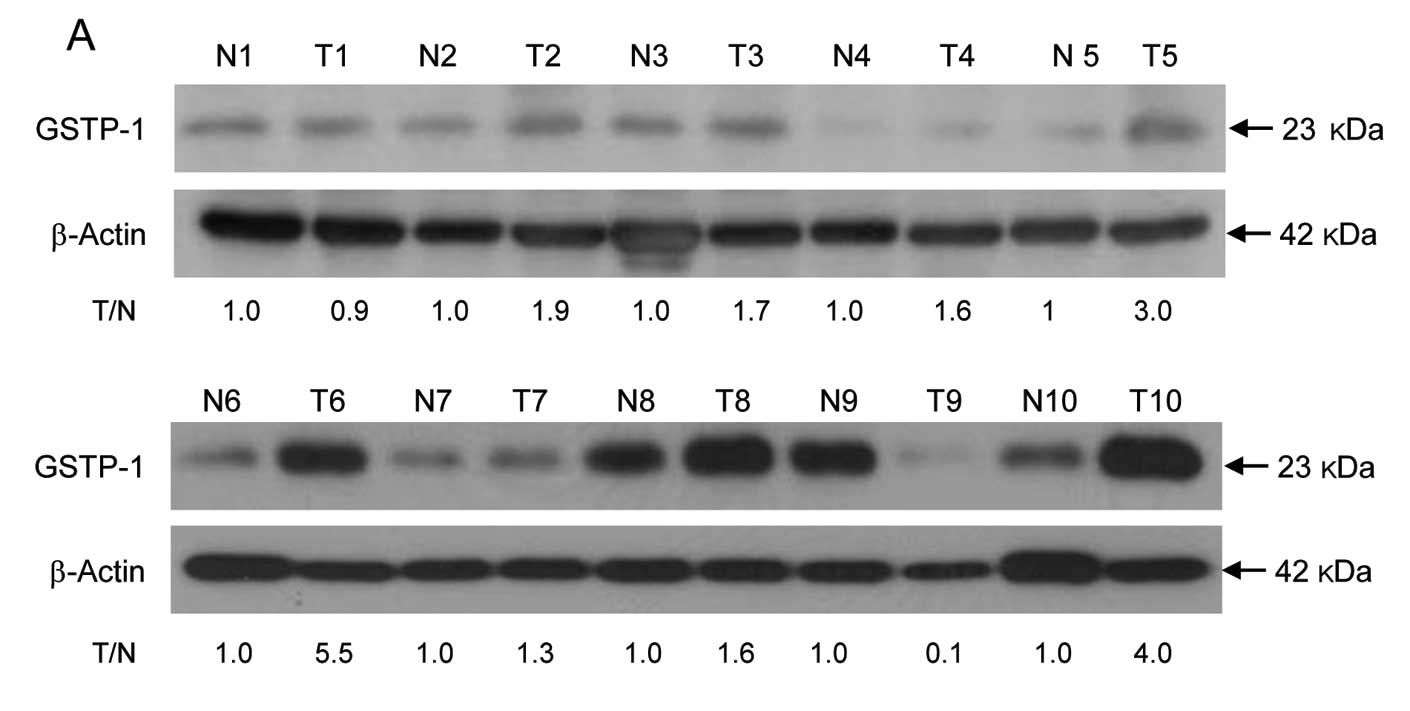
result, GSTP-1 protein level and enzyme activity were assessed in
colon cancer tissues from 10 patients by western blotting and an
enzyme activity assay, respectively. The expression level of GSTP-1
protein that showed >1.5-fold higher than in corresponding
normal tissues was detected in 70% (7/10) of tumor tissues from
colon cancer patients (Fig. 2A).
Immunohistochemical analysis revealed faint expression of GSTP-1
protein in normal mucosa (Fig.
2Ba). The carcinoma cells exhibited increased GSTP-1 expression
in the cytoplasm, which was distributed in a diffuse pattern
(Fig. 2Bb–d), and some cells were
strongly positive (Fig. 2Bd).
Neuroendocrine carcinoma cells in specimens exhibited no GSTP-1
expression (Fig. 2Be). GST enzyme
activity was higher in tumor tissues than in normal tissues from
colon cancer patients, consistent with the western blotting data
(Fig. 2C). These results
demonstrate that GSTP-1 is highly expressed in tumor tissues from
Korean colon cancer patients, suggesting that this protein might be
used as a biomarker for cancer detection.
