Introduction
Cervical cancer is the third most common type of
cancer diagnosed and the fourth leading cause of female cancer
mortality worldwide, accounting for ~529,000 new cancer cases and
275,000 cancer deaths in 2008, >85% of which occurred in
developing countries (1). It is
widely accepted that infection by an oncogenic type of human
papillomavirus (HPV) is a necessary but not sufficient risk factor
for cervical carcinogenesis (2).
Various genetic and environmental factors may also play roles in
the pathogenesis of or predisposition to cervical cancer, as only a
small percentage of infected females develop the cancer (3). Evidence that first-degree relatives
of females with cervical cancer have a doubled risk of tumor
development distinctly indicates a strong genetic predisposition to
the disease (4). The first known
cervical cancer susceptibility gene was human leukocyte antigen
(HLA)-DQw3(5). With the
development of new technology and invention of new genomic tools, a
number of genes outside the HLA region have been examined in
patients with cervical cancer across populations of various
ethnicities. These include cytotoxic T-lymphocyte-associated
antigen-4 (CTLA-4) (6),
interleukin-10 (7) and cyclin D1
(8), among other genes.
CTLA-4, also known as cluster of
differentiation (CD)152, is a member of the immunoglobulin super
family and plays a crucial role in the negative regulation of
T-lymphocyte activation and proliferation, indirectly controlling
effector T cells (9). The
CTLA-4 gene is situated on chromosome 2q33, close to genes
of other regulatory molecules, including CD28 and inducible
costimulator (10). It consists of
four exons that encode separate functional domains: Leader
sequence, and extracellular, transmembrane and cytoplasmic domains
(11). In the early stages of
tumorigenesis, CTLA-4 may elevate the activation threshold
of T-cells, thereby weakening the antitumor response and increasing
susceptibility to cancer (12).
Studies have shown that CTLA-4 blockade results in
enhancement of the immune response (13), rejection (14) or in certain cases, cure of tumors
in mice treated with a combination of tumor vaccines (15). Moreover, recent studies have
demonstrated that CTLA-4 polymorphisms were associated with
susceptibility to cancer (16–19).
Thus, it is extremely likely that CTLA-4 polymorphisms are
involved in the pathogenesis of cervical cancer. The most
frequently studied CTLA-4 polymorphisms are +49 A/G
(rs231775), −318 C/T (rs5742909) and CT60 G/A (rs3087243) (6,20–22).
A number of studies have been previously performed to determine
whether these polymorphisms confer susceptibility to cervical
cancer in various populations (6,23–29),
however, results have been inconclusive. The discrepancy may result
from limitations of individual studies, publication bias (30) or yet-unknown effects of the
CTLA-4 molecule.
Therefore, with the aim of deriving a more precise
assessment of the correlation between CTLA-4 +49 A/G, −318
C/T and CT60 G/A polymorphisms and cervical cancer, a meta-analysis
of 15 published case-control studies was performed. To the best of
our knowledge, this is the most comprehensive evaluation method
with regards to the associations between CTLA-4
polymorphisms and cervical cancer risk.
Subjects and methods
Study identification and selection
To identify all the published studies that have
examined the association of CTLA-4 gene polymorphisms with
cervical cancer, the electronic databases of PubMed, Embase and the
Chinese Biomedical Database were searched, with the last search
update being performed on January 1, 2013. Combinations of
keywords, including ‘cytotoxic T-lymphocyte associated antigen-4’,
‘CTLA-4’, ‘CD152’, ‘polymorphism’ and ‘uterine cervical neoplasms’
were entered as Medical Subject Headings and text words. No
language restriction was applied. References of retrieved articles
were also screened. All of the eligible studies were case-control
in design, based on unrelated individuals and had available data
(distribution of alleles and genotypes for cases and controls) to
estimate the odds ratios (ORs) with the 95% confidence intervals
(CIs). Furthermore, genotype distributions in the control groups
were required to be in Hardy-Weinberg equilibrium (HWE), according
to an exact test.
Data extraction
Data were extracted by two reviewers independently.
A consensus was reached following discussion if there was
disagreement. The following information was extracted from each
eligible study: Author, year of publication, ethnicity of the study
population, sample size, genotyping method and genotype number in
cases and controls.
Statistical analysis
HWE was tested by an internet-based HWE calculator
(http://ihg.gsf.de/cgi-bin/hw/hwa1.pl;
accessed March 5, 2013). The strength of associations between the
CTLA-4 polymorphisms and cervical cancer risk were evaluated
by ORs with the corresponding 95% CIs. The genetic models that were
assessed for pooled ORs of these polymorphisms were dominant models
(GG+GA vs. AA for +49 A/G, TT+TC vs. CC for −318 C/T and AA+AG vs.
GG for CT60 G/A). For each CTLA-4 polymorphism, other
genetic models (+49 A/G, GG vs. GA+AA, GG vs. AA, GA vs. AA and G
vs. A; −318 C/T, TT vs. TC+CC, TT vs. CC, TC vs. CC and T vs. C;
and CT60 G/A, AA vs. AG+GG, AA vs. GG, AG vs. GG and A vs. G) were
used to evaluate the association with cervical cancer risk.
Statistical heterogeneity among studies was tested
with Cochran’s Q-statistic, where P<0.10 was considered to
indicate a statistically significant difference. The random-effects
model (DerSimonian-Laird method) or the fixed-effects model
(Mantel-Haenszel method) was used to summarize the combined OR
according to the heterogeneity. When P≥0.10, the fixed-effects
model was used to calculate pooled OR, whereas the random-effects
model was used if P<0.10. Significance of the pooled OR was
estimated using a Z-test. P<0.05 was considered to indicate a
statistically significant difference. Subgroup analyses were
performed by ethnic group for the +49 A/G and −318 C/T
polymorphisms.
Publication bias was checked by the Begg’s funnel
plot and Egger’s test (31). If
publication bias existed, the trim and fill method was applied to
adjust the results. Statistical manipulations were carried out
using Review Manager 5.0 (Cochrane Collaboration, 2008; www.cc-ims.net/RevMan; accessed March 20, 2013) and
Stata 12.0 software (StataCorp LP, College Station, TX, USA).
Results
Study selection and subject
characteristics
A total of eight relevant articles investigating
CTLA-4 polymorphisms (+49 A/G, −318 C/T and CT60 G/A) and
cervical cancer risk met the study inclusion criteria (6,23–29).
