Introduction
Breast cancer is the most frequently diagnosed
cancer worldwide, with >2.3 million new cases diagnosed in 2020
(1), and its management has
improved over the past decade (2).
Cyclin-dependent kinase 4/6 inhibitors (CDK4/6is) are the mainstay
of treatment of hormone receptor (HR)+/human epidermal
growth factor receptor 2 (HER2)− patients with advanced
breast cancer (ABC) in first- and second-line settings. Despite
improvements in progression-free survival (PFS) and overall
survival (OS), as well as quality of life, most patients develop
disease progression due to drug resistance, resulting in poor
prognosis. Drug resistance is a common phenomenon; thus, effective
breast cancer therapy depends on appropriately monitoring the
patient response to treatment (3).
In ABC trials, recurrent diagnostic imaging has been
used as often as every 6–12 weeks (4). Response Evaluation Criteria in Solid
Tumors (RECIST) 1.1, in which lesions are described with imaging,
is widely accepted as a standardized measure of tumor response to
therapy (5). Nonetheless, disease
may progress with an ineffective therapy for a considerable period
before further imaging is performed. Furthermore, given prolonged
PFS in patients with ABC, imaging is time-consuming and toxicity of
iodine-based and gadolinium contrast may be an issue (6).
Tools based on tissue biopsies are not appropriate
for permanent monitoring, due to their invasive nature. Liquid
biopsies overcome these obstacles as sampling is quick, minimally
invasive and associated with low-risk complications. Furthermore,
circulating biomarkers can be collected more often than imaging or
tissue sampling, theoretically enabling more informed management.
Liquid biopsy comprises circulating tumor cells and DNA and
microRNA (miRNA/miR). Serum biomarkers are appealing for the
potential to detect resistance to treatment and disease progression
earlier than computed tomography (CT) imaging and circulating
miRNAs, such as miR-96 and miR-125, reflect presence of breast
tumors (7).
miRNAs are small (18–25 nucleotides) non-coding,
single-stranded RNA molecules that interact with specific target
messenger (m)RNAs, thus instigating their translational repression
or degradation. miRNAs modulate gene expression by binding to a
complementary sequence in the 3′untranslated region of the mRNA
(8). miRNAs may be dysregulated in
cancer due to genetic (genomic amplification, chromosomal
rearrangements, deletions or mutation) and epigenetic changes
(aberrant hypermethylation, global DNA hypomethylation and
post-translational histone modification) (9). Genome-wide analyses have demonstrated
that dysregulated expression of miRNAs contributes to the
pathogenesis of almost all types of human malignant diseases, and
>30% of genes are direct targets of miRNAs (10,11). A
single miRNA molecule can regulate hundreds of mRNAs. The
association between miRNA and cancer is not entirely understood.
Multiple feedback loops, numerous targets of single miRNA and the
fact that several miRNAs control the same mRNA, result in an
intricate web of relationships (9).
miRNAs are key regulators of cancer-related
pathways, and there is growing evidence that miRNAs serve an
essential role in response to CDK4/6is (12,13).
Certain miRNAs are associated with sensitivity to CDK4/6is, whereas
others confer resistance to treatment with CDK4/6is. Let-7a is
upregulated in luminal breast cancer (14), whereas circulating miR-17 and
miR-34a levels differ between patients who are HR− and
HR+ (15).
Hyperactivation of the PI3K/AKT/mTOR pathway is common in T cell
acute lymphoblastic leukemia (ALL)/lymphoma. CDK6 is one of the
most downregulated targets of let-7 and miR-21 in mTOR knockdown
tumors and treatment with an mTOR inhibitor (rapamycin) combined
with palbociclib is effective (16). Epigenetic downregulation of miR-9
induces upregulation of CDK6, while treatment of ALL cells with
palbociclib decreases proliferation and increases apoptosis
(17). The miR-17-92 family
includes miR-17, −19a and −20a (18). The E2F1-regulated miR-17-92 cluster
is highly expressed in proneural glioblastoma, which exhibits
increased vulnerability to CDK4/6 inhibition, and the E2F cell
cycle pathway may be a key driver. Palbociclib decreases expression
of the miR-17-92 family in sensitive glioblastoma cell-like lines
by suppressing E2F1 transcription factor (19). miR-29b inhibits breast cancer cell
proliferation and increases sensitivity to palbociclib (13). miR-106b expression is efficiently
suppressed by CDK4/6 inhibition in an E2F and
retinoblastoma-dependent manner (20). Silencing of the tumor suppressor
miR-124a regulates CDK6 expression and confers poor prognosis in
ALL (21). Palbociclib decreases
ALL cell proliferation in vitro, whereas overexpression of
pre-miR124a in a mouse model leads to decreased tumorigenicity
(21). miR-126 is involved in cell
cycle regulation, particularly M phase, and improves the effects of
ribociclib in vitro (22).
In the aforementioned study, miR-326 was reported to have a
non-significant anti-proliferative effect as a single agent and
conferred sensitivity to ribociclib. Furthermore, miR-223 may serve
as an oncosuppressor and an oncopromoter. miR-223 expression is
decreased in luminal breast cancer and inversely correlated with
survival of patients. Moreover, E2F1 is a suppressor of miR-223
transcription. Therefore, CDK4/6i, by inhibiting E2F1 activity, may
reinstate miR-223 expression and breast cancer resistance to CDK4/6
inhibition induced by miR-223 abrogation, both in vitro and
in vivo (23,24). Additionally, serum-based miRNA
signature, composed of miR451a, miR-16-5p, miR-17-3p and miR-940,
effectively distinguishes patients who respond to the first-line
combination of chemotherapy and trastuzumab from patients who are
resistant (25).
However promising, most studies concerning miRNAs in
the context of CDK4/6is are preclinical (21,22)
and data on clinical results from patients with breast cancer
treated with CDK4/6is are lacking. Thus, the present study assessed
the value of miRNAs in patients with ABC treated with CDK4/6is.
Materials and methods
Study design
Eligible patients were those treated for ABC with
CDK4/6is between June and August 2022. A total of 80 female
patients (median age, 59.5 year; age range, 33–84 years) were
followed-up at the Breast Cancer Centre at the Maria
Sklodowska-Curie National Research Institute of Oncology (Gliwice,
Poland). Patients were monitored for ≥7 months after miRNA
assessment until the data cut-off in March 2023.
