Introduction
Breast cancer is the most commonly occurring
malignancy in women worldwide. Various histological and molecular
subtypes of breast cancer have been identified with different
biologic implications (1–3). Apocrine differentiation (metaplasia)
is frequently observed in the mammary epithelium; however, invasive
apocrine carcinomas are rare, constituting less than 5% of all
breast carcinomas (1,4). Apocrine lesions of the breast are
characterized by over-expression of the androgen receptor (AR)
along with loss of estrogen receptor-α (ER-α) and the progesterone
receptor (PR) (5–7). Molecular studies of invasive apocrine
breast carcinoma have shown a specific molecular apocrine profile
based on the AR expression that divides ER-negative breast
carcinomas into two different clusters: ER−/AR− (basal) and ER−/AR+
(molecular apocrine cluster) (8).
Breast cancer cell lines are widely used for
experimental research as models of various subtypes of breast
carcinomas (9–12). The MDA-MB-453 breast cancer cell
line, obtained from a malignant pleural effusion of a 48-year-old
female, has been suggested as a model for the molecular apocrine
breast subtype (13–15). The cell line exhibits a
characteristic apocrine carcinoma steroid receptor profile:
ER-α-negative, PR-negative, and AR-positive (8,13).
Increased proliferation in response to androgens is the key feature
of the cell line, which can be blocked by anti-androgens, such as
flutamide (13,16,17).
Her-2/neu activity has been well documented in this cell line, as
well as the existence of a functional cross-talk between AR and
Her-2/neu, involving the MAPK/ERK1/2 pathway (13,14).
These features also tend to characterize a substantial proportion
of invasive apocrine carcinomas of the breast (18–20).
The aim of the present study was to further
characterize the MDA-MB-453 cell line and to correlate it with the
results obtained from the apocrine breast carcinoma samples of
patients.
Materials and methods
Breast cancer cell lines and
treatment
Human breast cancer cell line MDA-MB-453 was
obtained from the American Type Culture Collection (ATCC, Manassas,
VA, USA) and used for analysis and experiments, while MDA-MB-231,
MCF-7 and BT-474 cell lines from the ATCC served as controls.
MDA-MB-453, MDA-MB-231 and BT-474 were cultured in Dulbecco’s
modified Eagle’s medium (DMEM, Gibco-BRL, Invitrogen, Carlsbad, CA,
USA) supplemented with 10% fetal bovine serum (FBS) at 37°C in a
humidified CO2 incubator. The MCF-7 cell line was
cultured in Improved modified Eagle’s medium (IMEM, Invitrogen)
supplemented with 10% FBS. Prior to the experiments, the cells were
cultured in phenol-red-free DMEM (Invitrogen) for 24 h, and then in
serum-free DMEM overnight. U0126 (MEK1/2 inhibitor, Cell Signaling
Technology, Danvers, MA, USA), a highly selective inhibitor of
MEK1/2, was used.
Breast tissue samples
Formalin-fixed paraffin-embedded blocks of 8
cases of apocrine carcinoma (seven invasive and one in situ
case) that corresponded to the cell line profile (ER-α-, PR−, AR+
and Her-2/neu −/+) were selected from the previously
well-characterized cohort (18) and
used for comparative analysis.
Protein expression analysis
Protein expression was analyzed by Western blot
analysis in cell lysates or by immunocytochemistry (ICC) and
immunohistochemistry (IHC) on formalin-fixed and paraffin-embedded
cell blocks prepared from the MDA-MB-453 cell line and tissue
blocks from breast tissue samples (the list of antibodies is shown
in Table I). For ICC and IHC
analysis, commercially available detection kits and automated
staining procedures were employed (18). Cell lysates, immunoprecipitations
and Western blot analysis were carried out using standard
procedures as previously described (21,22).
Signals were detected by enhanced chemiluminiscence (ECL; Amersham,
GE Healthcare Biosciences, Pittsburgh, PA, USA).
 | Table IList of the antibodies used in the
study. |
Table I
List of the antibodies used in the
study.
| Antibody | Manufacturer |
|---|
| ER-α (clone 6F11),
PR (clone 16), Her-2/neu (Clone CB11), cyclin D1 | Ventana Medical
Systems |
| EGFR, MAPK1/2
(ERK1/2), pERK1/2 (Thr202/Tyr204) | Cell Signaling
Technology |
| p16 (mouse
monoclonal IgG1κ) | Cell Marque |
| Cyclin D1 (clone
SP4 rabbit IgG), GCDFP-15 (clone 23A3 mouse IgG2A κ), ER-α (Cat no.