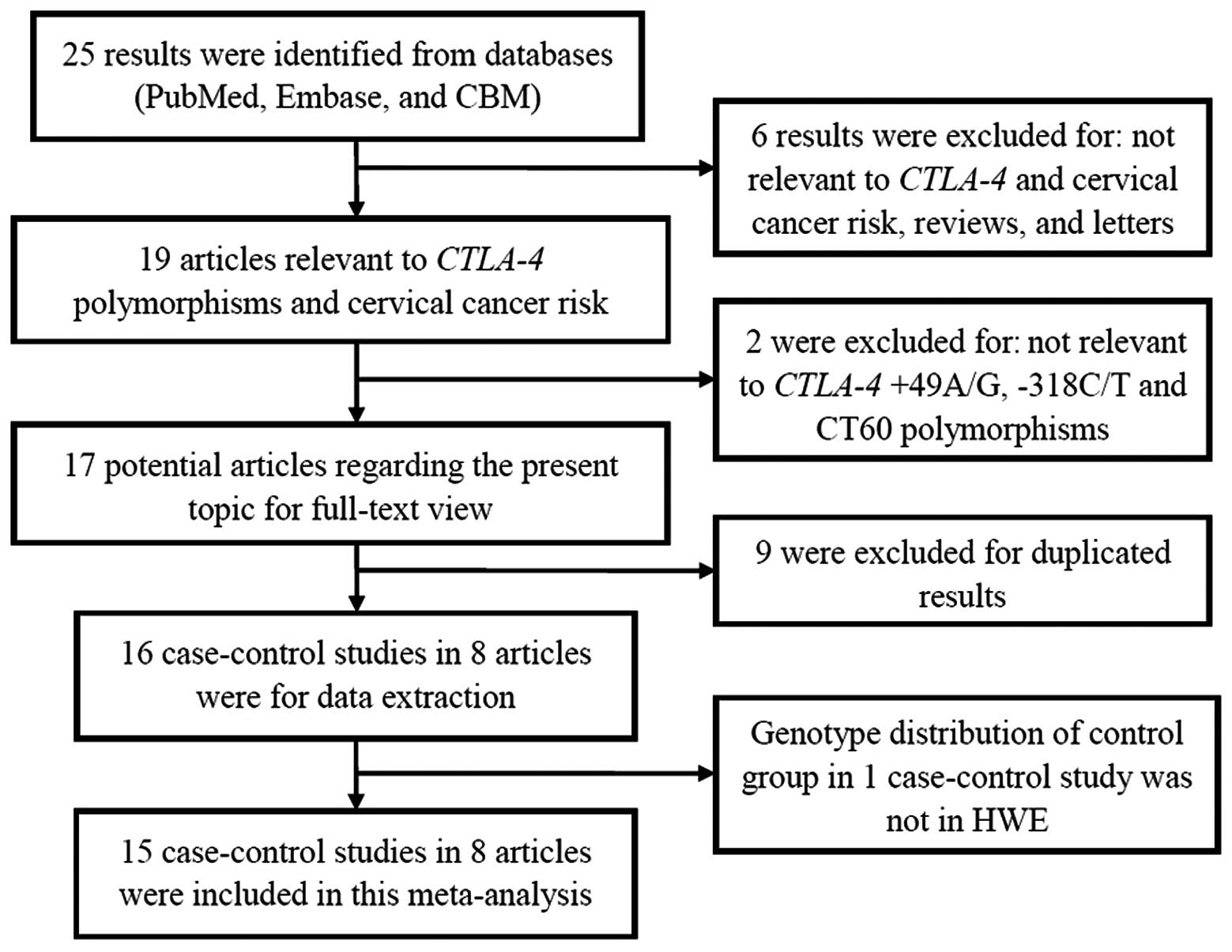
Fig. 1 shows the detailed
procedure for selecting eligible articles. The studies included in
the meta-analysis contained 3,684 cervical cancer cases and 4,110
controls. Among the eight articles, seven focused on the +49 A/G
polymorphism (6,23–25,27–29),
six on the −318 C/T polymorphism (6,24,26–29)
and two focused on the CT60 G/A polymorphism (27,29).
Furthermore, four articles were of Caucasian origin (6,26–28)
and four were from an Asian population (23–25,29).
In the eight articles, genomic DNA was extracted from peripheral
blood samples. For genotyping, various methods were used, including
restriction fragment length polymorphism, TaqMan,
amplification-refractory mutation system and multiplex polymerase
chain reaction with hybridization and Sequenom MassArray.
Characteristics of each article included in this meta-analysis are
summarized in Table I and the
genotype numbers are listed in Table
II.
 | Table ICharacteristics of the individual
studies included in the meta-analysis. |
Table I
Characteristics of the individual
studies included in the meta-analysis.
| Study | Year | Country
(ethnicity) | Sample size,
case/control | Genotype
method | Polymorphisms |
|---|
| Li et
al(23) | 2011 | China (Asians) | 314/320 | RFLP | +49 A/G |
| Jiang et
al(24) | 2011 | China (Asians) | 100/100 | Sequenom
MassArray | +49 A/G, −318
C/T |
| Rahimifar et
al(6) | 2010 | Iran
(Caucasians) | 55/110 | RFLP, PCR-ARMS | +49 A/G, −318
C/T |
| Hu et
al(25) | 2010 | China (Asians) | 696/709 | TaqMan | +49 A/G |
| Ivansson et
al(26) | 2010 | Sweden
(Caucasians) | 1,281/554 | TaqMan | −318 C/T |
| Pawlak et
al(27) | 2010 | Poland
(Caucasians) | 141/224 | RFLP | +49 A/G, −318 C/T,
CT60 G/A |
| Castro et
al(28) | 2009 | Sweden
(Caucasians) | 953/1,715 | Multiplex PCR with
hybridization | +49 A/G, −318
C/T |
| Su et
al(29) | 2007 | Taiwan
(Asians) | 144/378 | RFLP | +49 A/G, −318 C/T,
CT60 G/A |
 | Table IIDistribution of CTLA-4
polymorphism genotypes and alleles among cervical cancer patients
and controls. |
Table II
Distribution of CTLA-4
polymorphism genotypes and alleles among cervical cancer patients
and controls.
| A, Polymorphism +49
A/G |
|---|
|
|---|
| Case | Control | Case | Control | |
|---|
|
|
|
|
| |
|---|
| Study | AA | AG | GG | AA | AG | GG | G | A | G | A | HWE |
|---|
| Li et
al(23) | 30 | 144 | 140 | 18 | 129 | 173 | 424 | 204 | 475 | 165 | Yes |
| Jiang et
al(24) | 13 | 42 | 45 | 19 | 39 | 42 | 132 | 68 | 123 | 77 | Yes |
| Rahimifar et
al(6) | 28 | 27 | 0 | 58 | 45 | 7 | 27 | 83 | 59 | 161 | Yes |
| Hu et
al(25) | 80 | 290 | 326 | 56 | 300 | 353 | 942 | 450 | 1,006 | 412 | Yes |
| Pawlak et
al(27) | 43 | 72 | 26 | 71 | 103 | 43 | 124 | 158 | 189 | 245 | Yes |
| Castro et
al(28) | 252 | 449 | 252 | 456 | 825 | 434 | 953 | 953 | 1,693 | 1,737 | Yes |
| Su et
al(29) | 17 | 62 | 60 | 42 | 155 | 178 | 182 | 96 | 511 | 239 | Yes |
|
| B, Polymorphism
−318 C/T |
|
| Case | Control | Case | Control | |
|
|
|
|
| |
| Study | CC | TC | TT | CC | TC | TT | C | T | C | T | HWE |
|
| Jiang et
al(24) | 75 | 24 | 1 | 92 | 8 | 0 | 174 | 26 | 192 | 8 | Yes |
| Rahimifar et
al(6) | 51 | 3 | 0 | 89 | 20 | 1 | 105 | 3 | 198 | 22 | Yes |
| Ivansson et
al(26) | 1,044 | 228 | 9 | 458 | 92 | 4 | 2,316 | 246 | 1,008 | 100 | Yes |
| Pawlak et
al(27) | 99 | 38 | 3 | 180 | 35 | 1 | 236 | 44 | 395 | 37 | Yes |
| Castro et
al(28) | 5 | 124 | 819 | 6 | 223 | 1,471 | 134 | 1,762 | 235 | 3,165 | Yes |
| Su et
al(29) | 105 | 38 | 1 | 306 | 67 | 5 | 248 | 40 | 679 | 77 | Yes |
|
| C, Polymorphism
CT60 G/A |
|
| Case | Control | Case | Control | |
|
|
|
|
| |
| Study | GG | GA | AA | GG | GA | AA | G | A | G | A | HWE |
|
| Pawlak et
al(27) | 41 | 58 | 15 | 77 | 104 | 43 | 140 | 88 | 258 | 190 | Yes |
| Su et
al(29) | 87 | 45 | 7 | 238 | 123 | 17 | 219 | 59 | 599 | 157 | Yes |
Quantitative synthesis
+49 A/G polymorphism
In total, 2,398 cervical cancer cases and 3,546
controls from 7 case-control studies were included in the
meta-analysis of the association between CTLA-4 +49 A/G
polymorphism and cervical cancer (6,23–25,27–29).
Of these, 4 case-control studies were from an Asian population
(23–25,29)
and 3 from a Caucasian population (6,27,28).