GSTP-1 mRNA and protein expression and
enzyme activity in human colon cancer cell lines
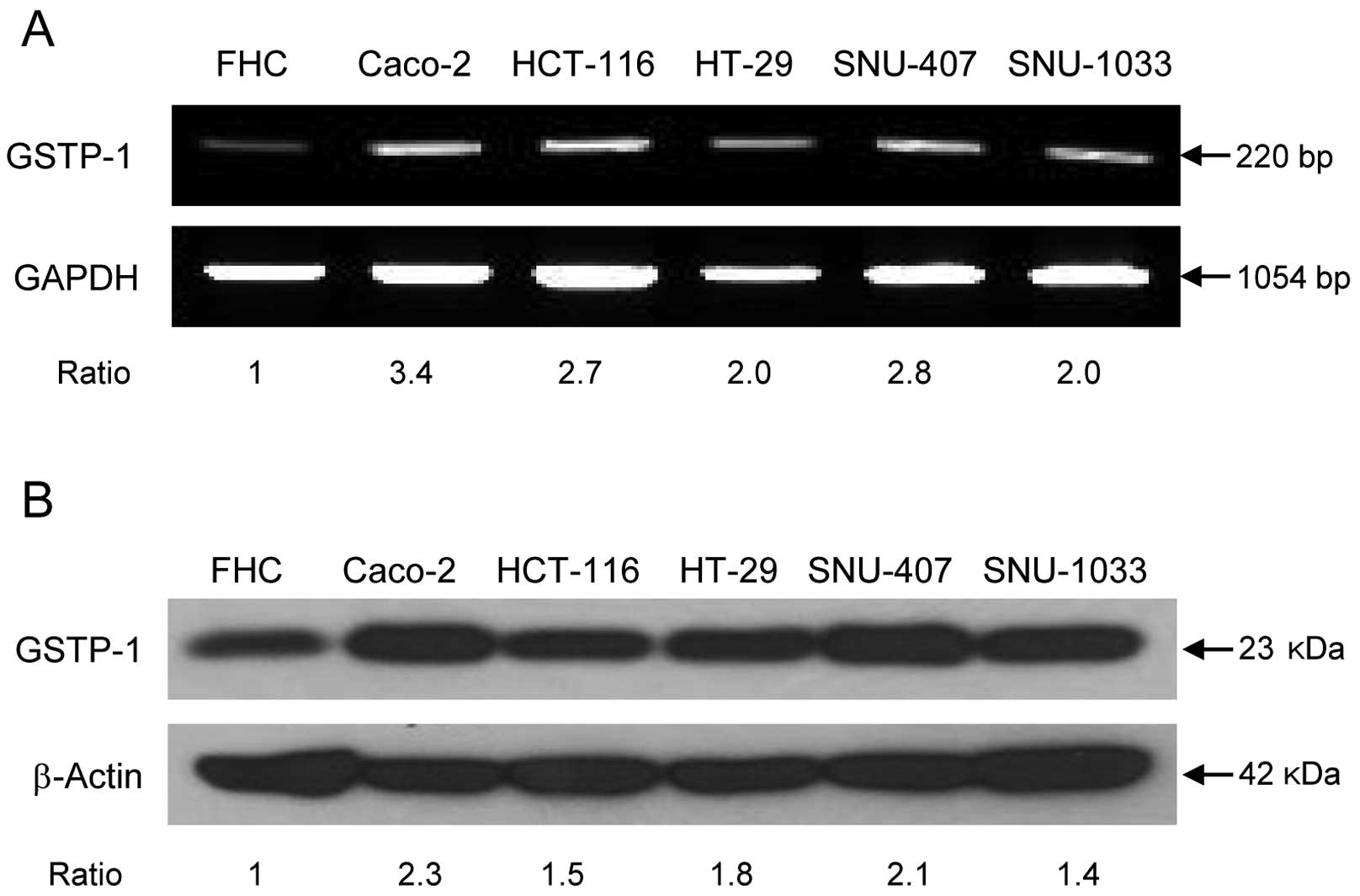
The assessment of GSTP-1 expression in colon cancer
cell lines by RT-PCR and western blotting revealed that GSTP-1 mRNA
(Fig. 3A) and protein levels
(Fig. 3B) were higher in the colon
cancer cell lines Caco-2, HCT-116, HT-29, SNU-407 and SNU-1033 than
in the normal colon cell line FHC. The protein expression and
cytoplasmic localization of GSTP-1 was assessed by confocal
microscopy (Fig. 3C), yielding
data consistent with the western blotting results. GST enzyme
activity was also higher in cancer cell lines, consistent with the
pattern of GSTP-1 protein expression (Fig. 3D), although both GSTP-1 expression
and activity levels differed among the cell lines.