RB-9016-P) | Neomarkers |
| EGFR (PharmDX
diagnostic kit), Topoisomerase-IIα (Clone Ki-S1), Cytokeratin 5/6
(D5/16B4), p63 (clone 4A4) | DakoCytomation |
| Rb (IF8): sc-102,
pRb(Ser795), p16 (N-20: sc-467), p53 (DO-1): sc-126, Actin [(I-19):
sc-1616], AR (clone AR441: sc-7305), pEGFR (Tyr845), Her-2 (Neu
(CB11): sc-52349), p-Neu (Tyr 1248)-R: sc-12352-R | Santa Cruz
Biotechnology |
A positive p16INK4A expression was
defined as the presence of nuclear and/or cytoplasmic staining, and
the percentage of positive cells was recorded (23,24).
For ER-α, PR, AR, cyclin D1, p53, p63 and topoisomerase-IIα
expression, only nuclear labeling was scored (24,25).
For Her-2/neu and EGFR proteins, only membranous staining was
considered positive. The scoring was carried out according to the
manufacturer’s (EGFR, Dako, Carpinteria, CA, USA) and American
Society of Clinical Oncology/College of American Pathologists’
guideline recommendations (Her-2/neu protein) (26). GCDFP-15 and CK5/6 proteins were
considered positive if any membranous/cytoplasmic staining was
observed.
Conventional cytogenetics
G-banding procedures were performed on metaphase
cells. Metaphase chromosomes were banded with Wright trypsin and
karyotypes were described following the established international
guidelines (27).
Fluorescent in situ hybridization
(FISH)
FISH was performed to evaluate copy numbers at
EGFR, TOP2A and HER-2/neu loci. Chromosome
enumeration probes CEP7 and CEP17 were used as indicators of
chromosome copy numbers (Abbott Molecular Inc., Des Plaines, IL,
USA). A total of 100 nuclei were scored per sample. A ratio of
HER-2/CEP17 >2.2 was defined as gene
amplification; a ratio of 1.8–2.1 was interpreted as borderline,
and a ratio of <1.8 was defined as negative. The same criteria
were used for the interpretation of EGFR/CEP7 and TOP2A/CEP17
ratios, respectively. For statistical purposes, equivocal FISH
results (ratio of 1.8–2.1) were considered negative (27). Polysomy 7 and 17 were defined as ≥3
CEP signals per cell (18,28).
Flow cytometry
Flow cytometry (FC) was applied to measure the
S-phase fraction of MDA-MB-453 cells. Cells at ~50% confluence were
harvested and 1 ml cold 70% ethanol was slowly added to the cell
pellet while vortexing. Ethanol-fixed cells were treated with 100
μg/ml RNaseA and 50 μg/ml propidium iodide (PI) in PBS at room
temperature for 30 min. Flow cytometry of cell cycle distribution
was performed using a FACSCalibur flow cytometer (BD-Biosciences,
San Jose, CA, USA). The results were analyzed using Multicycle for
Windows.
Gene mutation analysis
These assays included the detection of 12 mutations
in codons 12 and 13 of the K-RAS gene as well as the V600E
mutation in exon 15 of the B-RAF gene. The analysis was
performed using a Mutector II assay, using proprietary Shift
Termination Assay (STA) technology (TrimGen Corporation, Sparks,
MD, USA) following the previously described protocol (29). For quality control of the
K-RAS mutation analysis, we used a colon cancer biopsy known
to be positive for K-RAS mutation, while a case of malignant
melanoma served as a positive control for B-RAF
mutation.
Statistical analysis
Quantitative data of the experimental studies were
expressed as the mean ± SD. The student’s t-test was used to test
the differences between responses in the exposed and control
groups. Statistical Package for the Social Sciences version 17.0
(SPSS, Chicago, IL, USA) was used for statistical analysis.
P<0.05 was considered to be statistically significant.