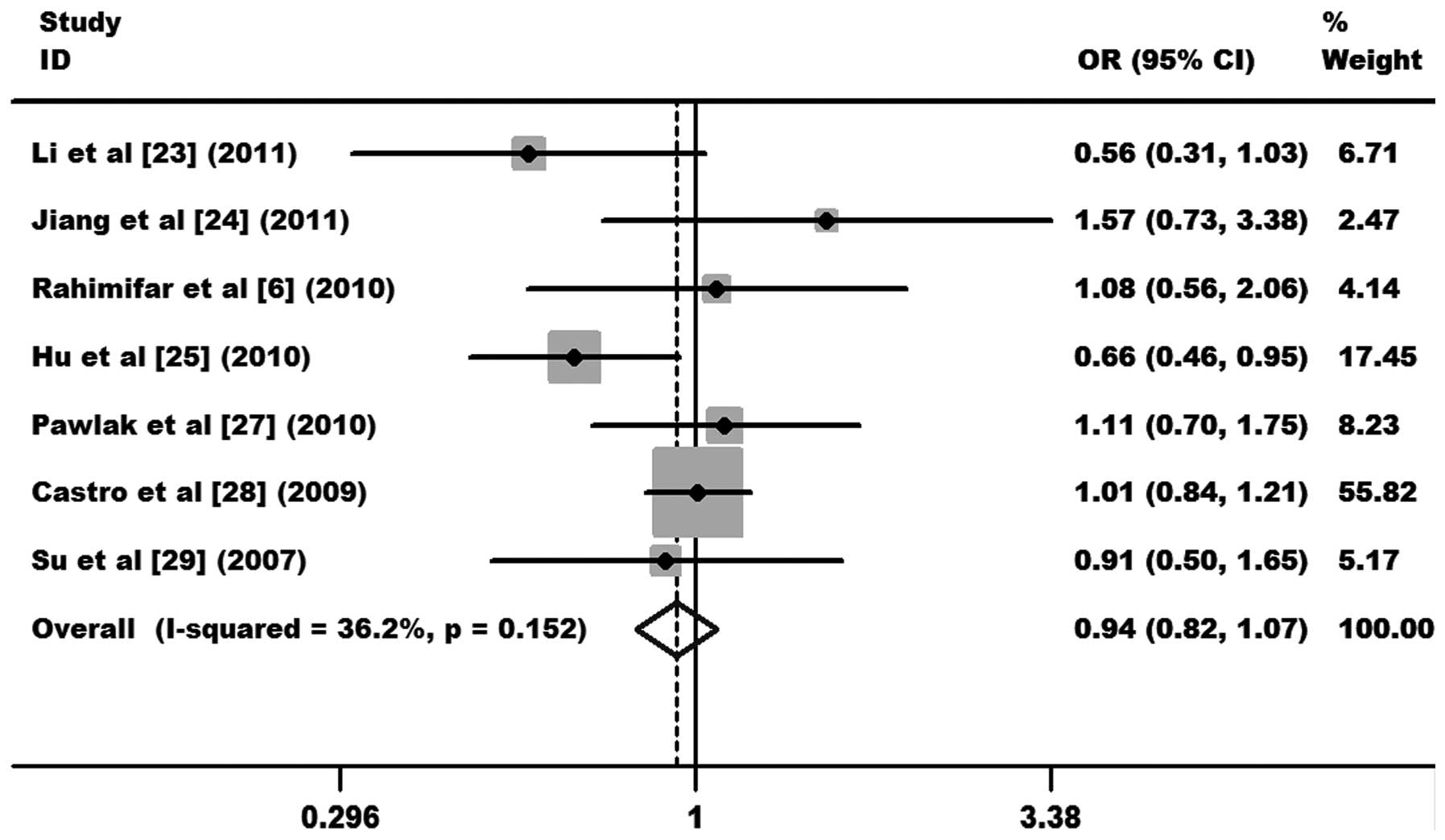
As shown in Fig. 2, the
heterogeneity of GG+AG vs. AA was tested for the 7 case-control
studies. Data from the meta-analysis were as follows:
χ2, 9.41, degrees of freedom (df), 6 and P=0.152 in a
fixed-effects model. I2 value, an additional
heterogeneity indicator, was 36.2%, indicating low heterogeneity.
Therefore, the fixed-effects model was used for the synthesis of
data. The results obtained showed no association between overall
cervical cancer risk and the CTLA-4 +49 A/G polymorphism
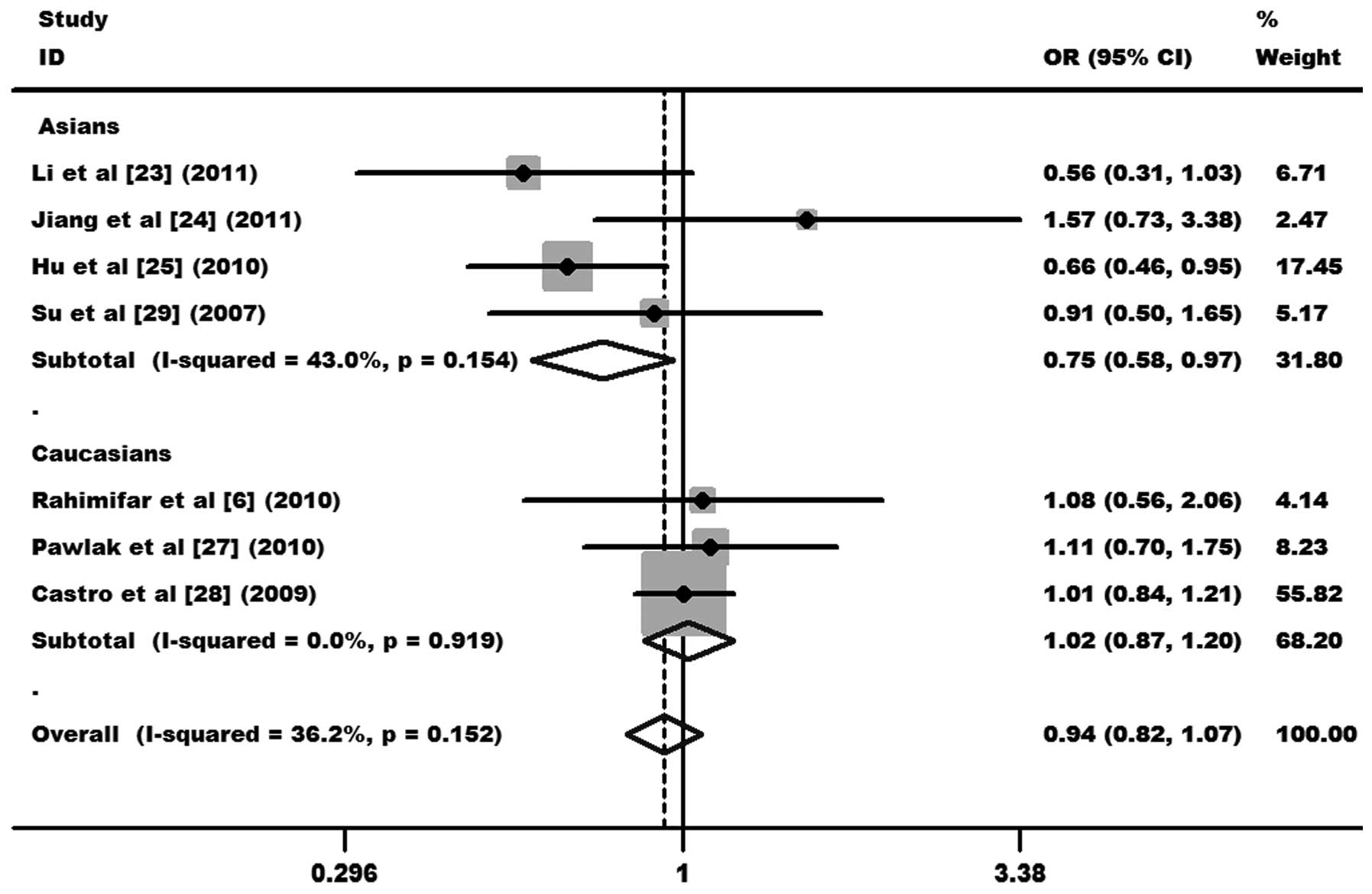
(OR, 0.94, 95% CI, 0.82–1.07 and P=0.349 for GG+AG vs. AA; Fig. 2). In subgroup analysis,
significantly decreased cervical cancer risks were found in Asians
(OR, 0.75, 95% CI, 0.58–0.97 and P=0.028 for GG+AG vs. AA), but not
in Caucasians (OR, 1.02, 95% CI, 0.87–1.20 and P=0.775 for GG+AG
vs. AA; Fig. 3). Other comparison
results are listed in Table III.
