Introduction
Hepatocellular carcinoma (HCC) is the third leading
cause of cancer-associated mortality worldwide, responsible for
~600,000 mortalities annually (1,2). The
majority of HCC cases occur in developing countries, with >50%
in China alone (3,4), and the burden of this cancer is
expected to increase further in coming years (5). To date, metastasis and recurrence are
major causes of the high mortality rate and poor prognosis of
tumors, particularly HCC.
Surgery remains the primary treatment for HCC, while
the incidence of metastatic recurrence following surgical resection
remains high (6). Thus, metastatic
recurrence is hypothesized to be the primary barrier to the
improvement of HCC treatment efficacy (1). Recently, several proteins that are
associated with the metastatic recurrence of HCC were reported
(7,8). However, the specific and sensitive
biomarkers remain to be elucidated.
In the current study, two human HCC cell lines,
HCCLM9 and MHCC97L, with high and low metastatic potentials,
respectively, were used to screen differentially expressed
proteins. These two cell lines were repeatedly selected in
vivo, and were maintained in a similar genetic background with
different metastatic behaviors (9–11),
providing an ideal cell model for evaluating the novel biomarker of
HCC metastasis in vitro (12,13).
The comparative proteomics approach provides a
powerful tool to simultaneously analyze hundreds of proteins that
are expressed in different cell and tissue samples. This approach
is capable of identifying cancer-associated proteins for
therapeutic intervention and establishing biomarkers for early
diagnosis (14,15).
In the present study, proteins from MHCC97L and
HCCLM9 cells were extracted and analyzed by sodium dodecyl
sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) coupled with
mass spectra (MS) technology. The aim of the present study was to
identify a novel candidate biomarker for HCC invasive
progression.
Materials and methods
Cell culture
MHCC97L and HCCLM9 cell lines, were cultured in
RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS;
both from Gibco-BRL, Carlsbad, CA, USA) and 100 units/ml
streptomycin-penicillin at 37°C in a humidified atmosphere of 95%
air and 5% CO2.
Migration assay
The invasion and migration activity of MHCC97L and
HCCLM9 cells were assayed using a transwell cell-culture chamber as
described previously (16).
Briefly, cells were seeded into a 6-well plate at a density of
105 cells/well. After 24 h of incubation, the monolayer
cells were wounded by scraping a line using a 10 μl pipette tip.
The real time images of the wound line were recorded using a
microscope (Olympus IX71; Olympus Optical Co., Ltd, Tokyo, Japan)
at 0, 24 and 48 h.
Protein extraction
The cells were digested with trypsin and collected
by centrifuging for 10 min at 1,500 × g three times. The cell
number was quantified using a hemocytometer (VWR Scientific, West
Chester, PA, USA). Lysis solution (×5) was added and the mixture
was repeatedly freeze-thawed three times in liquid nitrogen. The
mixture was sonicated (Model VC50; Sonics & Materials Inc.,
Danbury, CT, USA) for 1 sec × 30 on ice. The amplitude of vibration
was set as 22% at room-temperature for 30 min, and finally the
mixture was centrifuged at 18,000 × g for 1 h at 4°C. The
supernatant was collected and further stored at −70°C.
Bradford assay
The staining solution was obtained by diluting the
Bradford staining solution with water at a ratio of 1:4, and 0,
0.28, 0.56, 0.84, 1.12 and 1.4 mg/ml of bovine serum albumin were
prepared. Next, 100 μl diluted standard proteins and samples were
mixed with 5 ml diluted staining solution at room temperature for 5
min. Finally, the absorbance, at 595 nm, was detected using an
enzyme-linked immunosorbent assay reader (Bio-Rad, Richmond, CA,
USA). The standard curve was obtained, and the precise
concentration of samples was calculated.
SDS-PAGE analysis
The proteins were mixed with 5× and 1× protein
loading buffer. The mixture was blotted for 10 min and immediately
placed into ice for 5 min. Samples were loaded onto a 10% SDS-PAGE
gel, and the electrophoresis was performed at 60 V for 10 min and
100 V for 1.5 h. The gel was stained with coomassie brilliant blue
(CBB) for 2 h and then destained with destaining solution.