During CDK4/6i treatment, patients visited Maria
Sklodowska-Curie National Research Institute of Oncology every 28
days for a thorough history to identify potential symptoms,
physical examination and a routine blood test. A contrast-enhanced
CT (Somatom Definition Edge Plus, Siemens AG; IQon Spectral CT,
Philips Healthcare) was performed every 3 months. Additionally,
18-fluorodeoxyglucose positron emission tomography (PET)/CT or
magnetic resonance imaging was performed at the discretion of the
treating physician.
Tumor response was assessed according to RECIST 1.1
criteria and determined to be complete response (CR), partial
response (PR), stable disease (SD) or progressive disease (PD)
(26). CR, PR and SD comprised
clinical benefit. OS was defined as time from diagnosis of the
metastatic disease to time of death or last follow-up. PFS was
measured from the CDK4/6i commencement date to occurrence of PD or
death.
The primary objective of the present study was to
assess whether levels of circulating miRNAs differed between
patients with disease progression and patients deriving clinical
benefit (defined as SD, PR or CR) from CDK4/6i treatment. The
secondary objective was to assess whether levels of circulating
miRNAs differed between patients at the beginning of the CDK4/6i
treatment and patients exhibiting disease progression.
Blood processing and serum
isolation
All clinical samples were obtained from subjects who
provided written informed consent. Blood (10 ml) was collected on
day 1 of CDK4/6i treatment in collection tubes that maintained the
draw-time concentration of cell-free RNA (cfRNA; RNA Complete
BCT® CE; Streck LLC).
All laboratory procedures were performed in the
Department of Clinical and Molecular Genetic at Maria
Sklodowska-Curie National Research Institute of Oncology. Blood was
processed for plasma isolation within 1 h of collection. Blood was
centrifuged in cfRNA collection tubes at 1,800 × g at room
temperature in an Eppendorf 5810R for 15 min. Plasma was
transferred to a fresh tube and centrifuged at 2,800 × g at room
temperature for 15 min. Plasma was aliquoted, with inversion to mix
each aliquot, and stored at −80°C.
miRNA isolation and measurement of in
plasma using reverse transcription (RT)-quantitative (q)PCR
TaqMan™ miRNA ABC Purification Kit-Human
Panel A (cat. no. 4473087; Thermo Fisher Scientific, Inc.) was used
for miRNA isolation from plasma samples. TaqMan Advanced miRNA cDNA
Synthesis kit (cat. no. A28007; Thermo Fisher Scientific, Inc.) was
used for preparing the cDNA templates from miRNA. The kit enables
analysis of samples that are limited in quantity, including plasma.
Mature miRNAs from total RNA were modified by extending the 3′ end
of the mature transcript through poly(A) addition; the 5′ end was
lengthened by the 5′ end adaptor ligation. The modified miRNAs
underwent universal RT, followed by amplification to increase the
amount of cDNA uniformly for all miRNAs (miR-Amp reaction). The RT
reaction and cDNA amplification were carried out using Veriti Dx
96-well Fast Thermal Cycler (Thermo Fisher Scientific, Inc.). RT
was performed using a double-stage program, as follows: 15 min at
42°C and 5 min at 85°C. cDNA amplification was carried out using
the following conditions: 5 min at 95°C, followed by 14 cycles of 3
sec at 95°C and 30 sec at 60°C, and a final step for 10 min at
99°C.
Analysis of miRNA expression was performed using the
TaqMan Fast Advanced Master Mix for qPCR and Custom TaqMan Array
Advanced MicroRNA Cards-a pre-formulated primer and probe set (cat.
nos. 4444963 and A34722, respectively; Thermo Fisher Scientific,
Inc.). The assay can detect and quantify mature form of the miRNA
from 2 µl total RNA from serum or plasma. Based on the current
literature (12,13,16–25,27–31),
the following miRs were chosen: let-7a and miR-9, −17, −19a, −20a,
−21, −29b, −29c, −34a, −106b, −122, −124, −126, −128, −145, −193b,
−200a, −200b, −200c, −222, −223, −326 and −451.
qPCR was performed using the QuantStudio™
12K Flex Real-Time PCR System (Thermo Fisher Scientific, Inc.) with
a TaqMan Array Micro Fluidic Thermal Cycling Block (Thermo Fisher
Scientific, Inc.) with the following conditions: 20 sec at 95°C,
followed by 40 cycles of 1 sec at 95°C and 20 sec at 60°C. Nucleic
acid-free pipette tips were used to handle all reagents. All
procedures were performed according to the manufacturer's
protocol.
The Pfaffl method was used to calculate relative
gene expression values (32). These
values were divided by the normalization ratio, prepared with the
GeNorm VBA applet for Microsoft Excel (version 3.4) (33). miR-16-5p, miR-222-3p and miR-21-5p
were found the most stably expressed miRNAs across all samples
(34) and were used as the
control/housekeeping genes (35–39).
Statistical analysis
Continuous data are presented as median and
interquartile range (IQR, 25–75%). Wilcoxon rank-sum test was
performed to compare the expression levels of miRNAs (separately
for each miRNA; the number of miRNAs tested was 23) between groups
[patients with PD vs. patients with clinical benefit (SD, PR or CR)
and patients with PD vs. patients at the beginning of CDK4/6i
treatment separately]. P-values were adjusted using
Benjamini-Hochberg correction. OS and PFS were estimated using the
Kaplan-Meier method and 95% confidence intervals (CIs) for the
survival curves were calculated. Spearman's correlation coefficient
was used to assess correlation between miRNAs. The interpretation
of correlation coefficient was as follows: Negligible,
0.0≤r<0.1; weak, 0.1≤r≤0.39; moderate, 0.4≤r≤0.69; strong,
0.7≤r≤0.89 and very strong correlation, 0.9≤r≤1 (40). P<0.05 was considered to indicate
a statistically significant difference. All computational analysis
was performed in R Environment for Statistical Computing version
4.0.1 ‘See Things Now’ (R Foundation for Statistical Computing;
r-project.org).
Results
Patient characteristics
A total of 80 consecutive patients with ABC treated
with CDK4/6is in Maria Sklodowska-Curie National Research Institute
of Oncology were assessed. The median age was 59.5 (IQR, 50–68
years) and 19 (24%) patients were <50 years old. De novo
disease was present in 38 patients (47.5%), whereas 42 patients
(52.5%) had recurrent disease. Most patients were treated in the
first-line setting (n=63; 78.8%) and the rest were treated in the
second-line. The majority of patients had bone metastasis (n=73;
91.3%), including 28 patients (35%) with bone-only disease, whereas
seven patients (9%) had visceral metastases only. A total of 39
patients (49%) previously received chemotherapy, including 11
patients (14%) treated within 1 year of CDK4/6i commencement.