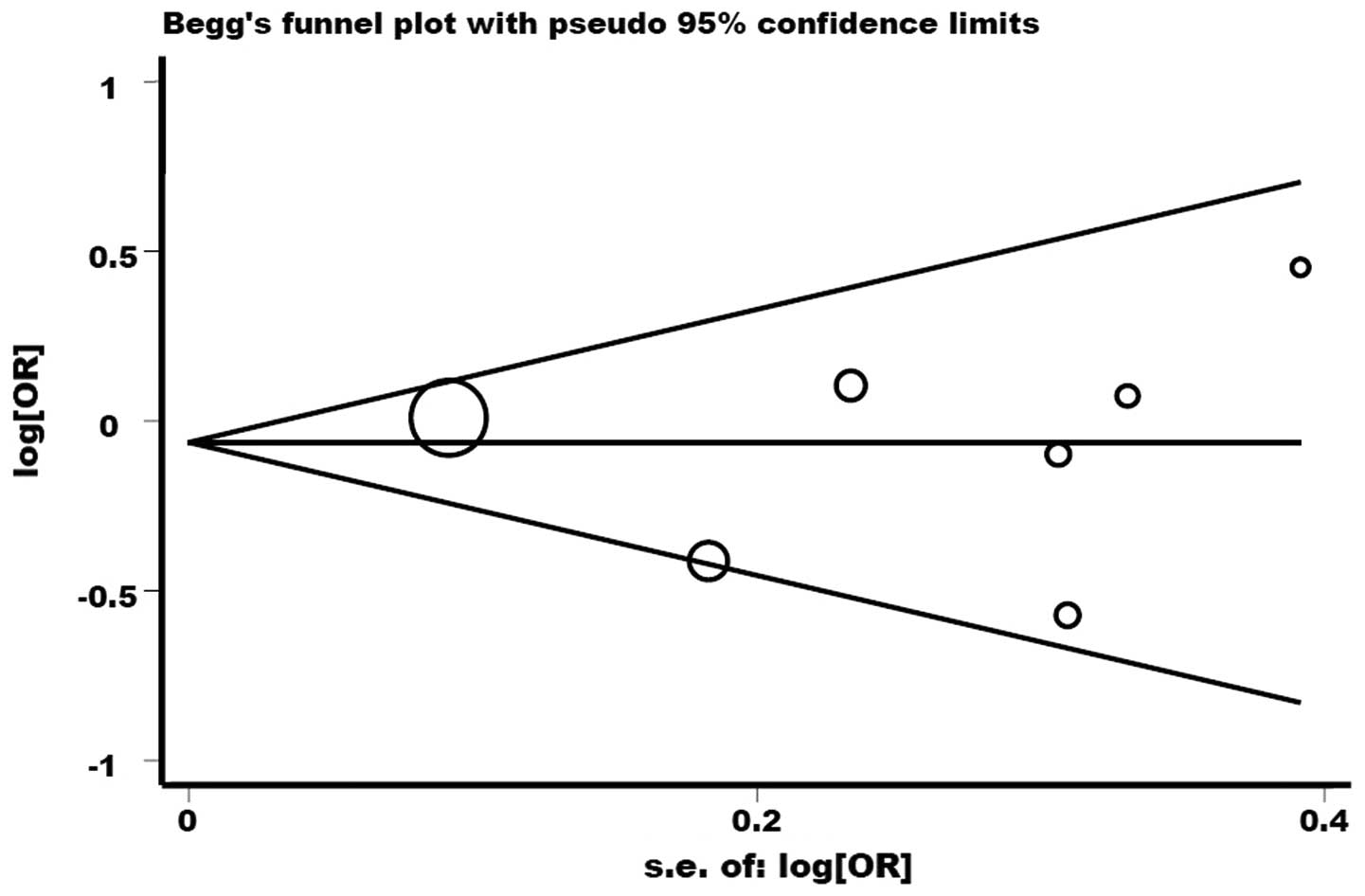
No publication bias was detected by Begg’s funnel plot or Egger’s
test (P>|t|=0.809; Fig. 8).
 | Table IIISummary of results of the
meta-analysis from various comparative genetic models. |
Table III
Summary of results of the
meta-analysis from various comparative genetic models.
| A, Polymorphism +49
A/G |
|---|
|
|---|
| | | Test of
heterogeneity |
|---|
| | |
|
|---|
| Genetic
comparison | Population | OR (95% CI);
P-value | P-value;
I2 (%) | Model |
|---|
| GG+AG vs. AA | All | 0.94 (0.82–1.07);
0.349 | 0.152; 36.2 | F |
| Asian | 0.75 (0.58–0.97);
0.028 | 0.154; 43.0 | F |
| Caucasian | 1.02 (0.87–1.20);
0.775 | 0.919; 0.0 | F |
| GG vs. AA+AG | All | 0.92 (0.82–1.03);
0.154 | 0.201; 29.8 | F |
| Asian | 0.84 (0.72–0.98);
0.028 | 0.389; 0.6 | F |
| Caucasian | 1.03 (0.87–1.22);
0.754 | 0.307; 15.3 | F |
| GG vs. AA | All | 0.84 (0.62–1.12);
0.234 | 0.057; 51.0 | R |
| Asian | 0.71 (0.54–0.92);
0.011 | 0.141; 45.1 | F |
| Caucasian | 1.02 (0.83–1.25);
0.833 | 0.384; 0.0 | F |
| AG vs. AA | All | 0.95 (0.82–1.10);
0.482 | 0.310; 15.7 | F |
| Asian | 0.79 (0.61–1.04);
0.093 | 0.257; 25.7 | F |
| Caucasian | 1.02 (0.86–1.21);
0.812 | 0.695; 0.0 | F |
| G vs. A | All | 0.94 (0.87–1.02);
0.133 | 0.120; 40.6 | F |
| Asian | 0.85 (0.76–0.96);
0.007 | 0.190; 37.0 | F |
| Caucasian | 1.02 (0.92–1.13);
0.716 | 0.870; 0.0 | F |
|
| B, Polymorphism
−318 C/T |
|
| | | Test of
heterogeneity |
| | |
|
| Genetic
comparison | Population | OR (95% CI);
P-value | P-value;
I2 (%) | Model |
|
| TT+TC vs. CC | All | 1.33 (0.82–2.16);
0.249 | 0.001; 74.8 | R |
| Asian | 2.28 (0.97–5.38);
0.060 | 0.070; 69.5 | R |
| Caucasian | 0.97 (0.51–1.85);
0.930 | 0.008; 74.6 | R |
| TT vs. TC+CC | All | 1.00 (0.80–1.25);