Results
Profiling of MDA-MB-453 cell line
A summary of the key findings and comparison with
the apocrine carcinoma tissue samples is shown in Table II. The MDA-MB-453 cell line was
negative for ER-α, PR and Her-2/neu protein on ICC, whereas AR was
found to be positive by Western blot analysis. Notably, the ICC
assay demonstrated no nuclear staining but predominantly
cytoplasmic distribution of AR, whereas the GCDFP-15 protein was
absent. Western blot analysis revealed low levels of Her-2/neu and
phosphorylation of the Her-2/neu protein at Tyr1248 in comparison
with BT-474 cells, an ER-positive breast cancer cell line with
HER-2/neu gene amplification. Western blot analysis revealed
that levels of total EGFR and phosphorylation of EGFR at Tyr845
were lower in MDA-MB-453 cells compared with EGFR- over-expressing
MDA-MB-231 cells. In contrast, the ICC assay revealed a strong EGFR
protein expression (score 3+). FISH analysis revealed neither
HER-2/neu (HER2/CEP17 was 3.96/3.68, ratio:
1.08) nor EGFR gene amplification (EGFR/CEP7 was 3.23/2.90,
ratio: 1.11). Gains of chromosomes 7 and 17 (CEP7 and 17) observed
with FISH were further confirmed using conventional cytogenetic
analysis, which revealed a hypertriploid clone characterized by
extensive numerical and structural abnormalities including gains of
chromosomes 7, 11, 17, 19 and 21 and losses of chromosomes X, 3, 4,
9, 13, 14, 16 and 18.
 | Table IIA summary of the key findings in the
cell line and apocrine carcinoma tissue samples. |
Table II
A summary of the key findings in the
cell line and apocrine carcinoma tissue samples.
| Feature | MDA-MB-453 | Apocrine carcinoma
tissue samples |
|---|
|
Estrogen/progesterone receptor | Negative | Negativeb |
| Androgen
receptor | Positivea | Positiveb |
| GCDFP-15
protein | Negative | Positivec |
| HER-2/neu
status | Low protein
expressiona/no gene
amplification | High protein
expression common/gene amplification b |
| EGFR status | Low protein
expressiona/no gene
amplification | High protein
expression common/no gene amplification b |
| KRAS gene
status | Mutated (Gly13Asp
GGC>GAC) | Not mutated |
| BRAF gene
status | Not mutated | Not mutated |
|
p16INK4A
protein | Lost due to gene
deletion at 9p.21/no protein detected | Present (variable
cytoplasmic/nuclear staining) |
| Cyclin D1
protein | Highly
over-expressed | Low expression |
| Polysomy 7 and 17
(CEP7 and 17) | Present | Frequently
observedb |
| Basal markers
(CK5/6, p63) | Low expression | Low
expressiond |
Of particular functional importance was the deletion
of the 9p21 locus that harbors the INK4A gene
(p16INK4A) and INK4B gene (p15), as evidenced by
the loss of the p16INK4A protein expression in Western
blot or ICC analysis (Fig. 1A).
CK5/6 was completely absent, whereas the p63 protein
expression with defined nuclear staining was observed in
approximately 10% of the cells.
MDA-MB-453 cells exhibit a high
proliferation rate
MDA-MB-453 cells exhibited an excessive
proliferation rate under optimal culture conditions as its S-phase
fraction measured up to 70% by flow cytometry. This rate was
consistent with the cyclin D1 over-expression (positive in ~70% of
the cells), inactivation of retinoblastoma protein (Rb), measured
by its phosphorylation at Ser795, and the loss of
p16INK4A protein expression. The cell line also
exhibited a high p53 protein expression measured by both Western
blot and ICC analysis. Consequently, topoisomerase-IIα was also
strongly expressed in approximately 70% of the cells (without
underlying TOP2A gene amplification, TOP2A/CEP17:
2.86/ 2.80, ratio: 1.02). When cultured in a serum-free medium for
24 h, the cell line remained highly proliferative with an S-phase
fraction of approximately 40%. The cell line was also capable of
sustained proliferation in a serum-free medium for up to 48 h, as
demonstrated by the S-phase fraction.