Ribociclib was administered to 39 patients (49%), 22 were treated
with palbociclib (27%) and 19 with abemaciclib (24%), and 58
patients (73%) received letrozole and 22 (27%) fulvestrant as an
endocrine compound. A total of 41 patients (51%) had an Eastern
Cooperative Oncology Group (ECOG) performance status (41) of 0, 27 had ECOG 1 (34%) and 12 had
ECOG 2 (15%). Elevated cancer antigen (CA)15-3 was found in 50
patients (62.5%, median 47.0 U/ml, IQR 23.1–169.2; reference range
<31.3 U/ml) (42). Blood was
collected from 11 patients at the beginning of CDK4/6i treatment (9
patients on day 1 of cycle 1 and 2 patients on day 1 of cycle 2).
Furthermore, blood was collected from 23 patients between cycles 3
and 10, 22 patients between cycles 11 and 20, 12 patients between
cycles 21 and 30, and 12 patients beyond 30 cycles of CDK4/6i
treatment. The median follow-up was 20.7 months (IQR, 12.1–29.3
months).
Treatment efficacy
At time of sampling, 14 patients were diagnosed with
PD, whereas 55 patients exhibited clinical benefit from CDK4/6i
treatment (including 34 patients with SD, 20 with PR and 1 with
CR). Patients diagnosed with disease progression were in the
following cycles of CDK4/6i treatment: Cycle 3 (n=1), 7 (n=2), 8
(n=1), 10 (n=1), 11 (n=1), 18 (n=2), 20 (n=1), 21 (n=2), 24 (n=1),
29 (n=1) and 32 (n=1). A total of 11 patients were at the beginning
of the treatment before the first radiological response evaluation,
46 patients had baseline PET/CT and three patients achieved a
complete metabolic response. The median PFS (Fig. 1) was not reached, whereas the
24-month PFS was 70.4% (95% CI, 59–84). The median OS (Fig. 2) was also not reached and the
36-month OS was 86.4% (95% CI, 76.9–97).
miRNA expression analysis
miRNA expression was measured in 76 patients (in 4
patients miRNA expression was not found), including 14 patients
with PD, 51 with clinical benefit and 11 at the beginning of
CDK4/6i treatment. Patients with disease progression had
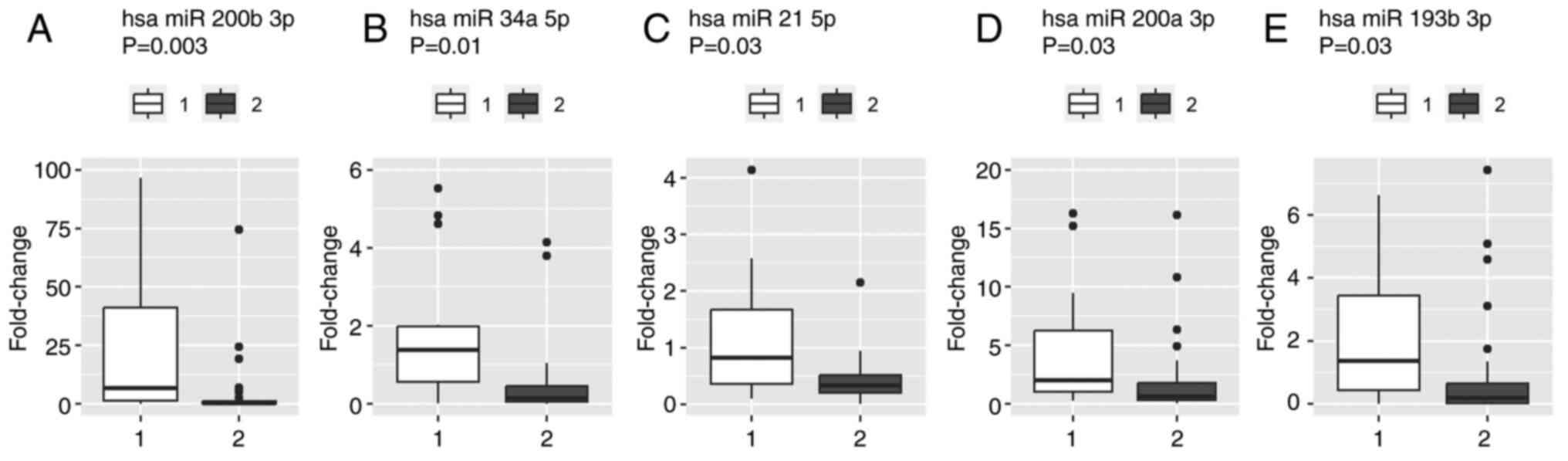
significantly higher levels of miR-21 (P=0.027), miR-34a (P=0.011),
miR-193b (P=0.032), miR-200a (P=0.027) and miR-200b (P=0.003)
compared with patients with clinical benefit of CDK4/6i treatment
(Fig. 3). Statistically significant
differences were not demonstrated for the remaining miRNAs.