0.982 | 0.752; 0.0 | F |
| Asian | 0.90 (0.18–4.45);
0.901 | 0.373; 0.0 | F |
| Caucasian | 1.00 (0.80–1.26);
0.967 | 0.603; 0.0 | F |
| TT vs. CC | All | 1.04 (0.53–2.03);
0.917 | 0.613; 0.0 | F |
| Asian | 1.05 (0.21–5.11);
0.955 | 0.351; 0.0 | F |
| Caucasian | 1.03 (0.49–2.18);
0.929 | 0.441; 0.0 | F |
| TC vs. CC | All | 1.34 (0.83–2.15);
0.232 | 0.003; 72.7 | R |
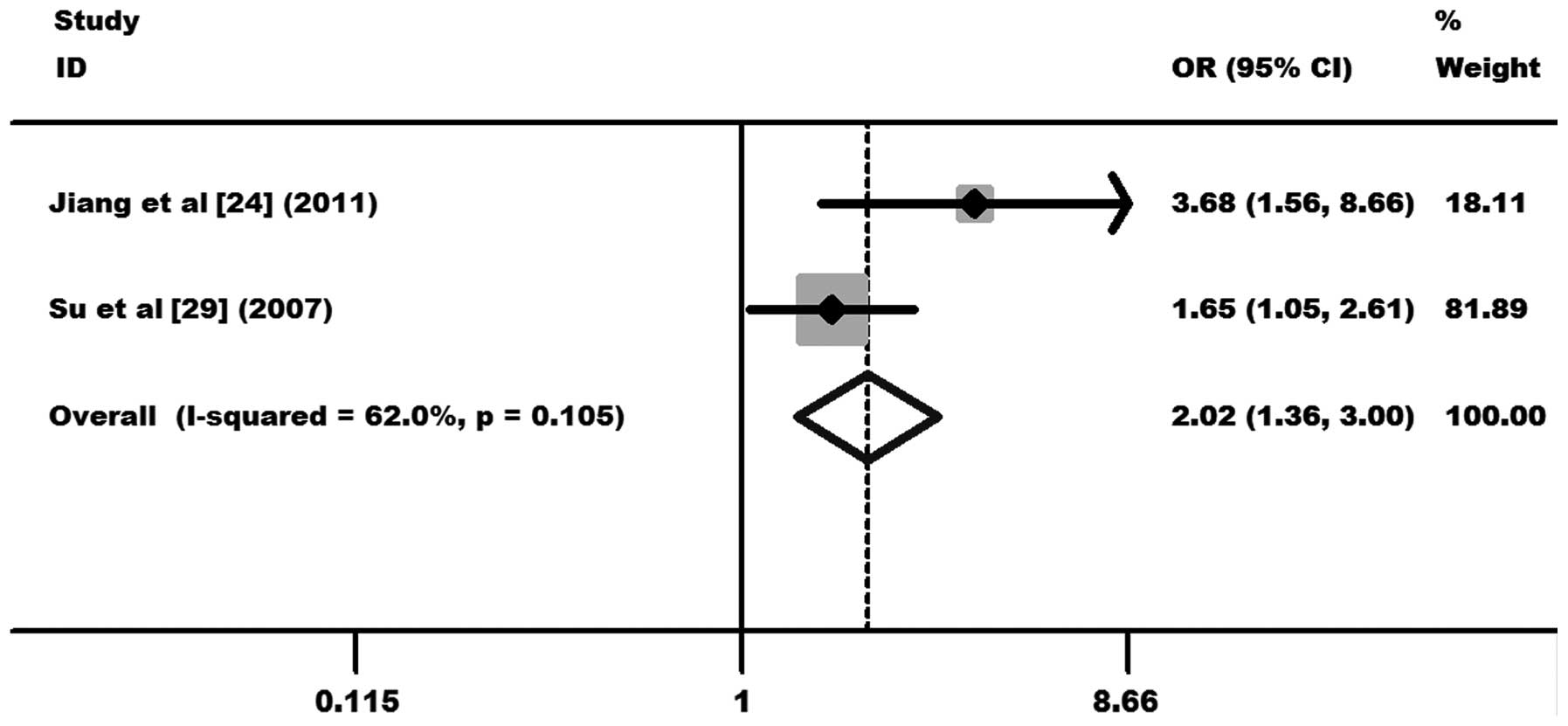
| Asian | 2.02 (1.36–3.00);
0.000 | 0.105; 62.0 | F |
| Caucasian | 0.98 (0.53–1.81);
0.950 | 0.015; 71.2 | R |
| T vs. C | All | 1.29 (0.90–1.83);
0.161 | 0.001; 77.2 | R |
| Asian | 2.11 (0.86–5.18);
0.105 | 0.047; 74.7 | R |
| Caucasian | 1.08 (0.74–1.59);
0.687 | 0.005; 76.3 | R |
|
| C, Polymorphism
CT60 G/A |
|
| | | Test of
heterogeneity |
| | |
|
| Genetic
comparison | Population | OR (95% CI);
P-value | P-value;
I2 (%) | Model |
|
| AA+AG vs. GG | All | 0.98 (0.72–1.33);
0.898 | 0.786; 0.0 | F |
| AA vs. AG+GG | All | 0.76 (0.45–1.28);
0.308 | 0.313; 1.9 | F |
| AA vs. GG | All | 0.80 (0.46–1.39);
0.420 | 0.356; 0.0 | F |
| AG vs. GG | All | 1.02 (0.74–1.41);
0.904 | 0.891; 0.0 | F |
| A vs. G | All | 0.93 (0.74–1.18);
0.565 | 0.437; 0.0 | F |
−318 C/T polymorphism
A total of 2,667 cases and 3,058 controls from 6
case-control studies were included in the meta-analysis of the
correlation between the −318 C/T polymorphism and cervical cancer
(6,24,26–29).
Two case-control studies were from an Asian population (24,29)
and 4 from a Caucasian population (6,26–28).
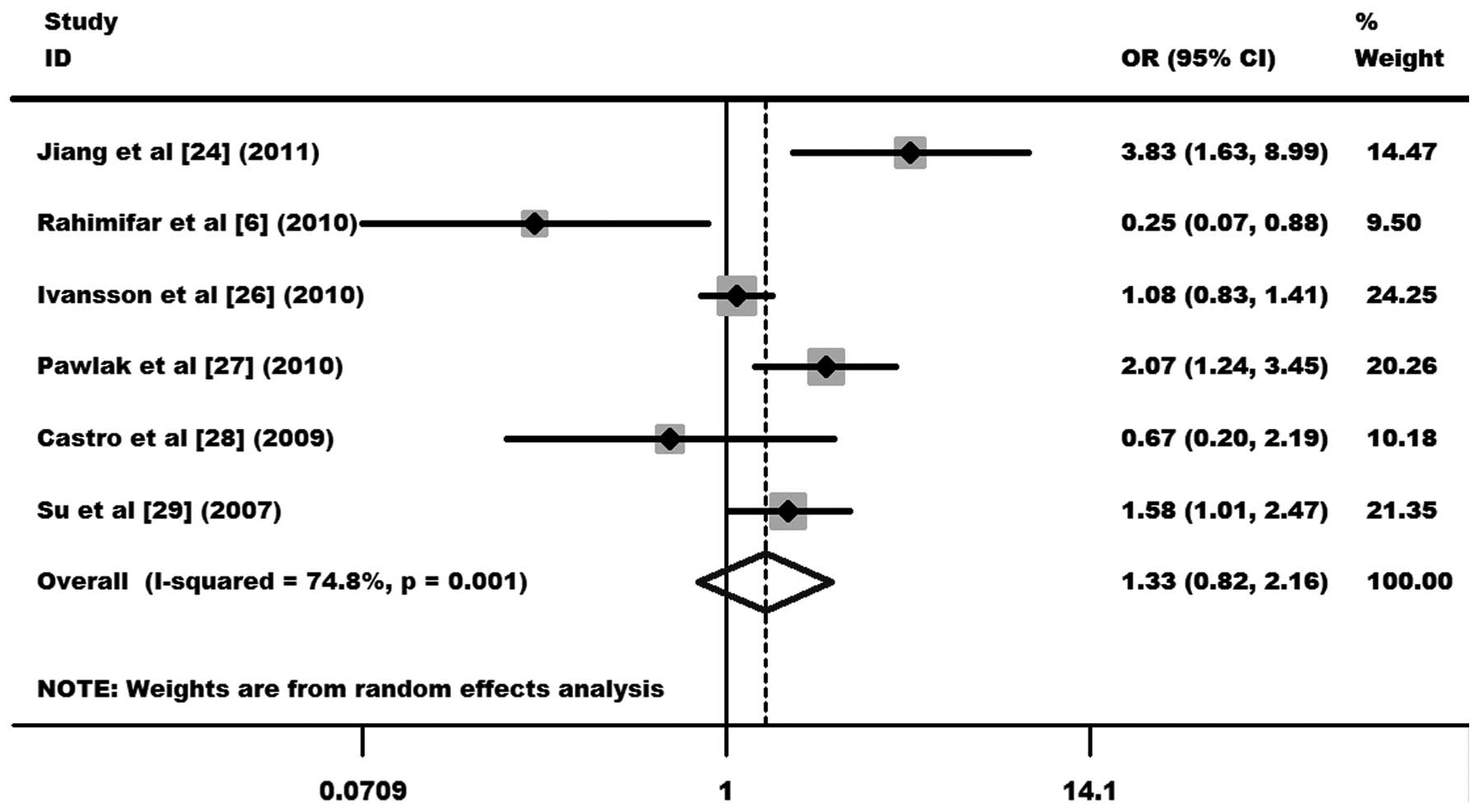
As shown in Fig. 4, the
heterogeneity of TT+TC vs. CC was evaluated for the 6 case-control
studies. Data from the meta-analysis were as follows:
χ2, 19.82, df, 5 and P=0.001 in a random-effects model.