Tryptic in-gel digestion
Protein spots were excised and transferred into
96-well plates. Gels were destained 1–2 times, and further
dehydrated with 100 μl acetonitrile (ACN) for 5 min. Next, 50 μl
DTT solution was incubated with the gel particles at 56°C for 30
min, and the gel was dehydrated with 100 μl ACN for 5 min.
Alkylation of the proteins was achieved by adding 50 μl
iodoacetamide (IOA), and dehydration was performed with 100 μl ACN
for 5 min. The gels were incubated with 15–20 μl trypsin (0.01
μg/μl) at 4°C for 30 min, and 15–20 μl trypsin buffer (25 mM
NH4HCO3) was added following complete
absorption of trypsin at 37°C overnight. Peptides were extracted
using solution I at 40°C for 1 h and solution II at 40°C for 1 h.
Finally, the extracted solution was combined and evaporated.
Electrospray ionization quadrupole
time-of-flight mass spectrometry (ESI-Q-TOF)
The MS data were obtained using a Q-TOF mass
spectrometer (Micromass, Manchester, UK) fitted with an ESI source
(Waters Corporation, Milford, MA, USA). The MS/MS data were
processed using MassLynx software (Waters Corporation), and Version
2.3 (Matrix Science, London, UK) was used to search the database. A
database search was performed using the following parameters:
Database, Swiss-Prot; mass tolerance, ±0.1 Dalton (Da); MS/MS
tolerance, ±0.05 Da; taxonomy, Homo sapiens; enzyme,
trypsin; and an allowance of one missed cleavage. In addition,
variable modifications of methionine oxidation and fixed
modifications of cysteine carbamidomethylation were allowed.
Semi-quantitative RT-PCR
Semi-quantitative RT-PCR was used to confirm the
differential expression of selected proteins at the mRNA level.
Total RNA was extracted with TRIzol reagent (Invitrogen Life
Technologies, Carlsbad, CA, USA), and RT-PCR was performed with a
PCR kit from Fermentas (Hanover, MD, USA). The primers used were as
follows: Sense: 5′-GGTGGTCGTTTCCCCAACAAA-3′ and antisense:
5′-GCCAGGTGTGGTAACTGCT-3′ for NCL (136 bp); and sense:
5′-GGAGTCCACTGGCGTCT -3′ and antisense: 5′-CATCATATTTGGCAGGTTTT-3′
for GAPDH (482 bp). GAPDH was used for normalization. The PCR
profile was as follows: 94°C for 2 min followed by 30 cycles of
94°C for 30 sec, 48°C for 30 sec, and 72°C for 30 sec, with a final
extension at 72°C for 7 min.
Statistical analysis
The data are presented as the mean ± standard
deviation. Statistical analysis was performed using Prism 5.0
software (GraphPad Software Inc., San Diego, CA, USA). Two-group
comparisons of continuous variables were performed using Student’s
t-test. P<0.05 was considered to indicate a statistically
significant difference.
Results
HCCLM9 cells exhibit a greater migration
ability compared with MHCC97L cells
The migration activity of MHCC97L and HCCLM9 cells
were evalated by a wound healing assay. The real time images of
cell migration were recorded using a microscope at 0, 24 and 48 h.
As shown in Fig. 1A, MHCC97L cells
did not present a marked migration ability, nevertheless, HCCLM9
cells primarily exhibited migration at 24 h, and the wound was
healed at ~48 h (Fig. 1B). These
findings suggested that the migration ability of HCCLM9 cells was
significantly stronger than MHCC97L cells.
Protein separation using SDS-PAGE
(Fig. 2)
Following confirmation of the differential migration
abilities of MHCC97L and HCCLM9 cells, the proteins that are
responsible for migration were investigated. Nuclear and
cytoplasmic proteins were, respectively, extracted from MHCC97L and
HCCLM9 cells, and the concentration of the proteins was obtained
using a Bradford assay (Fig. 2B).
Then, 50 μg proteins were loaded onto the 10% SDS-PAGE gel and the
proteins were separated at 60 V for 20 min and 100 V for 1.5 h. The
gel was stained with CBB and the differential protein bands were
analyzed (Fig. 2A). As shown in
Fig. 2C, these differentially
stained bands were quantified using gray scale. Finally, two bands
were cut out, and the peptides of these proteins were extracted and
identified using ESI-Q-TOF MS/MS, as described previously.