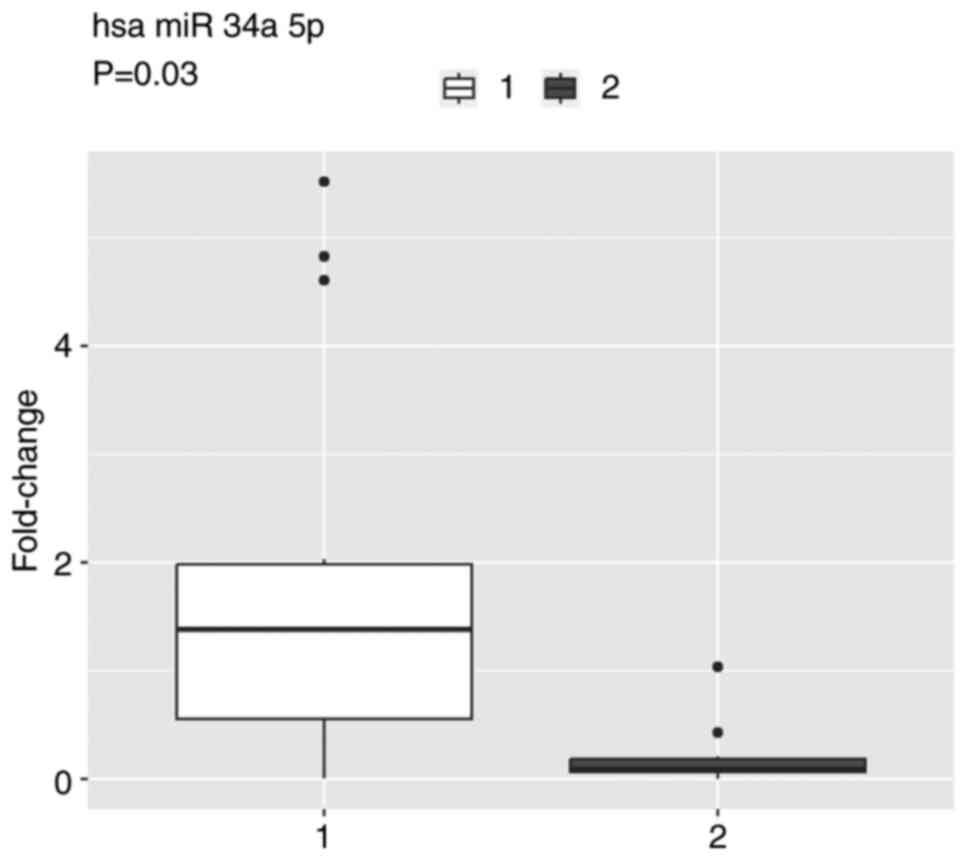
miRNA expression in patients with disease
progression differed from patients at the beginning of CDK4/6i
treatment. Significantly higher miR-34a expression was observed in
patients with PD (P=0.031; Fig. 4)
than in patients at the beginning of treatment; however,
statistically significant differences were not demonstrated for
other miRNAs assessed. Moreover, significantly higher miR-122
(P=0.070) and miR-193b (P=0.070) expression was observed in
patients with PD compared with the patients who were beginning
treatment, whereas expression of miR-17 and miR-20a was
significantly lower (P=0.070 for both) (data not shown).
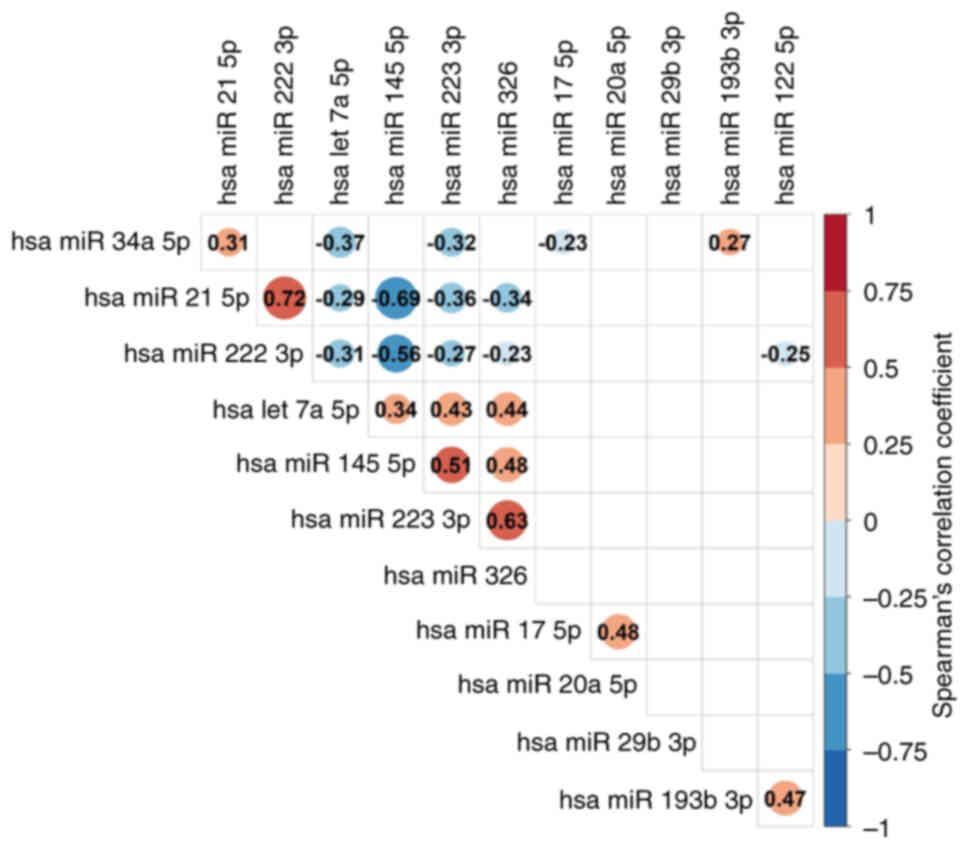
Correlations between miRs are presented in Fig. 5. A significant, strong positive
correlation was observed between miR-21 and miR-222 (r=0.72;
P<0.001). A significant, moderate positive correlation was
observed between miR-193b and miR-122 (r=0.47; P<0.001), miR-326
and miR-223 (r=0.63; P<0.001), let-7a and miR-223 (r=0.43;
P<0.001), miR-145 and miR-223 (r=0.51; P<0.001), miR-326 and
let-7a (r=0.44; P<0.001), miR-326 and miR-145 (r=0.48;
P<0.001), miR-20a and miR-17 (r=0.48; P<0.001), let-7a and
miR-223 (r=0.43; P<0.001). A significant, moderate negative
correlation was observed between miR-145 and miR-21 (r=−0.69;
P<0.001), and miR-145 and miR-222 (r=−0,56; P<0.001).
Levels of CA15-3 were also elevated in patients with
PD compared with patients exhibiting the clinical benefit of
CDK4/6i treatment (P<0.001) (data not shown).
Discussion
The present study demonstrated the role of
circulating miRNAs in patients with ABC treated with CDK4/6is.