Methylation status of GSTP-1 regulatory
regions in normal and colon carcinoma tissues and human colon
cancer cell lines
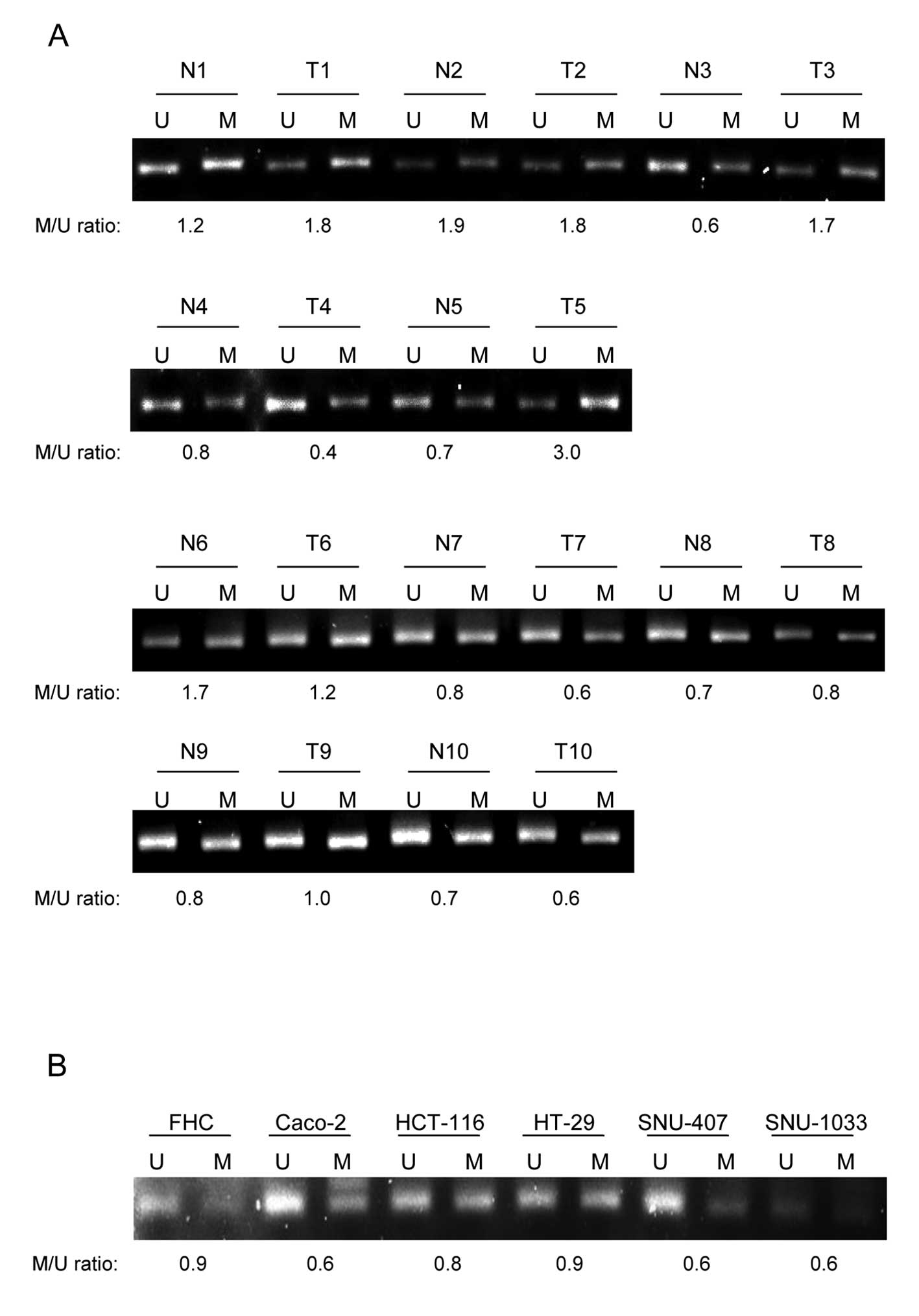
We next investigated why the expression of GSTP-1 is
higher in colorectal cancers than in normal tissues. To this end,
the methylation status of GSTP-1 regulatory regions was determined
by MS-PCR to elucidate the relationship between methylation status
and protein expression. The methylation status of GSTP-1 in patient
tissues was determined. While the relationship between methylation
degree and protein expression in tissues was not close enough
(Fig. 4A). Intriguingly, GSTP-1
was methylated to a lesser extent in the colon cancer cell lines
Caco-2, HCT-116, SNU-407 and SNU-1033, but not HT-29 (Fig. 4B), than in the FHC cell line,
almost concordant with GSTP-1 protein expression. These results
suggest that overexpression of GSTP-1 in human colon cancers is
only partially due to decreased methylation of the GSTP-1
regulatory region.