Additionally, I2 value was 74.8%, indicating moderated
heterogeneity. Thus, the random-effects model was used for the
synthesis of data. No statistical evidence of an association
between the −318 C/T polymorphism and cervical cancer risks (OR,
1.33, 95% CI, 0.82–2.16 and P=0.249 for TT+TC vs. CC; Fig. 4) was observed. In the subgroup
analysis, significantly increased cervical cancer risks were
observed in the Asian population (OR, 2.02, 95% CI, 1.36–3.00 and
P=0.000 for TC vs. CC; Fig. 5),
but not in the Caucasian population (OR, 0.98, 95% CI, 0.53–1.81
and P=0.950 for TC vs. CC; Fig.
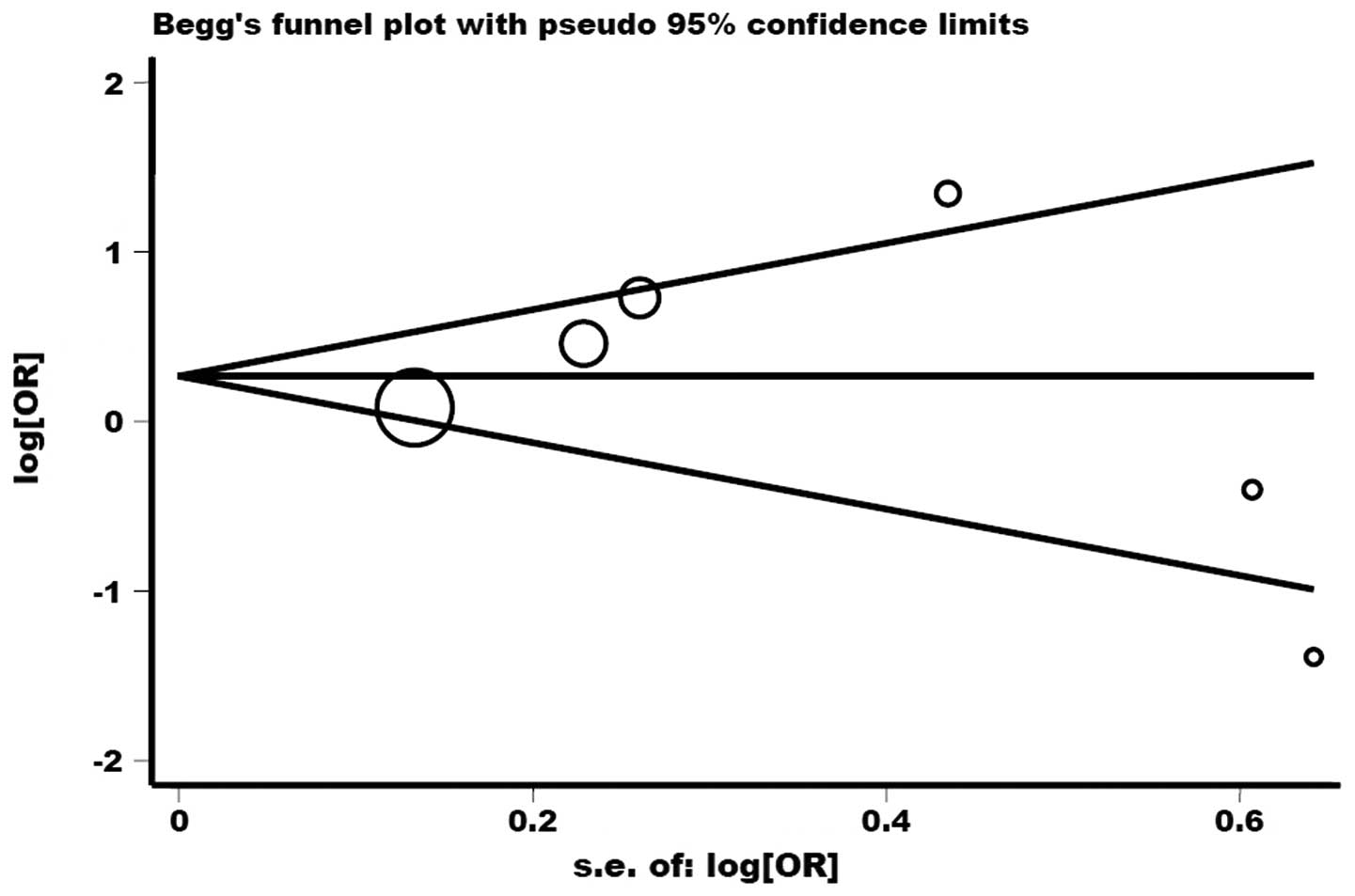
6). Other comparison results are listed in Table III. No publication bias was
detected by Begg’s funnel plot or Egger’s test (P>|t|=0.989;
Fig. 9).