Biomarkers may provide insight into the prognosis of patients with
ABC and predict the treatment response. Liquid biopsy with
blood-derived biomarkers is an appealing substitute for tissue
biomarkers, due to the non-invasive nature of this type of biopsy,
particularly in monitoring response to treatment. Currently, serum
biomarkers are less well-established than imaging in managing
treatment of patients with ABC. In the plethora of biomarkers
retrievable from liquid biopsy, miRNAs are promising predictive and
prognostic tools (43). miRNAs are
key regulators of breast cancer pathogenesis, progression and
response to therapy. Furthermore, emerging preclinical data have
confirmed the role of miRNAs as potential predictors of response to
CDK4/6i treatment (12). The
stability of miRNAs in plasma is an important prerequisite for
their use as biomarkers (44). The
present study assessed the role of a rationally selected subset of
miRNA in patients with ABC treated with CDK4/6i.
In the present study, miRNAs conferring resistance
to treatment with CDK4/6is were elevated in plasma derived from
patients with PD. miR-21 is a well-known oncogene associated with
protection of tumor cells from apoptosis, affecting metastasis and
invasion of breast cancer (45,46). A
meta-analysis of 1,629 breast cancer cases highlighted the
predictive value of miR-21 expression in both breast cancer tissue
and plasma samples (47).
Consistent with the aforementioned studies, the present study
demonstrated that miR-21 expression was elevated in patients with
disease progression. In a previous study, miR-193b targeted cyclin
D1 in prostate cancer and CDK4/6i inhibited proliferation of
prostate cancer cell lines expressing low levels of miR-193b but
did not affect the proliferation of cells with high miR-193b
expression (27). Another group of
miRNAs commonly dysregulated in breast cancer is the miR-200
family. miR-200a, a potential ‘cell cycle break’, decreases
response to CDK4/6i by decreasing CDK6 expression; low miRNA-200a
expression results in a more marked response to CDK4/6i in
metastatic melanoma (28). miR-200b
dysregulation is involved in chemoresistance by regulating
drug-associated cellular pathways (29). miR-200b is upregulated in various
types of cancer and downregulated by treatment with
antimetabolites, such as 5-fluorouracil (48).
The present study demonstrated that both miR-200a
and miR-200b were elevated in patients with PD compared with
patients responding to CDK4/6is. By contrast, the present study did
not observe a difference in expression of miR-200c. miR-200c serves
an antioncogenic role in renal cell cancer by controlling cell
proliferation and cell cycle progression by downregulating the G1-S
regulator CDK2 (30). It also
increases the sensitivity of breast cancer cells to doxorubicin
(31).
Most previous studies concerning miRNAs as
predictors of CDK4/6is treatment were based on cell lines, fresh
frozen tissue or animal models (13,22,49).
To the best of our knowledge, the present study is the first to
demonstrate the value of circulating miRNAs in patients with
HR+/HER2−ABC treated with CDK4/6is.
Nonetheless, in several studies concerning chemotherapy-based
treatment, circulating miRNAs have been reported to exhibit
prognostic and predictive value, suggesting distinctive signatures
associated with poor clinical outcomes (50,51).
Most of the aforementioned studies focused on the triple-negative
breast cancer subtype (52,53). In the HER2+ breast cancer
subtype, circulating miRNAs are putative biomarkers of response to
chemotherapy and anti-HER2 treatment (25). Circulating miR-451a, miR-16, miR-17
and miR-940 exhibit predictive value for response to trastuzumab
and serve as useful biomarkers for personalized therapy (25). To the best of our knowledge,
however, evidence of using miRNA as a predictor of response to
CDK4/6is is lacking. miR-940 was not included in the present
analysis as it is not available in the TaqMan miRNA ABC
Purification Kit-Human Panel A used. However, the results of the
present study suggested that miR-451a and miR-17 may be useful in
distinguishing patients responding to CDK4/6i and those refractory
to that treatment; however, following correction for multiple
testing, the difference was insignificant.
CA15-3 has value in the management of metastatic
disease; however, specificity remains low. CA15-3 is a
carbohydrate-containing protein antigen of the transmembrane
glycoprotein mucin-1, inhibiting tumor cell lysis and decreasing
cell-cell interactions (43).
Tampellini et al (54)
assessed use of the kinetics of CA 15-3 in patients with metastatic
breast cancer receiving anthracycline-based chemotherapy. Median
time to disease progression was longer in patients with CA15-3
within a normal range than in patients with increased levels.
CA15-3 elevation was reported in 50 patients (62.5%), which
complemented the findings of other studies (55–57).
Increasing evidence suggests that the hepatocyte
growth factor/mesenchymal-epithelial transition factor (c-MET)
signaling pathway serves a key role in carcinogenesis, regulation
of tumor microenvironment, metastasis and drug resistance (58,59).
Aberrations in c-MET have been described among several potential
mechanisms of resistance to CDK4/6is (60). Increased c-MET expression,
frequently observed in HR+/HER2− breast
cancer (61,62), is associated with disease stage,
progesterone receptor levels, Ki67 index and worse survival
(63).
A technology based on TaqMan low-density array
cards, used in the present study, is a recognized tool for
circulating miRNA analysis (64).
Nonetheless, the present study did not observe differences in
miR-9, miR-124 and miR-126 expression. miR-126 may be involved in
cell cycle regulation, particularly in the M phase, and improves
the effect of ribociclib in the MCF7 breast cancer cell line
(22).
One limitation of the present study is that the
origin of the identified miRNAs was not verified and the
association between miRNA expression with the corresponding breast
cancer tissue was not analyzed. The signature obtained by analyzing
circulating miRNA levels expression matches the corresponding tumor
tissue in a previous study (7),
whereas another reported differences between plasma and tissue
expression (65). Secondly, miRNA
expression was only assessed at a single time point. Serial
assessment, with blood collected at the beginning of and then
during treatment, may confer additional insight into miRNA
dynamics. To the best of our knowledge, the present study is the
first to assess circulating miRNA in patients with ABC treated with
CDK4/6is. Longitudinal studies are required to verify the impact of
a selected set of miRNAs on objective response. The results of the
present study provide a foundation for the design of such a
trial.
Although the field of biomarker-associated studies
in cancer continues to grow (66,67),
only a few are considered standard of care for clinical practice.
If confirmed in prospective clinical trials, the present miRNA
signature may be an important non-invasive tool to determine
treatment response, thus allowing timely treatment alternatives.
Various techniques of assessing plasma miRNAs, including those
presented in this study, are widely used in laboratories with a
reasonable cost of reagents. Thus, with the combination of liquid
biopsy and radiological assessment, personalized medicine could be
integrated into the standard of care.