Correlation of GSTP-1 transcriptional
activity with DNA methylation and histone modification
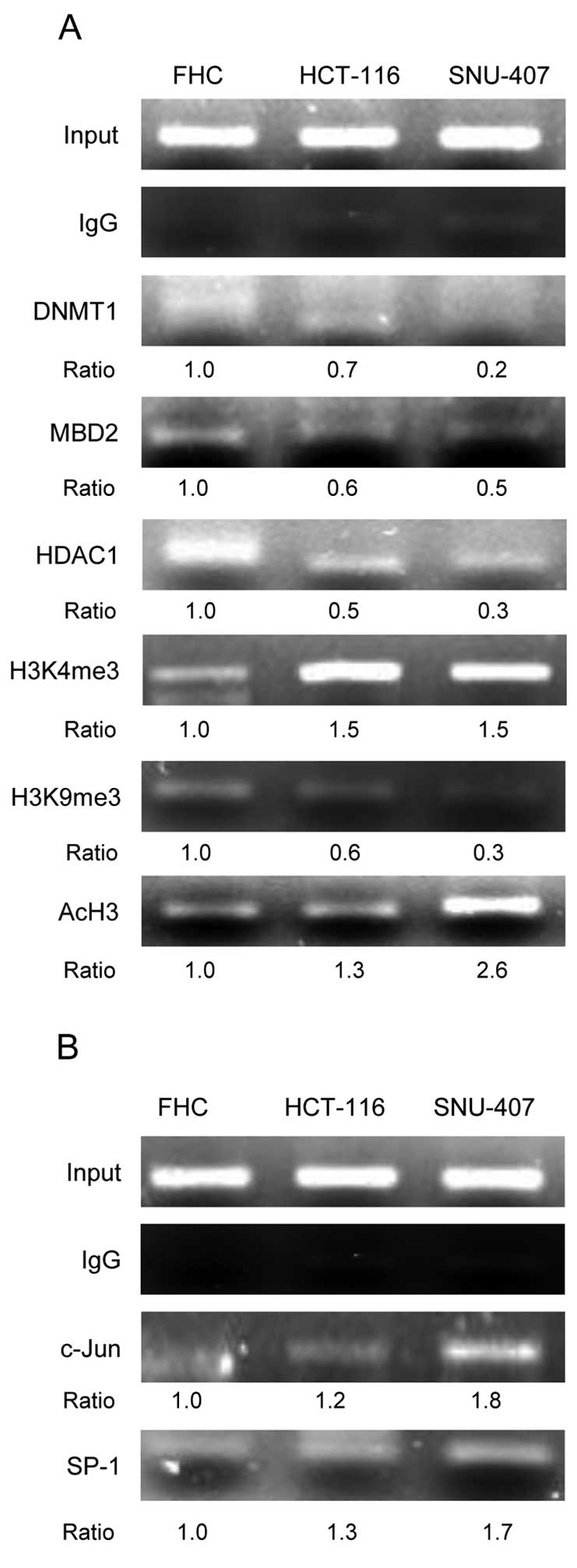
We next performed ChIP assays to investigate the
epigenetic-related proteins associated with the GSTP-1 promoter
region. The levels of DNA methylation- and
heterochromatin-associated proteins such as DNMT1, MBD2 and HDAC1
were lower in the HCT-116 and SNU-407 cell lines than in the FHC
cell line. In addition, the GSTP-1 promoter in HCT-116 and SNU-407
cells was highly enriched for trimethylation of lysine 4 of histone
H3 (H3K4me3) and for acetylation of histone H3 (AcH3), which induce
active transcription (Fig. 5A).