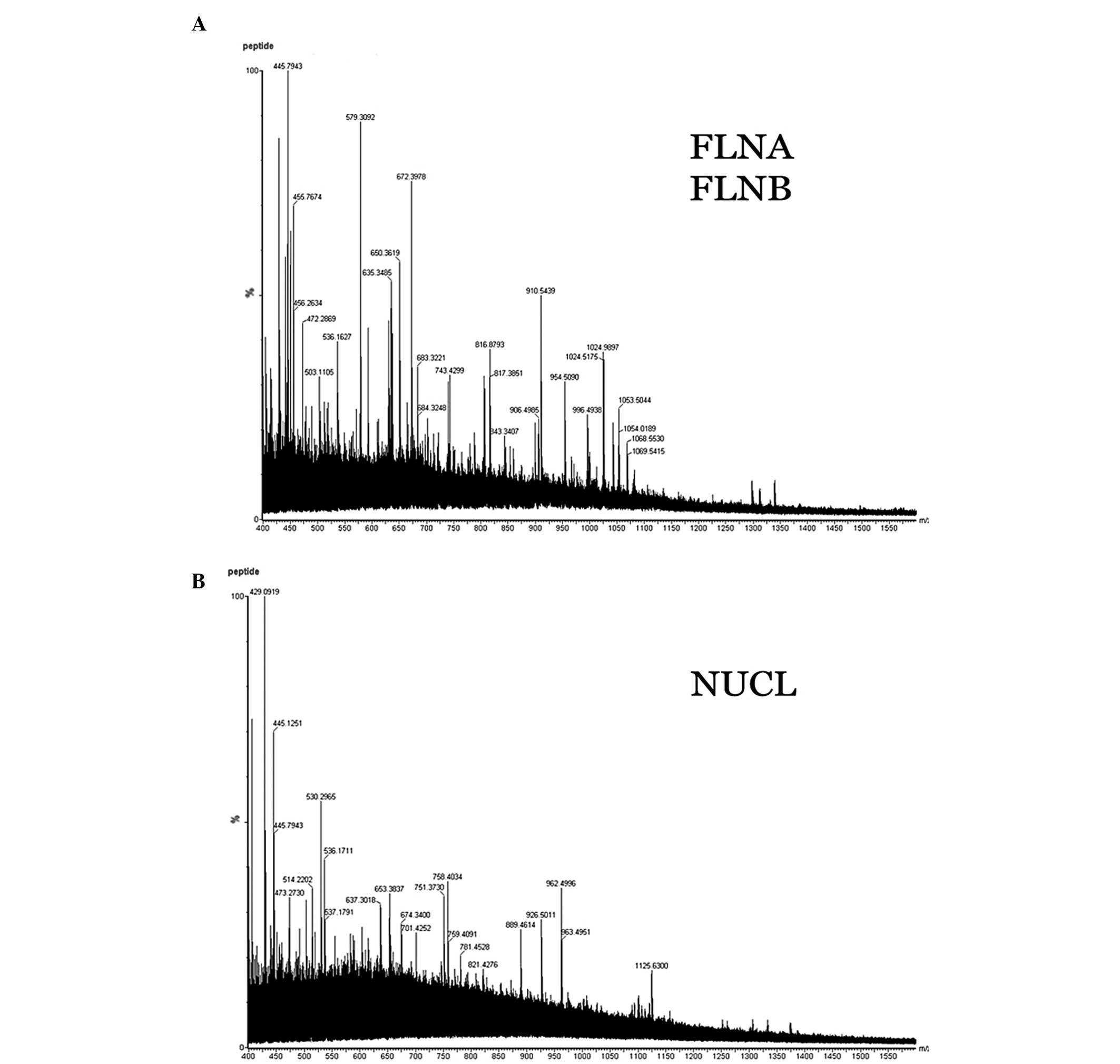
Differential protein identification using
ESI-Q-TOF MS/MS
The peptides of proteins were dissolved in 18 μl 50%
ACN. The top 10 most abundant ions for each MS scan were selected
for MS/MS analysis with a data-dependent mode. As shown in Fig. 3, the samples presented an excellent
MS/MS image, suggesting that these MS/MS data were of dependable
quality, thus, the proteins were identified accurately. In
addition, the peptides of trypsin and keratin were automatically
excluded. The MS/MS data were processed using MassLynx software
(Waters Corporation). The identified proteins are shown in Fig. 3.
