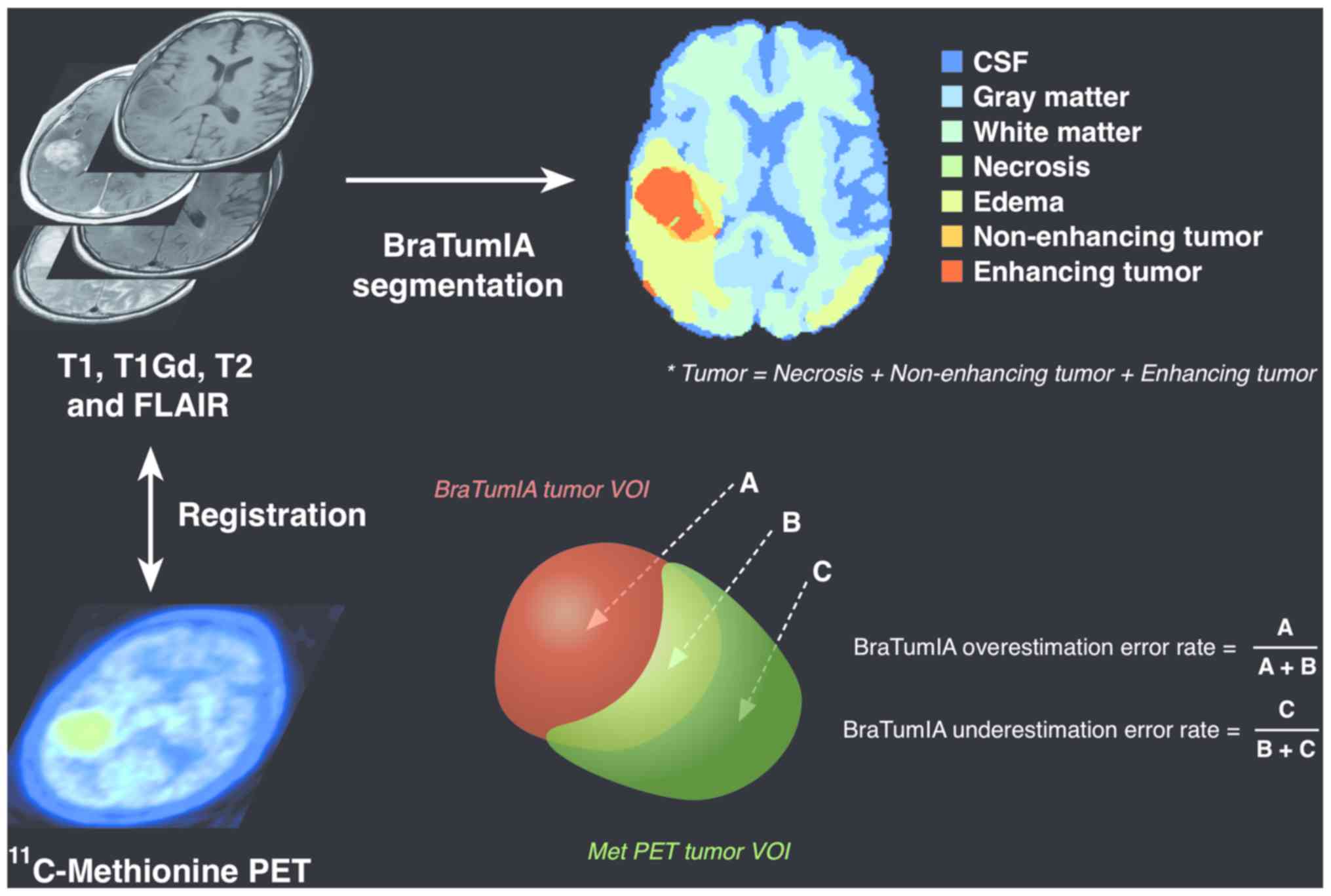
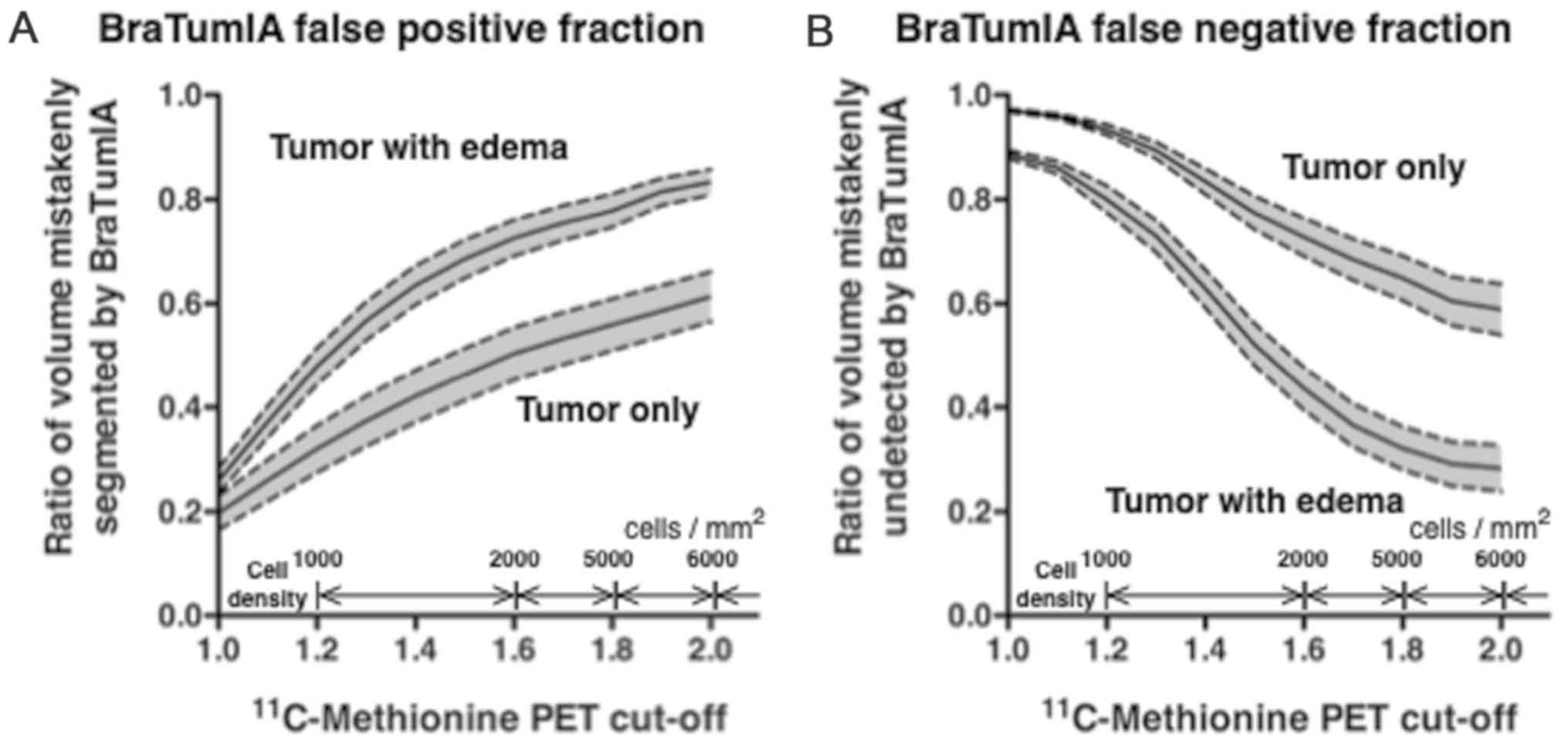
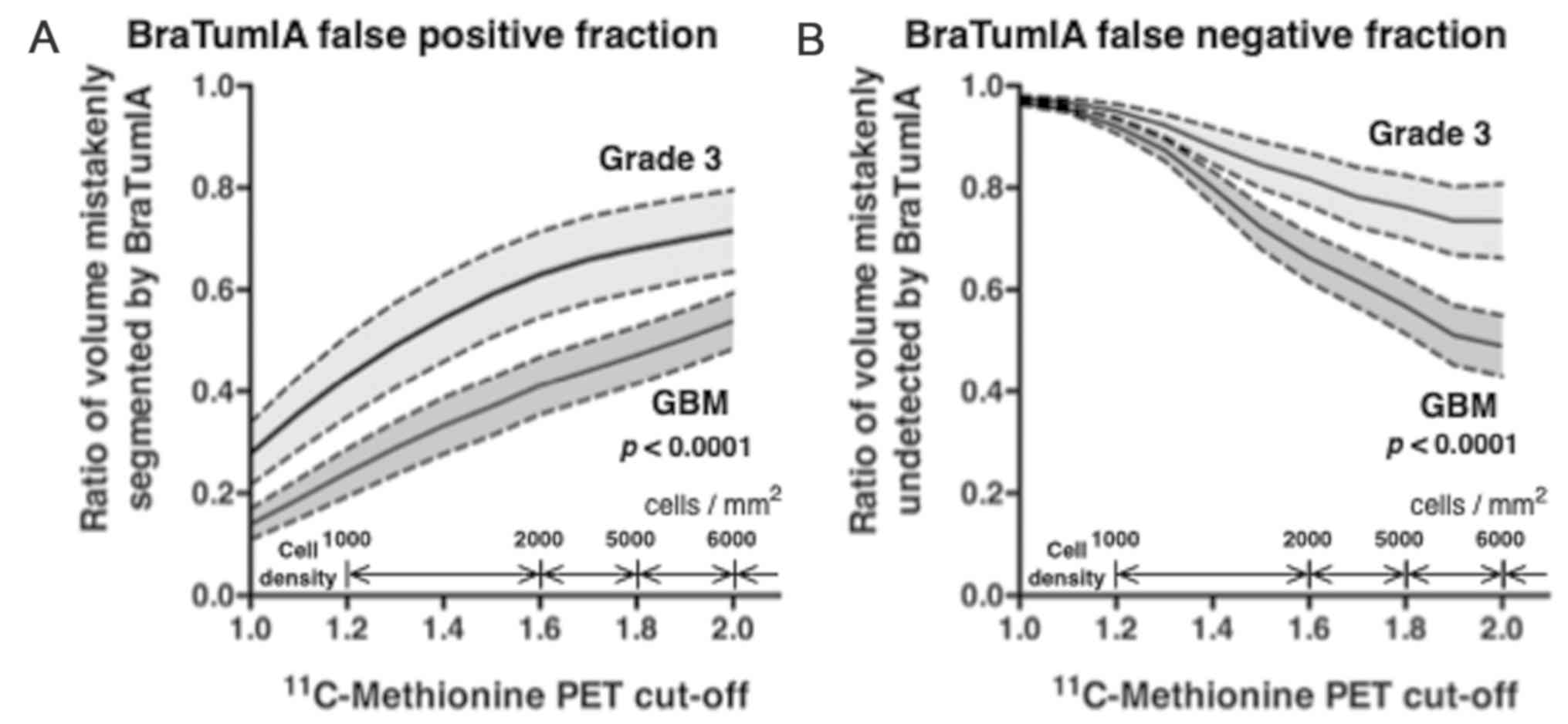
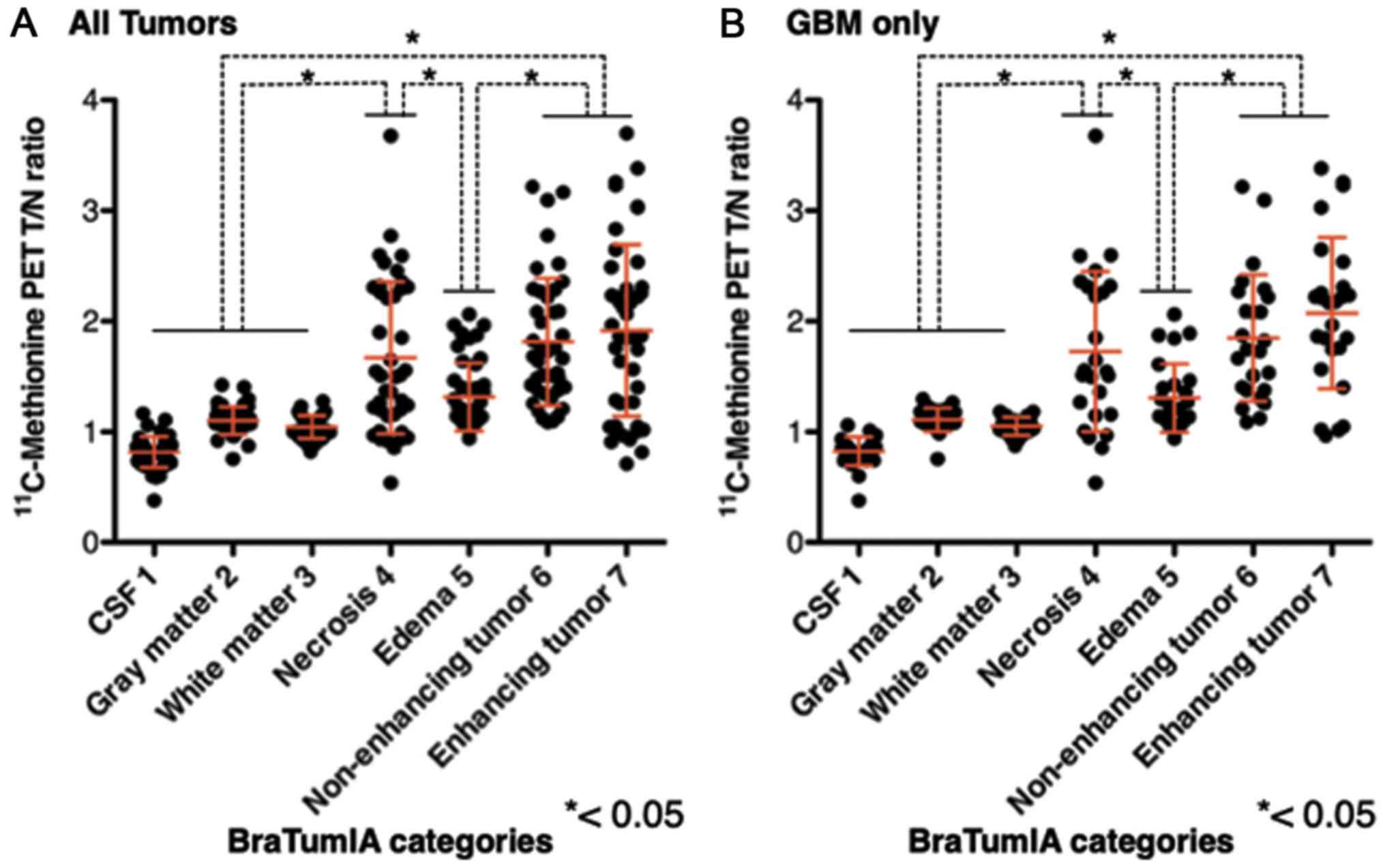
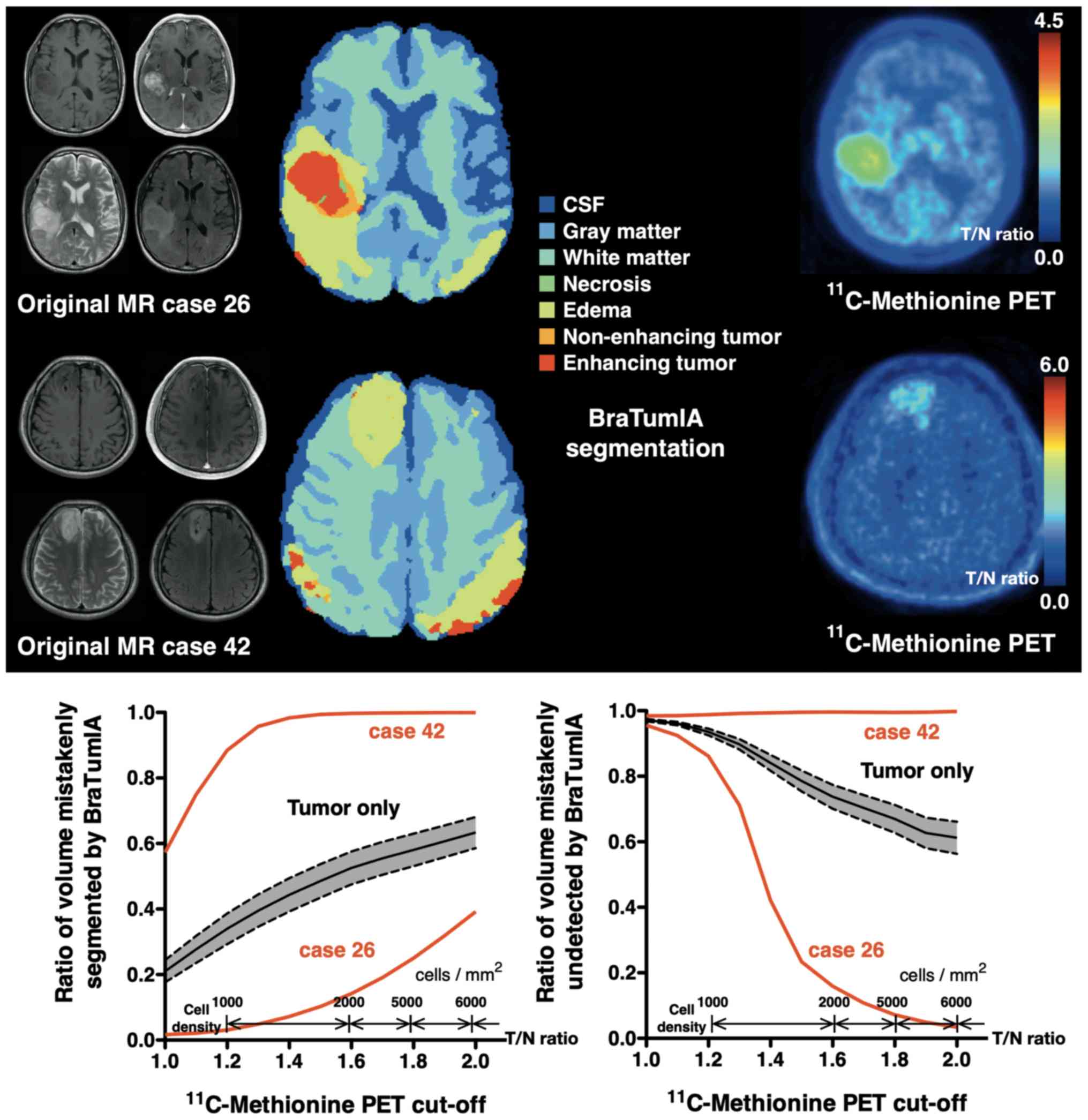
|
1
|
Ostrom QT, Gittleman H, Farah P, Ondracek
A, Chen Y, Wolinsky Y, Stroup NE, Kruchko C and Barnholtz-Sloan JS:
CBTRUS statistical report: Primary brain and central nervous system
tumors diagnosed in the United States in 2006–2010. Neuro Oncol. 15
(Suppl 2):ii1–ii56. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gigineishvili D, Shengelia N, Shalashvili
G, Rohrmann S, Tsiskaridze A and Shakarishvili R: Primary brain