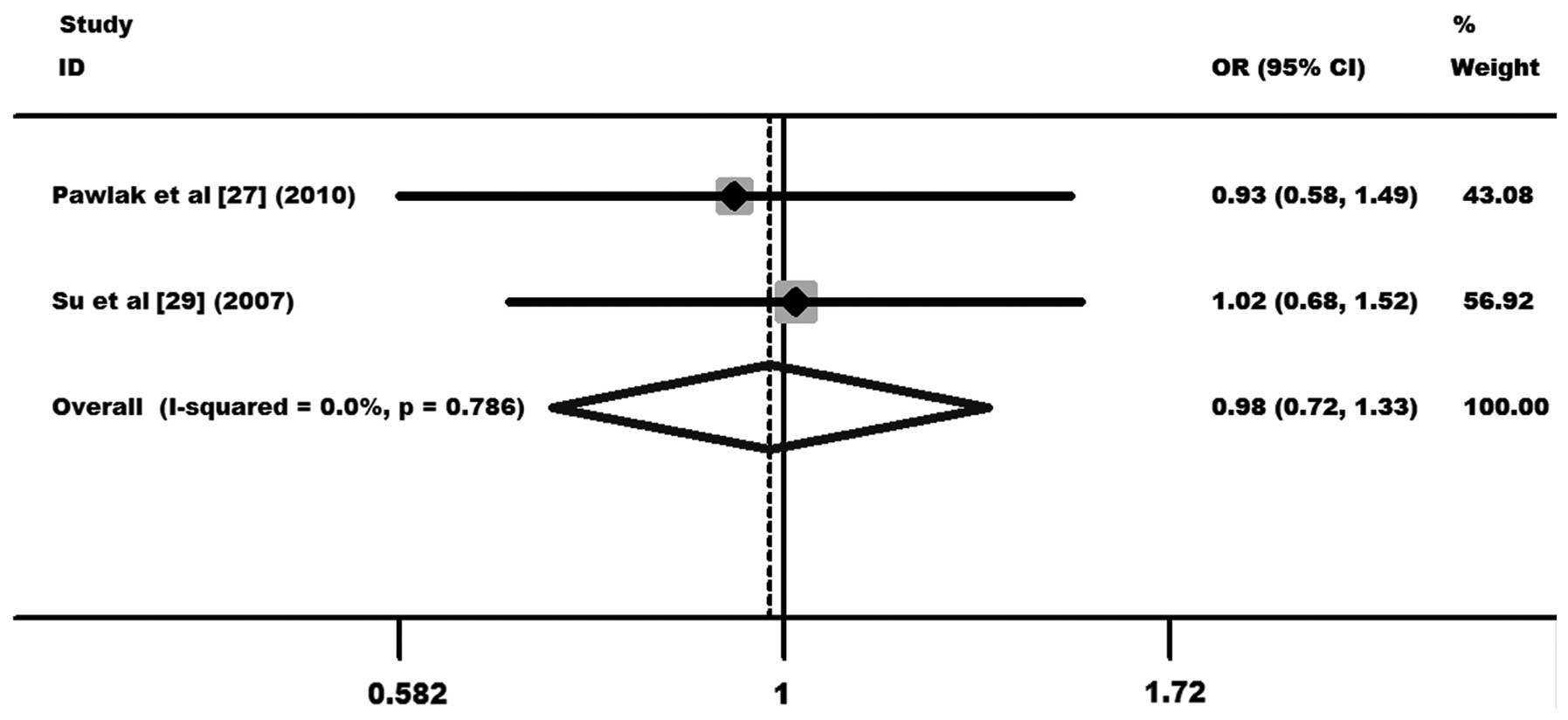
CT60 G/A polymorphism
A total of 253 cases and 602 controls from 2
case-control studies were included in the meta-analysis of the
correlation between the CT60 G/A polymorphism and cervical cancer
(27,29). One case-control study was from a
Caucasian population (27) and the
other from an Asian population (29). The results showed no statistical
evidence of an association between the CT60 G/A polymorphism and
the overall cervical cancer risk (OR, 0.98, 95% CI, 0.72–1.33 and
P=0.898 for AA+GA vs. GG; Fig. 7).
Further subgroup analysis was not performed due to the limited data
for this polymorphism. Other comparison results are listed in
Table III.
Discussion
Cervical cancer is a complex malignant tumor that
affects female reproductive organs and has a number of genetic and
environmental determinants. Specific oncogenic HPV is an important
etiologic agent in cervical cancer. However, HPV infection alone is
insufficient in inducing malignant changes (2). Host genetic factors may be important
in cervical cancer susceptibility. CTLA-4, one of the key
mediators for inhibiting activated T-lymphocytes, plays a pivotal
role in cancer immunosurveillance (13). Considering the importance of
CTLA-4, variations in this gene may affect the risk of
developing cervical cancer. In addition, the effect of gene
polymorphisms involved in tumorigenesis or susceptibility to
cervical cancer has gained increasing interest in previous years.
Specific studies have reported an association between genetic risk
factors and cervical cancer (6–8,23,24,32).
Rahimifar et al(6) observed
that at the −318 locus in CTLA-4, higher C allele frequency,
as well as increased frequency of −318 CC genotype, was found in
patients with cervical cancer when compared with controls. However,
this association was from a study with a small sample size and was
restricted to females in Iran. Li et al(23) showed that Chinese females with the
+49 AA genotype have a 2.06-fold higher risk of developing cervical
cancer compared with GG carriers. In addition, Jiang et
al(24) observed that a
single-nucleotide polymorphism in the promoter region of the
CTLA-4 gene may increase susceptibility to cervical cancer.
However, negative results were also obtained in certain studies,
due to conflicting observations and the limited sample size of
individual studies. Alternatively, meta-analysis is a strategy to
reduce the limitations of individual studies and is often applied
in genetic association studies. Thus, meta-analysis was used in the
present study to assess whether an association exists between the
most commonly studied polymorphisms of the CTLA-4 gene, +49
A/G, −318 C/T and CT60 G/A and the risk of developing cervical
cancer.
A total of 15 case-control studies from 8 articles,
comprising 3,684 cervical cancer cases and 4,110 controls, were
included in the meta-analysis. The results indicated no association
of CTLA-4 +49 A/G, −318 C/T and CT60 G/A polymorphisms with
overall cervical cancer risk. Furthermore, stratification by
ethnicity showed that Asian individuals with GG/AG genotypes had a
significantly decreased cervical cancer risk compared with AA
carriers for the +49 A/G polymorphism. By contrast, for the −318
C/T polymorphism, an increased cervical cancer risk was observed
for the TC genotype, compared with CC carriers. However, the
results should be interpreted with caution, as only two
case-control studies were included in an Asian population for the
−318 C/T polymorphism, which may have limited the statistical power
to reveal a reliable association. Therefore, future studies are
required to validate the association. The present meta-analysis
found that the +49 A/G polymorphism correlated with a decreased
risk for cervical cancer among Asian but not Caucasian individuals,
while the −318 C/T polymorphism correlated with an increased risk
among Asian but not Caucasian individuals. These observations
suggest that interactions between genetic diversity in various
ethnicities and genetic variants may contribute to various risks of
cervical cancer.
Since Su et al(29) reported that the −318 C/T variant in
the promoter region of the CTLA-4 gene was associated with
HPV-16-associated cervical squamous cell carcinoma in Taiwanese
females in 2007, more studies have focused on the association
between CTLA-4 polymorphisms and cervical cancer. However,
certain results have been conflicting. In the current
meta-analysis, eight eligible articles published up to January 1,
2013 were considered, comprising a total of 7,794 subjects. Thus,
the statistical analysis of the present study may provide more
powerful evidence of an association. Moreover, it was found that
the CTLA-4 +49 A/G and −318 C/T polymorphisms may play
various roles in cervical cancer susceptibility across various
populations, indicating that the associations may be
ethnicity-specific. In the future, a large number of studies are
required to analyze these associations in diverse ethnicities.
Heterogeneity and publication bias are two important
issues that should be addressed, as they may have affected the
results of the meta-analysis. Heterogeneity was observed between
studies for the +49 A/G and −318 C/T polymorphisms, in overall
comparisons in the dominant model. However, when stratification by
ethnicity was employed, heterogeneity decreased or was removed in
specific subgroups, indicating various roles for genetic
backgrounds, even for the same polymorphism. Significant
publication bias was not detected for the three polymorphisms,
indicating the reliability of the results from this
meta-analysis.
To a certain extent, several limitations may have
affected the results of the present study and should be considered
when interpreting the results. Firstly, the limited study sample
size of certain participants may have weakened the statistical
power to evaluate the association between CTLA-4
polymorphisms and cervical cancer. Secondly, the number of studies
included in the meta-analysis was relatively small, which prevented
further subgroup analysis for the CT60 G/A polymorphism.
Furthermore, on account of the small amount of data available for
each included study, it was not possible to conduct a subgroup
analysis by other covariates, including HPV subtype and status,
grade of differentiation of cervical cancer, lifestyle and
environmental factors. Some of these variables are documented as
important risk factors for cervical cancer (33). Thirdly, since all studies for data
analysis were from Asian and Caucasian populations, the results may
only be applicable to these two ethnic groups. Additionally,
eligible articles were identified from the selected databases;
thus, specific published articles concerning the current topic or
unpublished articles which had negative observations were missed,
which may have distorted the analysis.
In conclusion, in spite of the several
aforementioned limitations, results from the meta-analysis suggest
that the CTLA-4 +49 A/G and −318 C/T polymorphisms, but not
the CT60 G/A polymorphism, may be risk factors for cervical cancer.
In the future, more intensive studies based on various ethnicities
are required to further the understanding of gene-gene and
gene-environment interactions between CTLA-4 polymorphisms
and cervical cancer risk.