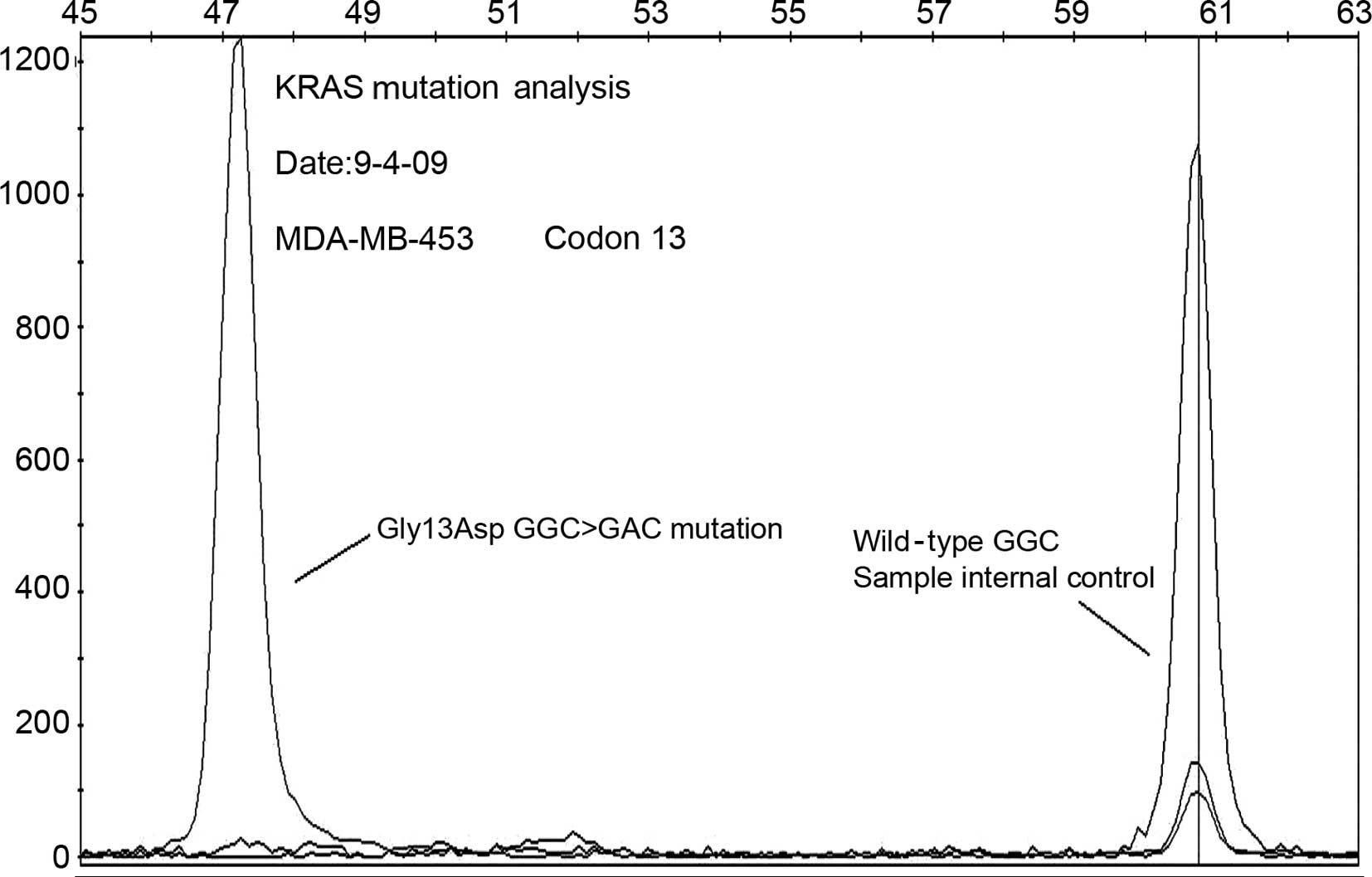
MDA-MB-453 cells harbor a K-RAS mutation
and exhibit constitutive activation of the MAPK/ERK pathway
Mutational analysis of the K-RAS gene
revealed codon 13 mutation (Gly 13 Asp GGC>GAC), a mutation that
constitutively activates K-Ras in the MDA-MB-453 cell line
(Fig. 2), whereas there is no gene
alteration in the B-RAF gene. Western blot analysis
confirmed that ERK1/2 is highly phosphorylated in cells
irrespective of the concentration of the fetal calf serum, and
culture conditions. In addition, MDA-MB-453 retained high ERK
activity even after 48 h in serum-free medium, and was highly
sensitive to the specific MEK1/2 inhibitor U0126. Low
concentrations of U0126 (0.1 μM) led to a partial inhibition of
ERK1/2 phosphorylation after 12 h. The exposure of MDA-MB-453 cells
to U0126 at a concentration of 10 μM for various time periods (from
15 min to 24 h) led to a markedly reduced S-phase fraction as
measured by flow cytometry (Fig. 3A and
B). Thus, 12 h treatment of the MDA-MB-453 cells with U0126
resulted in a significantly reduced proliferation rate in the
experimental group in comparison with the control group (22.6 vs.