tumour epidemiology in Georgia: First-year results of a
population-based study. J Neurooncol. 112:241–246. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dobes M, Khurana VG, Shadbolt B, Jain S,
Smith SF, Smee R, Dexter M and Cook R: Increasing incidence of
glioblastoma multiforme and meningioma, and decreasing incidence of
Schwannoma (2000–2008): Findings of a multicenter Australian study.
Surg Neurol Int. 2:1762011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gousias K, Markou M, Voulgaris S, Goussia
A, Voulgari P, Bai M, Polyzoidis K, Kyritsis A and Alamanos Y:
Descriptive epidemiology of cerebral gliomas in northwest Greece
and study of potential predisposing factors, 2005–2007.
Neuroepidemiology. 33:89–95. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Larjavaara S, Mäntylä R, Salminen T,
Haapasalo H, Raitanen J, Jääskeläinen J and Auvinen A: Incidence of
gliomas by anatomic location. Neuro Oncol. 9:319–325. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Arora RS, Alston RD, Eden TO, Estlin EJ,
Moran A and Birch JM: Age-incidence patterns of primary CNS tumors
in children, adolescents, and adults in England. Neuro Oncol.
11:403–413. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lee CH, Jung KW, Yoo H, Park S and Lee SH:
Epidemiology of primary brain and central nervous system tumors in
Korea. J Korean Neurosurg Soc. 48:145–152. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Stupp R, Mason WP, van den Bent MJ, Weller
M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn
U, et al: Radiotherapy plus concomitant and adjuvant temozolomide
for glioblastoma. N Engl J Med. 352:987–996. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Stupp R, Taillibert S, Kanner AA, Kesari
S, Steinberg DM, Toms SA, Taylor LP, Lieberman F, Silvani A, Fink
KL, et al: Maintenance therapy with tumor-treating fields plus
temozolomide vs temozolomide alone for glioblastoma: A randomized
clinical trial. JAMA. 314:2535–2543. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Stummer W, Pichlmeier U, Meinel T,
Wiestler OD, Zanella F and Reulen HJ; ALA-Glioma Study Group, :
Fluorescence-guided surgery with 5-aminolevulinic acid for
resection of malignant glioma: A randomised controlled multicentre
phase III trial. Lancet Oncol. 7:392–401. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sanai N and Berger MS: Glioma extent of
resection and its impact on patient outcome. Neurosurgery.