Whereas the level of H3K9me3, which represses transcription, was
lower in two cancer cell lines than in the FHC line (Fig. 5A). The GSTP-1 promoter region
contains binding sites for the transcription factors SP-1 and AP-1.
We performed ChIP assays to determine the roles of c-Jun (a
component of AP-1) and SP-1 in the regulation of GSTP-1 expression.
The c-Jun and SP-1 were associated with the GSTP-1 promoter region
in the HCT-116 and SNU-407 cell lines (Fig. 5B). These data reveal that the
upregulation of GSTP-1 in human colorectal cancers is probably due
to reduced methylation of the GSTP-1 promoter and active form of
histone modification and resulting in the increased activity of
specific transcription factors.
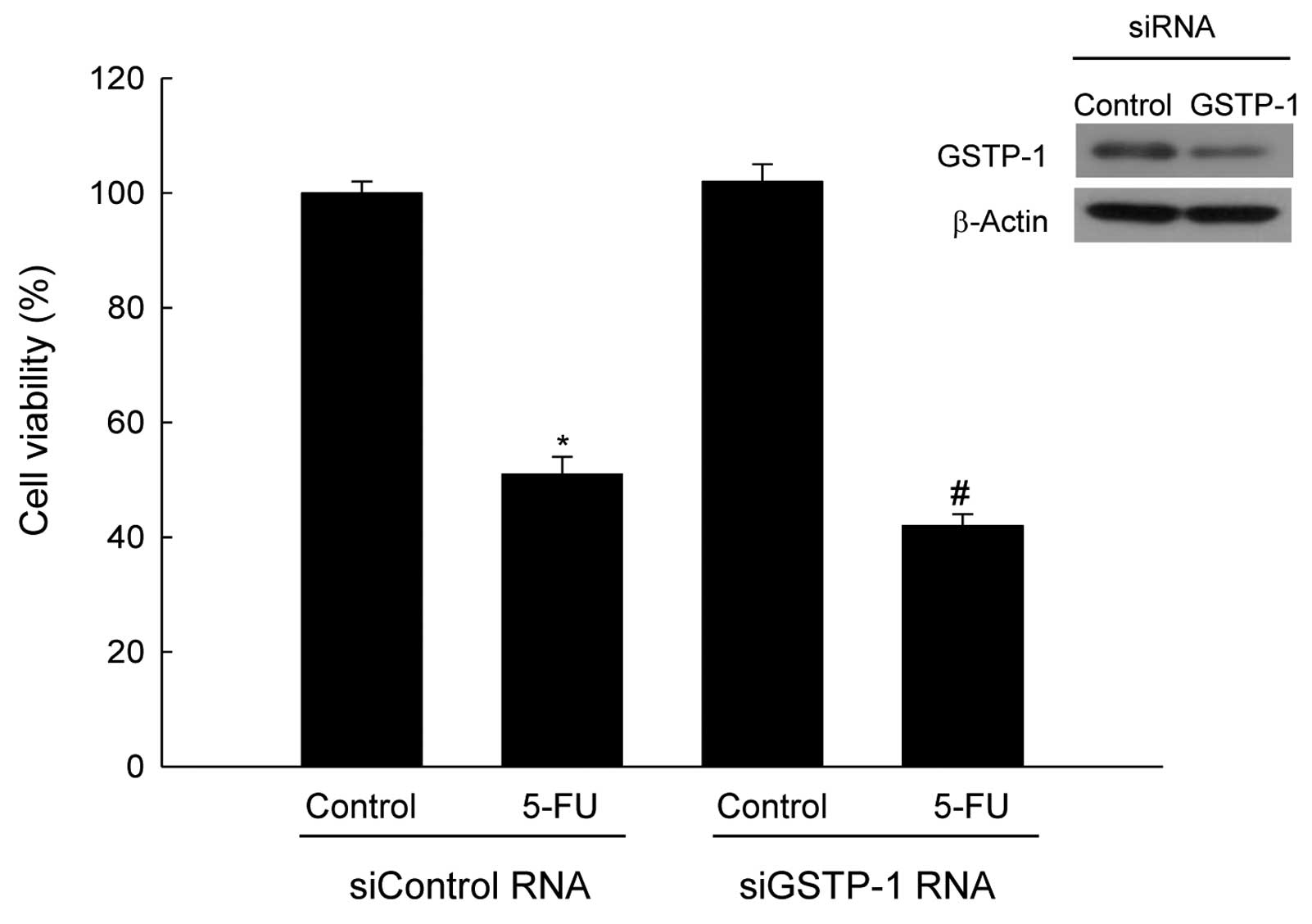
Depletion of GSTP-1 induces the
sensitivity of colorectal cancer cell lines to anticancer
agent
To investigate whether GSTP-1 is involved in drug
resistance, SNU-407 cells transfected with GSTP-1 siRNA were
treated with the anticancer agent 5-FU, at the IC value of 800 μM,
for 48 h. siRNA-mediated suppression of GSTP-1 expression increased
the sensitivity of SNU-407 cells to 5-FU (Fig. 6). These results indicate that
knockdown of GSTP-1 in SNU-407 cells increases sensitivity to the
anticancer drug.
Discussion
GSTP-1 is a phase-II detoxifying enzyme that
catalyzes the nucleophilic attack of glutathione on electrophilic
compounds (34). Recently, there
has been considerable clinical interest in GSTP-1 as a tumor marker
and therapeutic target. Overexpression of GSTP-1 in cancer cells is
associated with increased resistance to anticancer agents (35). Furthermore, a number of studies
have shown that GSTP-1 is the predominant GST isozyme found in
human cancer.
In this study, we examined GSTP-1 protein expression
and activity in normal and tumor specimens obtained from Korean
colon cancer patients. GSTP-1 protein expression and enzyme
activity were higher in colon cancer tissues than in the
surrounding normal tissues, and overexpression of GSTP-1 in colon
cancer tissues was confirmed by histological analysis. This pattern
was also observed in colon cancer cell lines. To explore the
precise mechanism underlying the upregulation of GSTP-1 in human