38.3%, reduction ~42%, p<0.001, Student’s t-test). Western blot
analysis with phospho-specific anti-ERK1/2 antibody also showed
that U0126 abolished ERK1/2 phosphorylation in MDA-MB-453
cells.
Breast tissue sample analysis
Of 5 tested patient samples of the apocrine
carcinomas, 1 exhibited an absence of p16INK4A protein
expression, 3 samples (two invasive and one in situ
carcinoma) had predominantly cytoplasmic p16INK4A
expression (observed in 20–80% of the tumor cells), whereas only 1
case retained a nuclear p16INK4A expression intensity,
compared with the positive control (Fig. 1B). Cyclin D1 expression was observed
in 3 out of 5 cases. However, only 2 of these cases reached more
than 10% of the positive cells with predominantly weak to moderate
nuclear intensity.
K-RAS and B-RAF mutational analysis
performed on 6 invasive apocrine carcinoma samples showed no gene
alterations in any of the studied cases. Notably, these cases had a
strong EGFR protein over-expression on IHC (scores 2+/3+ on
IHC) without underlying EGFR gene amplification (18).
Discussion
The present study describes complex and multiple
cytogenetic and molecular alterations in the MDA-MB-453 cell line
that is currently considered to be a model for breast apocrine
carcinoma. Conventional cytogenetic analysis revealed that
MDA-MB-453 is a hypertriploid cell line that corresponds closely to
its metastatic origin and high malignant potential (10). We also observed considerable
discrepancies between published cytogenetic findings regarding the
cell line and our own results, although certain previously reported
cytogenetic findings, including gains of chromosomes 7 and 17 and
loss of the 9p.21 locus (CDKN2 gene) (30), are also observed in our study. Thus,
previous studies (31,32) reported HER-2/neu gene
amplification in the MDA-MB-453 cell line, whereas our analysis did
not support those findings. FISH and conventional cytogenetic
analysis revealed a gain (polysomy) of CEP17 in only a small
population of cells (~5% of the counted cells) that carried
HER-2/neu gene amplification. Levels of total Her-2/neu
expression and phosphorylation of Her-2/neu at Tyr1248, often
associated with HER-2/neu gene amplification (33), were lower in comparison with the
positive cell control (BT-474 cell line). Belsches-Jablonski et
al (34) also revealed a low
Her-2/neu protein expression in the MDA-MB-453 cell line, whereas
Kao et al (11) found low
copy numbers of the HER-2/neu gene in the cell line using a
quantitative polymerase chain reaction, and thus classified
MDA-MB-453 cells as an HER-2-negative cell line. Consequently, the
classification of the cell line varies from luminal (12,31,32) or
‘weakly luminal’ (10) to an
apocrine cell line model (8,13–15).
We propose that the observed disparities are caused by the
prolonged cell culture, which may modify genetic/phenotypic
properties of the cell line (9,10), and
further stress the importance of a laboratory self validation of
the cell line.
Our study reports for the first time a codon 13
mutation in the K-RAS gene followed by constitutive MAPK/ERK
activation in the MDA-MB-453 cell line (35,36).
The K-RAS gene is an integral part of the MAPK signaling
pathway, whose activation strongly correlates with the degree of
the K-RAS gene activation (36). The MAPK/ERK signaling pathway is
crucial in the regulation of cell proliferation, differentiation,
survival and metastasis through ultrasensitive switch-like
responses to various stimuli depending on the strength and duration
of stimulation (35,37–40).
Some of these effects, such as proliferation, may be abolished by
the targeted inhibition, as we demonstrated using the specific
MEK1/2 inhibitor U0126 in our study.
K-RAS gene mutations (usually restricted to
codons 12, 13 and 61) are frequently observed in human tumors
(~15%) (41) and established breast
cancer cell lines (35). However,
Hollestelle et al (35)
reported PTEN but not K-RAS gene mutation in this
cell line, although these authors found that 13% of the studied
breast cancer cell lines harbored K-RAS gene mutations.
Furthermore, results obtained from the cell line were not in
concordance with the data from the breast cancer samples, which is
consistent with findings of previous reports that have confirmed
the lack of K-RAS mutations in breast carcinomas (36,42–45).
Notably, the lack of K-RAS mutations in apocrine carcinomas
may become a potential therapeutic benefit, since the tested cases
were EGFR-positive and thus are potentially amenable to targeted
anti-EGFR therapy (44).