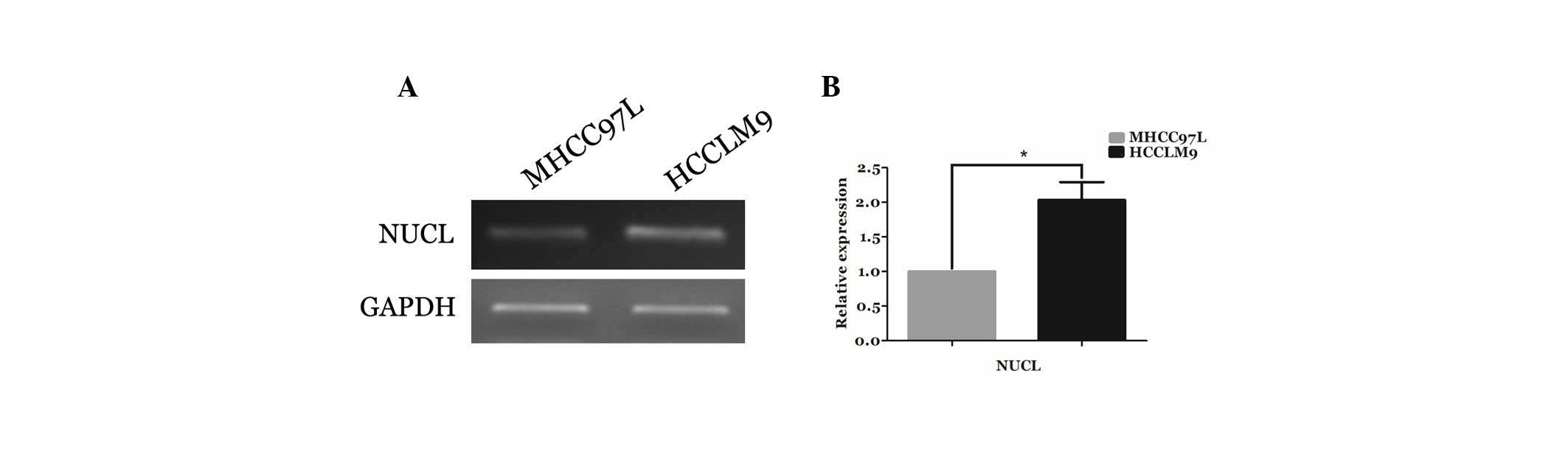
Validation using semi-quantitative
RT-PCR
To confirm the differential expression of an
identified protein at the mRNA level, nucleolin mRNA expression was
detected using semi-quantitative RT-PCR, and GAPDH was used as an
internal control. As shown in Fig.
4A, the mRNA expression of nucleolin significantly increased in
HCCLM9 cells, and the relative expression of nucleolin at mRNA
level was indicated in Fig. 4B.
These findings indicated that nucleolin is also overexpressed in
HCCLM9 cells at the mRNA level, suggesting that nucleolin may be a
candidate biomarker for the progression of HCC invasion.
Discussion
Hepatocellular carcinoma (HCC) is the most
predominant type of liver cancer in most countries, and the
majority of cases of HCC are associated with chronic hepatitis B
virus (HBV) or hepatitis C virus (HCV) infections (17). There are >500,000 new cases
diagnosed worldwide annually and the incidence of HCC is expected
to increase further in the next decade (5). Thus, accurate diagnosis and treatment
are critical for HCC patients, particularly for patients with
metastatic HCC. A previous study demonstrated that α-fetoprotein
(AFP) determination lacks adequate sensitivity and specificity for
effective surveillance of HCC (18). Indeed, the lack of good early
diagnostic markers has rendered this disease a major challenge.
Previous studies have investigated certain molecules associated
with the progression and metastasis of HCC, including transforming
growth factor-β1 (TGFβ) (19),
insulin-like growth factor II (IGF-II) (20), the human cervical cancer oncogene
(HCCR) (21), FLNA and PGK1
(8). However, these biomarkers are
not adequately specific and sensitive. Thus, the specific
diagnostic biomarkers for metastatic progression of HCC require
further investigation.