colon cancers, we investigated GSTP-1 promoter methylation status
and the contributions of transcriptional regulators such as DNA
methylation, histone modifications and promoter-associated proteins
to GSTP-1 expression levels. Promoter methylation status and GSTP-1
expression patterns somewhat correlated in colon cancer cell lines.
However, the relationship between promoter methylation status and
the pattern of GSTP-1 expression in colon tissues from patients
remains speculative. Therefore, we hypothesized that GSTP-1
expression is not regulated exclusively by DNA methylation, but is
also synergistically regulated by histone modifications, changes in
promoter-associated proteins and recruitment of transcription
factors. Indeed, the levels of DNA methylation- and
heterochromatin-associated proteins were reduced in human
colorectal cell lines in this study. Furthermore, the level of the
repressive histone modification H3K9me3 was reduced in human
colorectal cells. In addition, the levels of the permissive histone
modification H3K4me3 and acetylation of histone H3 were enriched in
human colorectal cells.
The transcriptional regulation of GST proteins of
the π class has been well characterized in the context of rodent
GSTP-1 (36,37). The upstream region of the GSTP-1
gene contains a TPA response element and a related enhancer element
termed GSTP-1 enhancer 1, which has characteristics similar to
those of antioxidant response elements and electrophile response
elements. GSTP-1 enhancer 1 is a specific enhancer of the rat GSTπ
gene, but is not present in the π class GST genes of mouse or human
or in genes of the other GST isoenzymes (38). Therefore, we speculated that the
regulatory mechanisms of the human GSTP-1 gene expression might
differ from those of rodents. The analysis of the human GSTP-1
proximal promoter in human leukemia has revealed the presence of
two putative SP-1 binding sites, located from −57 to −49 and from
−47 to −39, and an AP-1 response element from −69 to −63 (28,39).
We detected an association between these transcription factors and
the GSTP-1 promoter region. Binding of SP-1 and AP-1 to the GSTP-1
promoter was markedly enhanced in human colorectal cancer cells.