B-RAF gene mutations, mainly involving the V600E locus, have
also been described in a wide range of human tumors with the
highest frequency in malignant melanoma (41). These mutations are rarely observed
in breast carcinoma (41). Neither
the cell line nor apocrine breast carcinoma samples harbored the
B-RAF gene mutations.
Another significant finding in this study was the
deletion of the 9p21–22 region, responsible for the complete loss
of the p16INK4A protein as confirmed by ICC and Western
blot analysis. This finding is consistent with a recent study that
reported an allelic loss at 9p21 loci in MDA-MB-453 cells with a
barely detectable p16INK4A protein expression (29). p16INK4A, the product of
the CDKN2 gene, one of the key cell cycle regulators,
inhibits phosphorylation of the retinoblastoma protein, and thus
acts as a negative regulator of the cell cycle (46). In contrast, loss of
p16INK4A was found in only 1 of 5 tested apocrine
carcinomas of the breast with loss p16INK4A protein
expression, whereas the remaining 4 exhibited pre-dominantly
cytoplasmic patterns of distribution of the p16INK4A
protein. Notably, several investigators previously reported a
cytoplasmic distribution of the p16INK4A protein in a
subset of breast carcinomas (47,48),
which correlated with a high histological grade, loss of ER and PR,
p53 protein over-expression and accelerated tumor proliferation,
the parameters frequently featured in apocrine carcinoma and the
MDA-MB-453 cell line. Similarly, the cell line exhibited a high
cyclin D1 expression, which was not observed in the breast tissue
samples.
In conclusion, the observed cytogenetic and
molecular alterations in the MDA-MB-453 cell line were not
consistently present in the apocrine tumor samples. Our cell
culture results also differ in certain respects from previously
published data. MDA-MB-453 is not an ideal model of apocrine
carcinomas as recently defined using molecular methods. Notably,
EGFR-positive apocrine carcinomas of the breast do not tend to
harbor K-RAS gene mutations observed in the cell line, which
indicates that targeted therapy with EGFR inhibitors may be a
viable alternative in some patients.
Acknowledgements
This study was supported by the National Institutes
of Health Grant, DK070016 (Z.-Y. Wang). Dr Semir Vranic was a
research fellow at Creighton University Medical Center, Omaha, NE,
USA, and was supported by a UICC American Cancer Society Beginning
Investigators Fellowship (ACSBI) (ACS/08/004) funded by the
American Cancer Society. We appreciate the technical assistance of
Kay Krogman, Deborah Jankovich, and Kristin Bonnstetter (Creighton
Medical Laboratories, Creighton University School of Medicine,
Omaha, NE, USA) and Dr Faruk Skenderi (Sarajevo University School
of Medicine, Bosnia and Herzegovina). We are also indebted to Dr
Warren Sanger and Ms. Marilu Nelson (The University of Nebraska
Medical Center, Omaha, NE) for their excellent cytogenetics
support.
References
|
1
|
Tavassoli FA and Devilee P: World Health
Organization Classification of tumours Pathology and genetics of
tumours of the breast and female genital organs. IARC Press; Lyon:
2003
|
|
2
|
Perou CM, Sørlie T, Eisen MB, et al:
Molecular portraits of human breast tumours. Nature. 406:747–752.
2000. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sørlie T, Perou CM, Tibshirani R, et al:
Gene expression patterns of breast carcinomas distinguish tumor
subclasses with clinical implications. Proc Natl Acad Sci USA.
98:10869–10874. 2001.PubMed/NCBI
|
|
4
|
Weigelt B, Horlings HM, Kreike B, et al:
Refinement of breast cancer classification by molecular
characterization of histological special types. J Pathol.
216:141–150. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gatalica Z: Immunohistochemical analysis
of apocrine breast lesions. Consistent over-expression of androgen
receptor accompanied by the loss of estrogen and progesterone
receptors in apocrine metaplasia and apocrine carcinoma in situ.
Pathol Res Pract. 193:753–758. 1997.
|
|
6
|
Tavassoli FA, Purcell CA, Bratthauer GL,
et al: Androgen receptor expression along with loss of bcl-2, ER,
and PR expression in benign and malignant apocrine lesions of the
breast: implications for therapy. Breast J. 2:261–269. 1996.