62:753–764. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Devaux BC, O'Fallon JR and Kelly PJ:
Resection, biopsy, and survival in malignant glial neoplasms: A
retrospective study of clinical parameters, therapy, and outcome. J
Neurosurg. 78:767–775. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zagzag D, Esencay M, Mendez O, Yee H,
Smirnova I, Huang Y, Chiriboga L, Lukyanov E, Liu M and Newcomb EW:
Hypoxia- and vascular endothelial growth factor-induced stromal
cell-derived factor-1alpha/CXCR4 expression in glioblastomas: One
plausible explanation of Scherer's structures. Am J Pathol.
173:545–560. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kotrotsou A, Elakkad A, Sun J, Thomas GA,
Yang D, Abrol S, Wei W, Weinberg JS, Bakhtiari AS, Kircher MF, et
al: Multi-center study finds postoperative residual non-enhancing
component of glioblastoma as a new determinant of patient outcome.
J Neurooncol. 139:125–133. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ellingson BM, Abrey LE, Nelson SJ,
Kaufmann TJ, Garcia J, Chinot O, Saran F, Nishikawa R, Henriksson
R, Mason WP, et al: Validation of postoperative residual
contrast-enhancing tumor volume as an independent prognostic factor
for overall survival in newly diagnosed glioblastoma. Neuro Oncol.
20:1240–1250. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Pirzkall A, McKnight TR, Graves EE, Carol
MP, Sneed PK, Wara WW, Nelson SJ, Verhey LJ and Larson DA:
MR-spectroscopy guided target delineation for high-grade gliomas.
Int J Radiat Oncol Biol Phys. 50:915–928. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Daumas-Duport C, Monsaigneon V, Blond S,
Munari C, Musolino A, Chodkiewicz JP and Missir O: Serial
stereotactic biopsies and CT scan in gliomas: Correlative study in
100 astrocytomas, oligo-astrocytomas and oligodendrocytomas. J
Neurooncol. 4:317–328. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kelly PJ, Daumas-Duport C, Kispert DB,
Kall BA, Scheithauer BW and Illig JJ: Imaging-based stereotaxic
serial biopsies in untreated intracranial glial neoplasms. J
Neurosurg. 66:865–874. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Susheela SP, Revannasiddaiah S,
Madhusudhan N and Bijjawara M: The demonstration of extension of
high-grade glioma beyond magnetic resonance imaging defined edema
by the use of (11) C-methionine positron emission tomography. J
Cancer Res Ther. 9:715–717. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Porz N, Bauer S, Pica A, Schucht P, Beck
J, Verma RK, Slotboom J, Reyes M and Wiest R: Multi-modal
glioblastoma segmentation: Man versus machine. PLoS One.
9:e968732014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Porz N, Habegger S, Meier R, Verma R,
Jilch A, Fichtner J, Knecht U, Radina C, Schucht P, Beck J, et al:
Fully automated enhanced tumor compartmentalization: Man vs.
machine reloaded. PLoS One. 11:e01653022016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Herholz K, Hölzer T, Bauer B, Schröder R,
Voges J, Ernestus RI, Mendoza G, Weber-Luxenburger G, Löttgen J,
Thiel A, et al: 11C-methionine PET for differential diagnosis of
low-grade gliomas. Neurology. 50:1316–1322. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kracht LW, Miletic H, Busch S, Jacobs AH,
Voges J, Hoevels M, Klein JC, Herholz K and Heiss WD: Delineation
of brain tumor extent with [11C]L-methionine positron emission
tomography: Local comparison with stereotactic histopathology. Clin
Cancer Res. 10:7163–7170. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kinoshita M, Arita H, Okita Y, Kagawa N,
Kishima H, Hashimoto N, Tanaka H, Watanabe Y, Shimosegawa E,
Hatazawa J, et al: Comparison of diffusion tensor imaging and
11C-methionine positron emission tomography for reliable
prediction of tumor cell density in gliomas. J Neurosurg.