In summary, the present study suggested that
plasma-based expression of miR-21, −34a, −193b, −200a and −200b
effectively distinguished patients with ABC who respond to CDK4/6i
treatment from patients who are resistant. However, the results
require confirmation in larger prospective trials and longitudinal
studies are required to verify use of miRNA in monitoring CDK4/6i
treatment.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Science Centre,
Poland (grant no. 2021/05/X/NZ5/00971).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
MK, MJ and MOW conceived the study. MK, MJ, MOW, PT
and TT designed experiments. MK, AK and TT analyzed data. AK
performed graphical presentation of results. MJ and MK developed
the clinical data. PT and TT carried out the laboratory work. MK
wrote the manuscript. MK, MJ, MOW, AK, PT and TT reviewed the
manuscript. MK, MJ and MOW supervised the study. PT and TT confirm
the authenticity of all the raw data. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
The present study was performed in accordance with
the 1964 Declaration of Helsinki and with the ethical standards of
the Institutional Ethics Committee at Maria Sklodowska-Curie
National Research Institute of Oncology (Gliwice, Poland), which
approved the study (approval no. KB/430-05/22). All data were
entered into an anonymized database following the general data
protection regulation of the Maria Sklodowska-Curie National
Research Institute of Oncology. All patients provided written
informed consent before enrollment.
Patient consent for publication
Not applicable.
Competing interests
MK declares conference fees for Pfizer, Roche,
Novartis, Teva, and Amgen; clinical trials for Roche, MSD,
Novartis, Seagen, and Gilead; speaker's honoraria from Novartis,
Roche, Lilly, Teva, and Amgen, Swixx Biopharma; advisory board for
Novartis; all outside the submitted work. MJ declares conference
fees for Gilead, Roche; clinical trials for Roche, MSD, Novartis,
Seagen, and Gilead; speaker's honoraria from Novartis, Roche,
Lilly, Pfizer, Teva, Exact Sciences, and Mammotome; advisory boards
for Novartis and Pfizer; all outside submitted work. All other
authors declare they have no competing interests.
References
|
1
|
Arnold M, Morgan E, Rumgay H, Mafra A,
Singh D, Laversanne M, Vignat J, Gralow JR, Cardoso F, Siesling S
and Soerjomataram I: Current and future burden of breast cancer:
Global statistics for 2020 and 2040. Breast. 66:15–23. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Miglietta F, Bottosso M, Griguolo G, Dieci
MV and Guarneri V: Erratum to ‘Major advancements in metastatic
breast cancer treatment: When expanding options means prolonging
survival’: [ESMO Open Volume 7, Issue 2, April 2022, 100409]. ESMO
Open. 7:1004722022. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cetin B, Wabl CA and Gumusay O: CDK4/6
inhibitors: Mechanisms of resistance and potential biomarkers of
responsiveness in breast cancer. Future Oncol. 18:1143–1157. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hortobagyi GN: Ribociclib for the
first-line treatment of advanced hormone receptor-positive breast
cancer: A review of subgroup analyses from the MONALEESA-2 trial.
Breast Cancer Res. 20:1232018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Eisenhauer EA, Therasse P, Bogaerts J,
Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S,
Mooney M, et al: New response evaluation criteria in solid tumours:
revised RECIST guideline (version 1.1). Eur J Cancer. 45:228–247.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Caschera L, Lazzara A, Piergallini L,
Ricci D, Tuscano B and Vanzulli A: Contrast agents in diagnostic
imaging: Present and future. Pharmacol Res. 110:65–75. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Matamala N, Vargas MT, González-Cámpora R,
Miñambres R, Arias J, Menéndez P, Andrés-León E, Gómez-López G,
Yanowsky K, Calvete-Candenas J, et al: Tumor microRNA expression
profiling identifies circulating microRNAs for early breast cancer
detection. Clin Chem. 61:1098–1106. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kim VN and Nam JW: Genomics of microRNA.
Trends Genet. 22:165–173. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Di Leva G and Croce CM: Roles of small
RNAs in tumor formation. Trends Mol Med. 16:257–267. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Croce CM: Causes and consequences of
microRNA dysregulation in cancer. Nat Rev Genet. 10:704–714. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Johnson SM, Grosshans H, Shingara J, Byrom
M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D and Slack
FJ: RAS is regulated by the let-7 microRNA family. Cell.
120:635–647. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Andrikopoulou A, Shalit A, Zografos E,
Koutsoukos K, Korakiti AM, Liontos M, Dimopoulos MA and Zagouri F:
MicroRNAs as potential predictors of response to CDK4/6 inhibitor
treatment. Cancers (Basel). 13:41142021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ji W, Zhang W, Wang X, Shi Y, Yang F, Xie
H, Zhou W, Wang S and Guan X: c-myc regulates the sensitivity of
breast cancer cells to palbociclib via c-myc/miR-29b-3p/CDK6 axis.
Cell Death Dis. 11:7602020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Qattan A, Intabli H, Alkhayal W, Eltabache
C, Tweigieri T and Amer SB: Robust expression of tumor suppressor
miRNA's let-7 and miR-195 detected in plasma of Saudi female breast
cancer patients. BMC Cancer. 17:7992017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Eichelser C, Flesch-Janys D, Chang-Claude
J, Pantel K and Schwarzenbach H: Deregulated serum concentrations
of circulating cell-free microRNAs miR-17, miR-34a, miR-155, and
miR-373 in human breast cancer development and progression. Clin
Chem. 59:1489–1496. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gary JM, Simmons JK, Xu J, Zhang S, Peat
TJ, Watson N, Gamache BJ, Zhang K, Kovalchuk AL, Michalowski AM, et
al: Hypomorphic mTOR downregulates CDK6 and Delays Thymic Pre-T LBL
tumorigenesis. Mol Cancer Ther. 19:2221–2232. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Rodriguez-Otero P, Román-Gómez J,
Vilas-Zornoza A, José-Eneriz ES, Martín-Palanco V, Rifón J, Torres
A, Calasanz MJ, Agirre X and Prosper F: Deregulation of FGFR1 and
CDK6 oncogenic pathways in acute lymphoblastic leukaemia harbouring
epigenetic modifications of the MIR9 family. Br J Haematol.
155:73–83. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sakai A, Saitow F, Maruyama M, Miyake N,
Miyake K, Shimada T, Okada T and Suzuki H: MicroRNA cluster
miR-17-92 regulates multiple functionally related voltage-gated
potassium channels in chronic neuropathic pain. Nat Commun.