View Article : Google Scholar
|
|
7
|
Bratthauer GL, Lininger RA, Man YH and
Tavassoli FA: Androgen and estrogen receptor mRNA status in
apocrine carcinomas. Diagn Mol Pathol. 11:113–118. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Farmer P, Bonnefoi H, Becette V, et al:
Identification of molecular apocrine breast tumours by microarray
analysis. Oncogene. 24:4660–4671. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Burdall SE, Hanby AM, Lansdown MRJ and
Speirs V: Breast cancer cell lines: friend or foe? Breast Cancer
Res. 5:89–95. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lacroix M and Leclercq G: Relevance of
breast cancer cell lines as models for breast tumours: an update.
Breast Cancer Res Treat. 83:249–289. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kao J, Salari K, Bocanegra M, et al:
Molecular profiling of breast cancer cell lines defines relevant
tumor models and provides a resource for cancer gene discovery.
PLoS One. 4:e61462009.PubMed/NCBI
|
|
12
|
Neve RM, Chin K, Fridlyand J, et al: A
collection of breast cancer cell lines for the study of
functionally distinct cancer subtypes. Cancer Cell. 10:515–27.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Naderi A and Hughes-Davies L: A
functionally significant cross-talk between androgen receptor and
erbB2 pathways in estrogen receptor negative breast cancer.
Neoplasia. 10:542–548. 2008.
|
|
14
|
Chia KM, Liu J, Francis GD and Naderi A: A
feedback loop between androgen receptor and ERK signaling in
estrogen receptor-negative breast cancer. Neoplasma. 13:154–66.
2011.PubMed/NCBI
|
|
15
|
De Longueville F, Lacroix M, Barbuto AM,
et al: Molecular characterization of breast cancer cell lines by a
low-density microarray. Int J Oncol. 27:881–892. 2005.PubMed/NCBI
|
|
16
|
Doane AS, Danso M, Lal P, et al: An
estrogen receptor-negative breast cancer subset characterized by a
hormonally regulated transcriptional program and response to
androgen. Oncogene. 25:3994–4008. 2006. View Article : Google Scholar
|
|
17
|
Hall RE, Birrell SN, Tilley WD and
Sutherland RL: MDA-MB-453, an androgen-responsive human breast
carcinoma cell line with high level androgen receptor expression.
Eur J Cancer. 30A:484–490. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Vranic S, Tawfik O, Palazzo J, et al: EGFR
and HER-2/neu expression in invasive apocrine carcinoma of the
breast. Mod Pathol. 23:644–653. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bhargava R, Beriwal S, Striebel JM and
Dabbs DJ: Breast cancer molecular class ERBB2: preponderance of
tumors with apocrine differentiation and expression of basal
phenotype markers CK5, CK5/6, and EGFR. Appl Immunohistochem Mol
Morphol. 18:113–118. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Varga Z, Zhao J, Ohlschlegel C, Odermatt B
and Heitz PU: Preferential HER-2/neu overexpression and/or
amplification in aggressive histological subtypes of invasive
breast cancer. Histopathology. 44:332–338. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang XT, Kang LG, Ding L, Vranic S,
Gatalica Z and Wang ZY: A positive feedback loop of ER-α36/EGFR
promotes malignant growth of ER-negative breast cancer cells.
Oncogene. 30:770–780. 2011.
|
|
22
|
Wang Z, Zhang X, Shen P, Loggie BW, Chang
Y and Deuel TF: A variant of estrogen receptor-{alpha},
hER-{alpha}36: transduction of estrogen- and antiestrogen-dependent
membrane-initiated mitogenic signaling. Proc Natl Acad Sci USA.
103:9063–9068. 2006.
|
|
23
|
Subhawong AP, Subhawong T, Nassar H, et
al: Most basal-like breast carcinomas demonstrate the same Rb−/p16+
immunophenotype as the HPV-related poorly differentiated squamous
cell carcinomas which they resemble morphologically. Am J Surg
Pathol. 33:163–75. 2009.PubMed/NCBI
|
|
24
|
Milde-Langosch K, Bamberger AM, Rieck G,
Kelp B and Löning T: Overexpression of the p16 cell cycle inhibitor
in breast cancer is associated with a more malignant phenotype.
Breast Cancer Res Treat. 67:61–70. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Elayat G, Selim AG and Wells CA:
Alterations of the cell cycle regulators cyclin D1, cyclin A, p27,
p21, p16, and pRb in apocrine metaplasia of the breast. Breast J.
15:475–482. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wolff AC, Hammond ME, Schwartz JN, et al:
American Society of Clinical Oncology/College of American
Pathologists guideline recommendations for human epidermal growth
factor receptor 2 testing in breast cancer. J Clin Oncol.