Acknowledgements
This work was funded by the National Natural Science
Foundation of China (31170857, 30972783; http://www.nsfc.gov.cn/Portal0/default166.htm).
The authors wish to thank Dr Haiyong Gu for technical guidance.
Abbreviations:
|
CI
|
confidence interval
|
|
OR
|
odds ratio
|
|
CTLA-4
|
cytotoxic T-lymphocyte associated
antigen-4
|
|
HWE
|
Hardy-Weinberg equilibrium
|
|
HPV
|
human papillomavirus
|
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar
|
|
2
|
Stanley M: HPV - immune response to
infection and vaccination. Infect Agent Cancer. 5:192010.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Moore EE, Wark JD, Hopper JL, Erbas B and
Garland SM; CeCaGeEn Study Group. The roles of genetic and
environmental factors on risk of cervical cancer: a review of
classical twin studies. Twin Res Hum Genet. 15:79–86. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zoodsma M, Sijmons RH, de Vries EG and Zee
AG: Familial cervical cancer: case reports, review and clinical
implications. Hered Cancer Clin Pract. 2:99–105. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wank R and Thomssen C: High risk of
squamous cell carcinoma of the cervix for women with HLA-DQw3.
Nature. 352:723–725. 1991. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rahimifar S, Erfani N, Sarraf Z and
Ghaderi A: ctla-4 gene variations may influence cervical cancer
susceptibility. Gynecol Oncol. 119:136–139. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Matsumoto K, Oki A, Satoh T, et al:
Interleukin-10 -1082 gene polymorphism and susceptibility to
cervical cancer among Japanese women. Jpn J Clin Oncol.
40:1113–1116. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Satinder K, Chander SR, Pushpinder K, Indu
G and Veena J: Cyclin D1 (G870A) polymorphism and risk of cervix
cancer: a case control study in north Indian population. Mol Cell
Biochem. 315:151–157. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Teft WA, Kirchhof MG and Madrenas J: A
molecular perspective of CTLA-4 function. Annu Rev Immunol.
24:65–97. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Buonavista N, Balzano C, Pontarotti P, Le
Paslier D and Golstein P: Molecular linkage of the human CTLA4 and
CD28 Ig-superfamily genes in yeast artificial chromosomes.
Genomics. 13:856–861. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ligers A, Teleshova N, Masterman T, Huang
WX and Hillert J: CTLA-4 gene expression is influenced by promoter
and exon 1 polymorphisms. Genes Immun. 2:145–152. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Egen JG, Kuhns MS and Allison JP: CTLA-4:
new insights into its biological function and use in tumor
immunotherapy. Nat Immunol. 3:611–618. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Leach DR, Krummel MF and Allison JP:
Enhancement of antitumor immunity by CTLA-4 blockade. Science.
271:1734–1736. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hurwitz AA, Foster BA, Kwon ED, et al:
Combination immunotherapy of primary prostate cancer in a
transgenic mouse model using CTLA-4 blockade. Cancer Res.
60:2444–2448. 2000.PubMed/NCBI
|
|
15
|
Curran MA and Allison JP: Tumor vaccines
expressing flt3 ligand synergize with ctla-4 blockade to reject
preimplanted tumors. Cancer Res. 69:7747–7755. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hou R, Cao B, Chen Z, Li Y, et al:
Association of cytotoxic T lymphocyte-associated antigen-4 gene
haplotype with the susceptibility to gastric cancer. Mol Biol Rep.
37:515–520. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sun T, Zhou Y, Yang M, et al: Functional
genetic variations in cytotoxic T-lymphocyte antigen 4 and
susceptibility to multiple types of cancer. Cancer Res.
68:7025–7034. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lang C, Chen L and Li S: Cytotoxic
T-lymphocyte antigen-4 +49G/A polymorphism and susceptibility to
pancreatic cancer. DNA Cell Biol. 31:683–687. 2012.
|
|
19
|
Karabon L, Pawlak-Adamska E, Tomkiewicz A,
et al: Variations in suppressor molecule ctla-4 gene are related to
susceptibility to multiple myeloma in a polish population. Pathol
Oncol Res. 18:219–226. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Karabon L, Pawlak E, Tomkiewicz A, et al:
CTLA-4, CD28, and ICOS gene polymorphism associations with
non-small-cell lung cancer. Hum Immunol. 72:947–954. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kamesh L, Heward JM, Williams JM, et al:
CT60 and +49 polymorphisms of CTLA 4 are associated with
ANCA-positive small vessel vasculitis. Rheumatology (Oxford).
48:1502–1505. 2009.
|
|
22
|
Heidari A, Noori Daloii MR, Keramatipour
M, Rashikinezhad A, Sahmani AA and Amirzargar AA: CTLA-4 gene
polymorphisms (−318C/T, +49A/G, +6230A/G) in Iranian patients with
multiple sclerosis. Iran J Allergy Asthma Immunol. 9:219–223.
2010.
|
|
23
|
Li H, Zhou YF, Guo HY, Sun T, Zhang WH and
Lin DX: Association between CTLA-4 gene polymorphism and
susceptibility to cervical cancer. Zhonghua Zhong Liu Za Zhi.
33:681–684. 2011.(In Chinese).
|
|
24
|
Jiang L, Luo RY, Zhang W, Wang LR, Wang F
and Cheng YX: Single nucleotide polymorphisms of CTLA4 gene and
their association with human cervical cancer. Zhonghua Yi Xue Yi
Chuan Xue Za Zhi. 28:313–317. 2011.(In Chinese).
|
|
25
|
Hu L, Liu J, Chen X, et al: CTLA-4 gene
polymorphism +49 A/G contributes to genetic susceptibility to two
infection-related cancers - hepatocellular carcinoma and cervical
cancer. Hum Immunol. 71:888–891. 2010.
|
|
26
|
Ivansson EL, Juko-Pecirep I and Gyllensten
UB: Interaction of immunological genes on chromosome 2q33 and IFNG
in susceptibility to cervical cancer. Gynecol Oncol. 116:544–548.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pawlak E, Karabon L, Wlodarska-Polinska I,
et al: Influence of CTLA-4/CD28/ICOS gene polymorphisms on the
susceptibility to cervical squamous cell carcinoma and stage of
differentiation in the Polish population. Hum Immunol. 71:195–200.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Castro FA, Haimila K, Sareneva I, et al:
Association of HLA-DRB1, interleukin-6 and cyclin D1 polymorphisms
with cervical cancer in the Swedish population - a candidate gene
approach. Int J Cancer. 125:1851–1858. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Su TH, Chang TY, Lee YJ, et al: CTLA-4
gene and susceptibility to human papillomavirus-16-associated
cervical squamous cell carcinoma in Taiwanese women.
Carcinogenesis. 28:1237–1240. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Vriens D, de Wilt JH, van der Wilt GJ,
Netea-Maier RT, Oyen WJ and de Geus-Oei LF: The role of
[18F]-2-fluoro-2-deoxy-d-glucose-positron emission
tomography in thyroid nodules with indeterminate fine-needle
aspiration biopsy: systematic review and meta-analysis of the
literature. Cancer. 117:4582–4594. 2011.
|
|
31
|
Pereira TV, Rudnicki M, Pereira AC,
Pombo-de-Oliveira MS and Franco RF: 5,10-Methylenetetrahydrofolate
reductase polymorphisms and acute lymphoblastic leukemia risk: a
meta-analysis. Cancer Epidemiol Biomarkers Prev. 15:1956–1963.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sousa H, Santos AM, Pinto D and Medeiros
R: Is the p53 codon 72 polymorphism a key biomarker for cervical
cancer development? A meta-analysis review within European
populations. Int J Mol Med. 20:731–741. 2007.PubMed/NCBI
|
|
33
|
de Freitas AC, Gurgel AP, Chagas BS,
Coimbra EC and do Amaral CM: Susceptibility to cervical cancer: an
overview. Gynecol Oncol. 126:304–311. 2012.
|