8:160792017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li M, Xiao A, Floyd D, Olmez I, Lee J,
Godlewski J, Bronisz A, Bhat KPL, Sulman EP, Nakano I and Purow B:
CDK4/6 inhibition is more active against the glioblastoma proneural
subtype. Oncotarget. 8:55319–55331. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Thangavel C, Boopathi E, Ertel A, Lim M,
Addya S, Fortina P, Witkiewicz AK and Knudsen ES: Regulation of
miR106b cluster through the RB pathway: Mechanism and functional
targets. Cell Cycle. 12:98–111. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Agirre X, Vilas-Zornoza A, Jiménez-Velasco
A, Martin-Subero JI, Cordeu L, Gárate L, San José-Eneriz E,
Abizanda G, Rodríguez-Otero P, Fortes P, et al: Epigenetic
silencing of the tumor suppressor microRNA Hsa-miR-124a regulates
CDK6 expression and confers a poor prognosis in acute lymphoblastic
leukemia. Cancer Res. 69:4443–4453. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Baldassari F, Zerbinati C, Galasso M,
Corrà F, Minotti L, Agnoletto C, Previati M, Croce CM and Volinia
S: Screen for microRNA and drug interactions in breast cancer cell
lines points to miR-126 as a modulator of CDK4/6 and PIK3CA
inhibitors. Front Genet. 9:1742018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Citron F, Segatto I, Vinciguerra GLR,
Musco L, Russo F, Mungo G, D'Andrea S, Mattevi MC, Perin T,
Schiappacassi M, et al: Downregulation of miR-223 expression is an
early event during mammary transformation and confers resistance to
CDK4/6 inhibitors in luminal breast cancer. Cancer Res.
80:1064–1077. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Favero A, Segatto I, Perin T and Belletti
B: The many facets of miR-223 in cancer: Oncosuppressor, oncogenic
driver, therapeutic target, and biomarker of response. Wiley
Interdiscip Rev RNA. 12:e16592021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li H, Liu J, Chen J, Wang H, Yang L, Chen
F, Fan S, Wang J, Shao B, Yin D, et al: A serum microRNA signature
predicts trastuzumab benefit in HER2-positive metastatic breast
cancer patients. Nat Commun. 9:16142018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Eisenhauer EA, Therasse P, Bogaerts J,
Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S,
Mooney M, et al: New response evaluation criteria in solid tumours:
Revised RECIST guideline (version 1.1). Eur J Cancer. 45:228–247.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kaukoniemi KM, Rauhala HE, Scaravilli M,
Latonen L, Annala M, Vessella RL, Nykter M, Tammela TL and
Visakorpi T: Epigenetically altered miR-193b targets cyclin D1 in
prostate cancer. Cancer Med. 4:1417–1425. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bustos MA, Ono S, Marzese DM, Oyama T,
Iida Y, Cheung G, Nelson N, Hsu SC, Yu Q and Hoon DSB: MiR-200a
Regulates CDK4/6 Inhibitor Effect by Targeting CDK6 in Metastatic
Melanoma. J Invest Dermatol. 137:1955–1964. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Jayaraj R, Nayagam SG, Kar A, Sathyakumar
S, Mohammed H, Smiti M, Sabarimurugan S, Kumarasamy C,
Priyadharshini T, Gothandam KM, et al: Clinical theragnostic
relationship between drug-resistance specific mirna expressions,
chemotherapeutic resistance, and sensitivity in breast cancer: A
systematic review and Meta-Analysis. Cells. 8:12502019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang X, Chen X, Han W, Ruan A, Chen L,
Wang R, Xu Z, Xiao P, Lu X, Zhao Y, et al: miR-200c Targets CDK2
and suppresses tumorigenesis in renal cell carcinoma. Mol Cancer
Res. 13:1567–1577. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Safaei S, Amini M, Najjary S, Mokhtarzadeh
A, Bolandi N, Saeedi H, Alizadeh N, Javadrashid D and Baradaran B:
miR-200c increases the sensitivity of breast cancer cells to
Doxorubicin through downregulating MDR1 gene. Exp Mol Pathol.
125:1047532022. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Pfaffl MW: A new mathematical model for
relative quantification in real-time RT-PCR. Nucleic Acids Res.
29:e452001. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hildyard JCW and Wells DJ: Identification
and validation of quantitative PCR reference genes suitable for
normalizing expression in normal and dystrophic cell culture models
of myogenesis. PLoS Curr.
6:ecurrents.md.faafdde4bea8df4aa7d06cd5553119a6. 2014.PubMed/NCBI
|
|
34
|
Rinnerthaler G, Hackl H, Gampenrieder SP,
Hamacher F, Hufnagl C, Hauser-Kronberger C, Zehentmayr F, Fastner
G, Sedlmayer F, Mlineritsch B and Greil R: miR-16-5p Is a
Stably-Expressed housekeeping MicroRNA in breast cancer tissues
from primary tumors and from metastatic sites. Int J Mol Sci.
17:1562016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Vandesompele J, De Preter K, Pattyn F,
Poppe B, Van Roy N, De Paepe A and Speleman F: Accurate
normalization of real-time quantitative RT-PCR data by geometric
averaging of multiple internal control genes. Genome Biol.
3:RESEARCH00342002. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Pfaffl MW, Tichopad A, Prgomet C and
Neuvians TP: Determination of stable housekeeping genes,
differentially regulated target genes and sample integrity:
BestKeeper-Excel-based tool using pair-wise correlations.
Biotechnol Lett. 26:509–515. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Radonić A, Thulke S, Mackay IM, Landt O,
Siegert W and Nitsche A: Guideline to reference gene selection for
quantitative real-time PCR. Biochem Biophys Res Commun.
313:856–862. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Andersen CL, Jensen JL and Ørntoft TF:
Normalization of real-time quantitative reverse transcription-PCR
data: A model-based variance estimation approach to identify genes
suited for normalization, applied to bladder and colon cancer data
sets. Cancer Res. 64:5245–5250. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Dheda K, Huggett JF, Bustin SA, Johnson
MA, Rook G and Zumla A: Validation of housekeeping genes for
normalizing RNA expression in real-time PCR. Biotechniques.
37:112–119. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Schober P and Schwarte LA: Correlation
coefficients: Appropriate use and interpretation. Anesth Analg.