125:1136–1142. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Louis DN, Ohgaki H, Wiestler OD, Cavenee
WK, Burger PC, Jouvet A, Scheithauer BW and Kleihues P: The 2007
WHO classification of tumours of the central nervous system. Acta
Neuropathol. 114:97–109. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hatakeyama T, Kawai N, Nishiyama Y,
Yamamoto Y, Sasakawa Y, Ichikawa T and Tamiya T: 11C-methionine
(MET) and 18F-fluorothymidine (FLT) PET in patients with newly
diagnosed glioma. Eur J Nucl Med Mol Imaging. 35:2009–2017. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Goldman S, Levivier M, Pirotte B, Brucher
JM, Wikler D, Damhaut P, Dethy S, Brotchi J and Hildebrand J:
Regional methionine and glucose uptake in high-grade gliomas: A
comparative study on PET-guided stereotactic biopsy. J Nucl Med.
38:1459–1462. 1997.PubMed/NCBI
|
|
28
|
Miwa K, Shinoda J, Yano H, Okumura A,
Iwama T, Nakashima T and Sakai N: Discrepancy between lesion
distributions on methionine PET and MR images in patients with
glioblastoma multiforme: Insight from a PET and MR fusion image
study. J Neurol Neurosurg Psychiatry. 75:1457–1462. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Grosu AL, Weber WA, Riedel E, Jeremic B,
Nieder C, Franz M, Gumprecht H, Jaeger R, Schwaiger M and Molls M:
L-(methyl-11C) methionine positron emission tomography for target
delineation in resected high-grade gliomas before radiotherapy. Int
J Radiat Oncol Biol Phys. 63:64–74. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Verburg N, Hoefnagels FWA, Barkhof F,
Boellaard R, Goldman S, Guo J, Heimans JJ, Hoekstra OS, Jain R,
Kinoshita M, et al: Diagnostic accuracy of neuroimaging to
delineate diffuse gliomas within the brain: A meta-analysis. AJNR
Am J Neuroradiol. 38:1884–1891. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tynninen O, Aronen HJ, Ruhala M, Paetau A,
Von Boguslawski K, Salonen O, Jääskeläinen J and Paavonen T: MRI
enhancement and microvascular density in gliomas. Correlation with
tumor cell proliferation. Invest Radiol. 34:427–434. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Meier R, Porz N, Knecht U, Loosli T,
Schucht P, Beck J, Slotboom J, Wiest R and Reyes M: Automatic
estimation of extent of resection and residual tumor volume of
patients with glioblastoma. J Neurosurg. 127:798–806. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Meier R, Knecht U, Loosli T, Bauer S,
Slotboom J, Wiest R and Reyes M: Clinical evaluation of a
fully-automatic segmentation method for longitudinal brain tumor
volumetry. Sci Rep. 6:233762016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Dextraze K, Saha A, Kim D, Narang S,
Lehrer M, Rao A, Narang S, Rao D, Ahmed S, Madhugiri V, et al:
Spatial habitats from multiparametric MR imaging are associated
with signaling pathway activities and survival in glioblastoma.
Oncotarget. 8:112992–113001. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ullrich RT, Kracht L, Brunn A, Herholz K,
Frommolt P, Miletic H, Deckert M, Heiss WD and Jacobs AH:
Methyl-L-11C-methionine PET as a diagnostic marker for malignant
progression in patients with glioma. J Nucl Med. 50:1962–1968.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kinoshita M, Arita H, Goto T, Okita Y,
Isohashi K, Watabe T, Kagawa N, Fujimoto Y, Kishima H, Shimosegawa
E, et al: A novel PET index, 18F-FDG-11C-methionine uptake
decoupling score, reflects glioma cell infiltration. J Nucl Med.
53:1701–1708. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wu R, Watanabe Y, Arisawa A, Takahashi H,
Tanaka H, Fujimoto Y, Watabe T, Isohashi K, Hatazawa J and Tomiyama
N: Whole-tumor histogram analysis of the cerebral blood volume map:
Tumor volume defined by 11C-methionine positron emission tomography
image improves the diagnostic accuracy of cerebral glioma grading.
Jpn J Radiol. 35:613–621. 2017. View Article : Google Scholar : PubMed/NCBI
|