Finally, knockdown of GSTP-1 reduced cell proliferation and
increased sensitivity to chemotherapy. These observations suggest
that GSTP-1 might be a survival factor for colon carcinoma cells.
In summary, this study indicates that GSTP-1 may serve as a
clinically useful biomarker of colon cancer and as a target for
anticancer drugs.
Acknowledgements
This study was supported by a grant from the
National R&D Program for Cancer Control, Ministry for Health
and Welfare, Korea (1120340).
References
|
1
|
Coles BF, Chen G, Kadlubar FF and
Radominska-Pandya A: Interindividual variation and organ-specific
patterns of glutathione S transferase alpha, mu, and pi expression
in gastrointestinal ltract mucosa of normal individuals. Arch
Biochem Biophys. 403:270–276. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hayes JD, Flanagan JU and Jowsey IR:
Glutathionetransferases. Annu Rev Pharmacol Toxicol. 45:51–88.
2005.
|
|
3
|
Board PG, Webb GC and Coggan M: Isolation
of a cDNA clone and localization of the human glutathione
S-transferase 3 genes to chromosome bands 11q13 and 12q13–14. Ann
Hum Genet. 53:205–213. 1989.PubMed/NCBI
|
|
4
|
Islam MQ, Platz A, Szpirer J, Szpirer C,
et al: Chromosomal localization of human glutathione transferase
genes of classes alpha, mu, and pi. Hum Genet. 82:338–342. 1989.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ranganathan S and Tew KD:
Immunohistochemical localization of glutathione S-transferases α,
μ, and π in normal tissue and carcinomas from human colon.
Carcinogenesis. 12:2383–2387. 1991.
|
|
6
|
Miyanishi K, Takayama T, Ohi M, et al:
Glutathione Stransferase-π overexpression is closely associated
with K-ras mutation during human colon carcinogenesis.
Gastroenterology. 121:865–874. 2001.
|
|
7
|
Adler V, Yin Z, Fuchs SY, et al:
Regulation of JNK signaling by GSTp. EMBO J. 18:1321–1334. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Townsend DM, Manevich Y, He L, Hutchens S,
et al: Novel role for glutathione S-transferase pi: regulator of
protein S-glutathionylation following oxidative and nitrosative
stress. J Bio Chem. 284:436–445. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lo HW and Ali-Osman F: Genetic
polymorphism and function of glutathione S-transferases in tumor
drug resistance. Curr Opin Pharmacol. 7:367–374. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Henderson CJ, Smith AG, Ure J, Brown K, et
al: Increased skin tumorigenesis in mice lacking pi class
glutathione S-transferases. Proc Natl Acad Sci USA. 95:5275–5280.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wright JL and Lange PH: Newer potential
biomarkers in prostate cancer. Rev Urol. 9:207–213. 2007.PubMed/NCBI
|
|
12
|
Chen CL, Sheen TS, Lou LU and Huang AC:
Expression of multidrug resistance 1 and
glutathione-S-transferase-Pi protein in nasopharyngeal carcinoma.
Hum Path. 32:1240–1244. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cullen KJ, Newkirk KA, Schumaker LM,
Aldosari N, et al: Glutathione-S-transferase pi amplification is
associated with cisplatin resistance in head and neck squamous cell
carcinoma cell lines and primary tumors. Cancer Res. 63:8097–8102.
2003.PubMed/NCBI
|
|
14
|
Li M and Gu J: Changing patterns of
colorectal cancer in China over a period of 20 years. World J
Gastroenterol. 11:4685–4688. 2005.PubMed/NCBI
|
|
15
|
Kinzler KW and Vogelstein B: Lessons from
hereditary colon cancer. Cell. 87:159–170. 1996. View Article : Google Scholar
|
|
16
|
Jones PA and Baylin SB: The fundamental
role of epigenetic events in cancer. Nat Rev Genet. 3:415–428.
2002.PubMed/NCBI
|
|
17
|
Cheung P, Allis CD and Sassone-Corsi P:
Signaling to chromatin through histone modifications. Cell.