126:1763–1768. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Oken MM, Creech RH, Tormey DC, Horton J,
Davis TE, McFadden ET and Carbone PP: Toxicity and response
criteria of the Eastern Cooperative Oncology Group. Am J Clin
Oncol. 5:649–656. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Duffy MJ: Biochemical markers in breast
cancer: Which ones are clinically useful? Clin Biochem. 34:347–352.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Seale KN and Tkaczuk KHR: Circulating
biomarkers in breast cancer. Clin Breast Cancer. 22:e319–e331.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Mitchell PS, Parkin RK, Kroh EM, Fritz BR,
Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant
KC, Allen A, et al: Circulating microRNAs as stable blood-based
markers for cancer detection. Proc Natl Acad Sci USA.
105:10513–10518. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Song B, Wang C, Liu J, Wang X, Lv L, Wei
L, Xie L, Zheng Y and Song X: MicroRNA-21 regulates breast cancer
invasion partly by targeting tissue inhibitor of metalloproteinase
3 expression. J Exp Clin Cancer Res. 29:292010. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wang H, Tan Z, Hu H, Liu H, Wu T, Zheng C,
Wang X, Luo Z, Wang J, Liu S, et al: microRNA-21 promotes breast
cancer proliferation and metastasis by targeting LZTFL1. BMC
Cancer. 19:7382019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Jinling W, Sijing S, Jie Z and Guinian W:
Prognostic value of circulating microRNA-21 for breast cancer: A
systematic review and meta-analysis. Artif Cells Nanomed
Biotechnol. 45:1–6. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Rossi L, Bonmassar E and Faraoni I:
Modification of miR gene expression pattern in human colon cancer
cells following exposure to 5-fluorouracil in vitro. Pharmacol Res.
56:248–253. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Yu Y, Liao H, Xie R, Zhang Y, Zheng R,
Chen J and Zhang B: Overexpression of miRNA-3613-3p enhances the
sensitivity of triple negative breast cancer to CDK4/6 inhibitor
palbociclib. Front Oncol. 10:5908132020. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
van Zweeden AA, Opperman RCM, Honeywell
RJ, Peters GJ, Verheul HMW, van der Vliet HJ and Poel D: The
prognostic impact of circulating miRNAs in patients with advanced
esophagogastric cancer during palliative chemotherapy. Cancer Treat
Res Commun. 27:1003712021. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Xiao W, Zhong Y, Wu L, Yang D, Ye S and
Zhang M: Prognostic value of microRNAs in lung cancer: A systematic
review and meta-analysis. Mol Clin Oncol. 10:67–77. 2019.PubMed/NCBI
|
|
52
|
Sahlberg KK, Bottai G, Naume B, Burwinkel
B, Calin GA, Børresen-Dale AL and Santarpia L: A serum MicroRNA
signature predicts tumor relapse and survival in triple-negative
breast cancer patients. Clin Cancer Res. 21:1207–1214. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Piña-Sánchez P, Valdez-Salazar HA and
Ruiz-Tachiquín ME: Circulating microRNAs and their role in the
immune response in triple-negative breast cancer. Oncol Lett.
20:224:2020. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Tampellini M, Berruti A, Bitossi R,
Gorzegno G, Alabiso I, Bottini A, Farris A, Donadio M, Sarobba MG,
Manzin E, et al: Prognostic significance of changes in CA 15-3
serum levels during chemotherapy in metastatic breast cancer
patients. Breast Cancer Res Treat. 98:241–248. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
De Cock L, Heylen J, Wildiers A, Punie K,
Smeets A, Weltens C, Neven P, Billen J, Laenen A and Wildiers H:
Detection of secondary metastatic breast cancer by measurement of
plasma CA 15.3. ESMO Open. 6:1002032021. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Yerushalmi R, Tyldesley S, Kennecke H,
Speers C, Woods R, Knight B and Gelmon KA: Tumor markers in
metastatic breast cancer subtypes: Frequency of elevation and
correlation with outcome. Ann Oncol. 23:338–345. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Yang Y, Zhang H, Zhang M, Meng Q, Cai L
and Zhang Q: Elevation of serum CEA and CA15-3 levels during
antitumor therapy predicts poor therapeutic response in advanced
breast cancer patients. Oncol Lett. 14:7549–7556. 2017.PubMed/NCBI
|
|
58
|
Fu J, Su X, Li Z, Deng L, Liu X, Feng X
and Peng J: HGF/c-MET pathway in cancer: From molecular
characterization to clinical evidence. Oncogene. 40:4625–4651.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Faiella A, Riccardi F, Cartenì G,
Chiurazzi M and Onofrio L: The Emerging Role of c-met in
carcinogenesis and clinical implications as a possible therapeutic
target. J Oncol. 2022:51791822022. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Zhou FH, Downton T, Freelander A, Hurwitz
J, Caldon CE and Lim E: CDK4/6 inhibitor resistance in estrogen
receptor positive breast cancer, a 2023 perspective. Front Cell Dev
Biol. 11:11487922023. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Yan S, Jiao X, Zou H and Li K: Prognostic
significance of c-Met in breast cancer: A meta-analysis of 6010
cases. Diagn Pathol. 10:622015. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Ho-Yen CM, Jones JL and Kermorgant S: The
clinical and functional significance of c-Met in breast cancer: A
review. Breast Cancer Res. 17:522015. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Iovino F, Diana A, Carlino F, Ferraraccio
F, Antoniol G, Fisone F, Perrone A, Zito Marino F, Panarese I,
Tathode MS, et al: Expression of c-MET in estrogen receptor
positive and HER2 negative resected breast cancer correlated with a
poor prognosis. J Clin Med. 11:69872022. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Zearo S, Kim E, Zhu Y, Zhao JT, Sidhu SB,
Robinson BG and Soon PSh: MicroRNA-484 is more highly expressed in
serum of early breast cancer patients compared to healthy
volunteers. BMC Cancer. 14:2002014. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Li M, Zou X, Xia T, Wang T, Liu P, Zhou X,
Wang S and Zhu W: A five-miRNA panel in plasma was identified for
breast cancer diagnosis. Cancer Med. 8:7006–7017. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Sarhadi VK and Armengol G: Molecular
biomarkers in cancer. Biomolecules. 12:10212022. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Khan H, Shah MR, Barek J and Malik MI:
Cancer biomarkers and their biosensors: A comprehensive review.
TrAC Trends Anal Chem. 158:1168132023. View Article : Google Scholar
|