It was previously reported that the metastatic
recurrence of HCC is accompanied by numerous molecular alterations
(22). Therefore, global profiling
of metastatic progression of HCC may aid the development of novel
therapeutic targets and identification of diagnostic biomarkers,
and is likely to improve the diagnosis and treatment of metastatic
HCC.
In the present study, SDS-PAGE-coupled MS/MS
technology was used to identify differentially expressed proteins
in MHCC97L and HCCLM9 cells. In total, three proteins were
identified in two bands, namely FLNA, FLNB and nucleolin. Among
these proteins, a novel nuclear protein, nucleolin, was identified
from band 2, and it was observed to be overexpressed in HCCLM9
cells. The differential expression of nucleolin was further
confirmed at the mRNA level. Thus, the current data suggested that
nucleolin was overexpressed in HCCLM9 cells at protein and mRNA
levels.
Notably, the present study identified differential
expression of FLNA and FLNB in the cytoplasmic proteins, which is
consistent with findings reported by Ai et al (8). Aside from the two proteins in the
cytoplasm, nucleolin was the only differentially expressed protein
among the proteins isolated from the nucleus, and it provided a
novel candidate for the diagnosis and treatment of HCC.
Furthermore, it was reported that nucleolin may be key in cell
proliferation (23–25). Nucleolin induced chromatin
decondensation by binding to histone H1, and it was hypothesized to
be involved in pre-rRNA transcription and ribosome assembly
(26). Additionally, nucleolin may
exhibit a role in the process of transcriptional elongation
(26).
In conclusion, to the best of our knowledge the
current study was the first to show that nucleolin was
overexpressed in HCCLM9 cells at the protein and mRNA level. It was
proposed that nucleolin is a novel potential biomarker for the
metastasis of HCC and a possible therapeutic target for the
treatment of HCC patients.
References
|
1
|
Ding SJ, Li Y, Tan YX, Jiang MR, Tian B,
Liu YK, Shao XX, Ye SL, Wu JR, Zeng R, et al: From proteomic
analysis to clinical significance: overexpression of cytokeratin 19
correlates with hepatocellular carcinoma metastasis. Mol Cell
Proteomics. 3:73–81. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Altekruse SF, McGlynn KA and Reichman ME:
Hepatocellular carcinoma incidence, mortality, and survival trends
in the United States from 1975 to 2005. J Clin Oncol. 27:1485–1491.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
El-Serag HB and Rudolph KL: Hepatocellular
carcinoma: epidemiology and molecular carcinogenesis.
Gastroenterology. 132:2557–2576. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar
|
|
5
|
Venook AP, Papandreou C, Furuse J and de
Guevara LL: The incidence and epidemiology of hepatocellular
carcinoma: a global and regional perspective. Oncologist. 15(Suppl
4): 5–13. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Llovet JM, Burroughs A and Bruix J:
Hepatocellular carcinoma. Lancet. 362:1907–1917. 2003. View Article : Google Scholar
|
|
7
|
Wu L, Peng CW, Hou JX, Zhang YH, Chen C,
Chen LD and Li Y: Coronin-1C is a novel biomarker for
hepatocellular carcinoma invasive progression identified by
proteomics analysis and clinical validation. J Exp Clin Cancer Res.
24: 29:172010.PubMed/NCBI
|
|
8
|
Ai J, Huang H, Lv X, Tang Z, Chen M, Chen
T, Duan W, Sun H, Li Q, Tan R, et al: FLNA and PGK1 are two
potential markers for progression in hepatocellular carcinoma. Cell
Physiol Biochem. 27:207–216. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tian J, Tang ZY, Ye SL, Liu YK, Lin ZY,
Chen J and Xue Q: New human hepatocellular carcinoma (HCC) cell
line with highly metastatic potential (MHCC97) and its expressions
of the factors associated with metastasis. Br J Cancer. 81:814–821.
1999. View Article : Google Scholar
|
|
10
|
Li Y, Tang ZY, Ye SL, Liu YK, Chen J, Xue
Q, Chen J, Gao DM and Bao WH: Establishment of cell clones with
different metastatic potential from the metastatic hepatocellular
carcinoma cell line MHCC97. World J Gastroenterol. 7:630–636.