103:263–271. 2000. View Article : Google Scholar
|
|
18
|
Wang Y, Wysocka J, Sayegh J, et al: Human
PAD4 regulates histone arginine methylation levels via
demethylimination. Science. 306:279–283. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Garinis GA, Patrinos GP, Spanakis NE and
Menounos PG: DNA hypermethylation: when tumour suppressor genes go
silent. Hum Gene. 111:115–127. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Craig JM: Heterochromatin - many flavours,
common themes. Bioessays. 27:17–28. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Robertson KD and Wolffe AP: DNA
methylation in health and disease. Nat Rev Gene. 1:11–19. 2000.
View Article : Google Scholar
|
|
22
|
Heard E, Rougeulle C, Arnaud D, Avner P,
et al: Methylation of histone H3 at Lys-9 is an early mark on the X
chromosome during X inactivation. Cell. 107:727–738. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Rountree MR, Bachman KE, Herman JG and
Baylin SB: DNA methylation, chromatin inheritance, and cancer.
Oncogene. 20:3156–3165. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Watt PM, Kumar R and Kees UR: Promoter
demethylation accompanies reactivation of the HOX11 proto-oncogene
in leukemia. Genes Chromosomes Cancer. 29:371–377. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Schübeler D, MacAlpine DM, Scalzo D, et
al: The histone modification pattern of active genes revealed
through genome-wide chromatin analysis of a higher eukaryote. Genes
Dev. 18:1263–1271. 2004.PubMed/NCBI
|
|
26
|
Berger SL: The complex language of
chromatin regulation during transcription. Nature. 447:407–412.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Moffat GJ, McLaren AW and Wolf CR:
Sp1-mediated transcriptional activation of the human Pi class
glutathione S-transferase promoter. J Bio Chem. 271:1054–1060.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Duvoix A, Schnekenburger M, Delhalle S, et
al: Expression of glutathione S-transferase P1-1 in leukemic cells
is regulated by inducible AP-1 binding. Cancer Lett. 216:207–219.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Morceau F, Duvoix A, Dehalle S,
Schnekenburger M, et al: Regulation of glutathione S-transferase
P1-1 gene expression by NF-kappa B in tumor necrosis factor
alpha-treated K562 leukemia cells. Biochem Pharma. 67:1227–1238.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Moon PG, Lee JE, You S, et al: Proteomic
analysis of urinary exosomes from patients of early IgA nephropathy
and thin basement membrane nephropathy. Proteomics. 11:2459–2475.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Silva JC, Denny R, Dorschel CA, et al:
Quantitative proteomic analysis by accurate mass retention time
pairs. Anal Chem. 77:2187–2200. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Karius T, Schnekenburger M, Ghelfi J,
Walter J, et al: Reversible epigenetic fingerprint-mediated
glutathione-S-transferase P1 gene silencing in human leukemia cell
lines. Biochem Pharma. 81:1329–1342. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Carmichael J, DeGraff WG, Gazdar AF, Minna
JD, et al: Evaluation of a tetrazolium-based semiautomated
colorimetric assay: assessment of chemosensitivity testing. Cancer
Res. 47:936–941. 1987.PubMed/NCBI
|
|
34
|
Rushmore TH and Pickett CB: Glutathione
S-transferases, structure, regulation, and therapeutic
implications. J Bio Chem. 268:11475–11478. 1993.PubMed/NCBI
|
|
35
|
Townsend DM and Tew KD: The role of
glutathione-S-transferase in anti-cancer drug resistance. Oncogene.
22:7369–7375. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Sakai M, Okuda A and Muramatsu M: Multiple
regulatory elements and phorbol 12-O-tetradecanoate-13-acetate
responsiveness of the rat placental glutathione transferase gene.
Proc Nat Acad Sci USA. 85:9456–9460. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Osada S, Takano K, Nishihara T, Suzuki T,
et al: CCAAT/enhancer-binding proteins alpha and beta interact with
the silencer element in the promoter of glutathione S-transferase P
gene during hepato-carcinogenesis. J Bio Chem. 270:31288–31293.
1995. View Article : Google Scholar
|
|
38
|
Dixon KH, Cowell IG, Xia CL, Pemble SE, et
al: Control of expression of the human glutathione S-transferase pi
gene differs from its rat orthologue. Biochem Biophy Res Commun.
163:815–822. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Duvoix A, Schmitz M, Schnekenburger M, et
al: Transcriptional regulation of glutathione S-transferase P1-1 in
human leukemia. Biofactors. 17:131–138. 2003. View Article : Google Scholar : PubMed/NCBI
|