25:118–145. 2007. View Article : Google Scholar
|
|
27
|
Shaffer LG and Tommerup N: ISCN 2005 An
International System for Human Cytogenetic Nomenclature. Karger;
Basel: 2005
|
|
28
|
Vranic S, Teruya B, Repertinger S, Ulmer
P, Hagenkord J and Gatalica Z: Assessment of HER2 gene status in
breast carcinomas with polysomy of chromosome 17. Cancer.
117:48–53. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shackelford W, Deng S, Murayama K and Wang
J: A new technology for mutation detection. Ann N Y Acad Sci.
1022:257–262. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hollestelle A, Nagel JH, Smid M, et al:
Distinct gene mutation profiles among luminal-type and basal-type
breast cancer cell lines. Breast Cancer Res Treat. 121:53–64. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Arriola E, Marchio C, Tan DS, et al:
Genomic analysis of the HER2/TOP2A amplicon in breast cancer and
breast cancer cell lines. Lab Invest. 88:491–503. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Mackay A, Tamber N, Fenwick K, et al: A
high-resolution integrated analysis of genetic and expression
profiles of breast cancer cell lines. Breast Cancer Res Treat.
118:481–498. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Taniyama K, Ishida K, Toda T, et al:
Tyrosine1248-phosphorylated HER2 expression and HER2 gene
amplification in female invasive ductal carcinomas. Breast Cancer.
15:231–240. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Belsches-Jablonski AP, Biscardi JS, Peavy
DR, Tice DA, Romney DA and Parsons SJ: Src family kinases and HER2
interactions in human breast cancer cell growth and survival.
Oncogene. 20:1465–1475. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hollestelle A, Elstrodt F, Nagel JHA,
Kallemeijn WW and Schutte M: Phosphatidylinositol-3-OH kinase or
RAS pathway mutations in human breast cancer cell lines. Mol Cancer
Ther. 5:195–201. 2007.PubMed/NCBI
|
|
36
|
Von Lintig FC, Dreilinger AD, Varki NM,
Wallace AM, Casteel DE and Boss GR: Ras activation in human breast
cancer. Breast Cancer Res Treat. 62:51–62. 2000.PubMed/NCBI
|
|
37
|
Torii S, Yamamoto T, Tsuchiya Y and
Nishida E: ERK MAP kinase in G1 cell cycle progression and cancer.
Cancer Sci. 97:697–702. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lu Z and Xu S: ERK1/2 MAP kinases in cell
survival and apoptosis. IUBMB Life. 58:621–631. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Roberts PJ and Der CJ: Targeting the
Raf-MEK-ERK mitogen-activated protein kinase cascade for the
treatment of cancer. Oncogene. 26:3291–3310. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
McCubrey JA, Steelman LS, Chappell WH, et
al: Role of the RAF/MEK/ERK pathway in cell growth, malignant
transformation and drug resistance. Biochim Biophysic Acta.
1773:1263–1284. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Davies H, Bignell GR, Cox C, et al:
Mutations of the BRAF gene in human cancer. Nature. 417:949–954.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Capela G, Cronauer-Mitra S, Peinado MA and
Perucho M: Frequency and spectrum of mutations at codons 12 and 13
of the C-K-Ras gene in human tumors. Environ Health Perspect.
93:125–131. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Myakis S, Sourvinos G and Spandidos DA:
Differential expression and mutation of the ras family genes in
human breast cancer. Biochem Biophys Res Commun. 251:609–612. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Sánchez-Muñoz A, Gallego E, de Luque V, et
al: Lack of evidence for KRAS oncogenic mutations in
triple-negative breast cancer. BMC Cancer. 10:1362010.PubMed/NCBI
|
|
45
|
Karnoub AE and Weinberg RA: Ras oncogenes:
split personalities. Nat Rev Mol Cell Biol. 9:517–531. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Geradts J and Wilson PA: High frequency of
aberrant p16(INK4A) expression in human breast cancer. Am J Pathol.
149:15–20. 1996.PubMed/NCBI
|
|
47
|
Emig R, Magener A, Ehemann V, et al:
Aberrant cytoplasmic expression of the p16 protein in breast cancer
is associated with accelerated tumour proliferation. Br J Cancer.
78:1661–1668. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Di Vinci A, Perdelli L, Banelli B, et al:
p16INK4a promoter methylation and protein expression in breast
fibroadenoma and carcinoma. Int J Cancer. 114:414–421.
2005.PubMed/NCBI
|