2001.PubMed/NCBI
|
|
11
|
Li Y, Tang Y, Ye L, Liu B, Liu K, Chen J
and Xue Q: Establishment of a hepatocellular carcinoma cell line
with unique metastatic characteristics through in vivo selection
and screening for metastasis-related genes through cDNA microarray.
J Cancer Res Clin Oncol. 129:43–51. 2003.
|
|
12
|
Li Y, Tian B, Yang J, Zhao L, Wu X, Ye SL,
Liu YK and Tang ZY: Stepwise metastatic human hepatocellular
carcinoma cell model system with multiple metastatic potentials
established through consecutive in vivo selection and studies on
metastatic characteristics. J Cancer Res Clin Oncol. 130:460–468.
2004.
|
|
13
|
Li Y, Tang ZY, Tian B, Ye SL, Qin LX, Xue
Q and Sun RX: Serum CYFRA 21–1 level reflects hepatocellular
carcinoma metastasis: study in nude mice model and clinical
patients. J Cancer Res Clin Oncol. 132:515–520. 2006.
|
|
14
|
Holly MK, Dear JW, Hu X, Schechter AN,
Gladwin MT, Hewitt SM, Yuen PS and Star RA: Biomarker and
drug-target discovery using proteomics in a new rat model of
sepsis-induced acute renal failure. Kidney Int. 70:496–506.
2006.PubMed/NCBI
|
|
15
|
Kabuyama Y, Resing KA and Ahn NG: Applying
proteomics to signaling networks. Curr Opin Genet Dev. 14:492–498.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang K, Ye C, Zhou Q, Zheng R, Lv X, Chen
Y, Hu Z, Guo H, Zhang Z, Wang Y, Tan R and Liu Y: PKD1 inhibits
cancer cells migration and invasion via Wnt signaling pathway in
vitro. Cell Biochem Funct. 25:767–774. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
El-Serag HB: Epidemiology of viral
hepatitis and hepatocellular carcinoma. Gastroenterology.
142:1264–1273. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lok AS, Sterling RK, Everhart JE, et al;
HALT-C Trial Group. Des-Gamma-carboxy prothrombin and
alpha-fetoprotein as biomarkers for the early dection of
hepatocellular carcinoma. Gastroenterology. 138:493–502. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Feng Y and Walsh CA: The many faces of
filamin: a versatile molecular scaffold for cell motility and
signalling. Nat Cell Biol. 6:1034–1038. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bedolla RG, Wang Y, Asuncion A, Chamie K,
Siddiqui S, Mudryj MM, Prihoda TJ, Siddiqui J, Chinnaiyan AM, Mehra
R, de Vere White RW and Ghosh PM: Nuclear versus cytoplasmic
localization of filamin A in prostate cancer: immunohistochemical
correlation with metastases. Clin Cancer Res. 15:788–796. 2009.
View Article : Google Scholar
|
|
21
|
Bourguignon LY, Gilad E and Peyrollier K:
Heregulin-mediated ErbB2-ERK signaling activates hyaluronan
synthases leading to CD44-dependent ovarian tumor cell growth and
migration. J Biol Chem. 282:19426–19441. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Dayan F, Roux D, Brahimi-Horn MC,
Pouyssegur J and Mazure NM: The oxygen sensor factor-inhibiting
hypoxia-inducible factor-1 controls expression of distinct genes
through the bifunctional transcriptional character of
hypoxia-inducible factor-1alpha. Cancer Res. 66:3688–3698. 2006.
View Article : Google Scholar
|
|
23
|
González V and Hurley LH: The C-terminus
of nucleolin promotes the formation of the c-MYC G-quadruplex and
inhibits c-MYC promoter activity. Biochemistry. 49:9706–9714.
2010.PubMed/NCBI
|
|
24
|
Abdelmohsen K and Gorospe M: RNA-binding
protein nucleolin in disease. RNA Biol. 9:799–808. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tajrishi MM, Tuteja R and Tuteja N:
Nucleolin: the most abundant multifunctional phosphoprotein of
nucleolus. Commun Integr Biol. 4:267–275. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Parada CA and Roeder RG: A novel RNA
polymerase II-containing complex potentiates Tat-enhanced HIV-1
transcription. Embo J. 18:3688–3701. 1999. View Article : Google Scholar : PubMed/NCBI
|