Introduction
Despite progress in cancer therapy, ovarian cancer
remains the most lethal gynecological cancer. Since there is no
reliable biomarker for early detection, and the early stages of the
disease are mostly asymptomatic, the majority of patients with
ovarian cancer are diagnosed when the disease is in advanced
stages, and the 5-year survival rate is less than 20% (1). The high mortality rate in ovarian
cancer is also linked to a high recurrence rate. Over 70% of
patients with ovarian cancer suffer from recurrence within 2 years
of primary standard treatment, which includes total hysterectomy
with bilateral salpingo-oophorectomy and subsequent chemotherapy
(2). All ovarian cancer recurrences
following primary treatment are metastatic recurrences.
Ovarian cancer metastasis has a unique biological
behavior that differs from the classical patterns of metastases
spreading through the vasculature. Ovarian cancer cells disseminate
primarily in the peritoneal cavity and subsequently implant onto
mesothelial surfaces (3). The
biological process of metastasis is complex at the molecular level,
involving adhesion, migration, invasion, growth, proliferation and
apoptosis. Understanding the molecular mechanisms of ovarian cancer
metastasis will likely lead to novel therapeutic targets and
biomarkers that will facilitate better predictions of
prognosis.
Interferon-induced transmembrane protein 1
(IFITM1) is a member of the interferon-induced transmembrane
protein family and is important for antiproliferative and
homotypic-adhesion signal transduction in lymphocytes (7–9).
IFITM1 also has antiviral functions, inhibiting influenza A
replication and enveloped virus infection (10). IFITM1 is upregulated in
diverse tumor tissues and cell lines (11–14)
and promotes migration and invasiveness in gastric cancer (15,16),
glioma (14) and head and neck
cancers (17). Furthermore,
IFITM1 is reportedly upregulated in response to the
anticancer drug paclitaxel in ovarian carcinoma xenografts;
however, the role of IFITM1 and the mechanisms that regulate
its expression in ovarian cancer have not yet been elucidated.
Human ovarian carcinoma xenografts are useful tools
for analyzing tumorigenicity and for testing the efficacy of newly
developed therapies in vivo (3). In particular, intraperitoneal or
orthotopic xenografts are useful for modeling the advanced stages
of ovarian carcinomas (3). These
xenografts produce carcinomatosis in the peritoneal cavity, with
large volumes of ascites resembling human ovarian metastatic
phenotypes (4–6). We established a mouse xenograft model
of human ovarian carcinoma and analyzed transcriptional expression
in metastatic implants from the xenografts. The expression pattern
of the metastatic implants reflected the pathophysiological
condition of ovarian metastatic phenotypes in humans. We selected
IFITM1 from among the upregulated genes in the metastatic
implants and investigated the mechanism regulating IFITM1
expression.
Materials and methods
Cell culture
The human ovarian cancer cell line SK-OV-3 was
purchased from the American Type Culture Collection (ATCC; no.
HTB-77) and cultured in McCoy’s 5A medium containing 10% fetal
bovine serum (FBS), 100 U/ml penicillin and 100 μg/ml streptomycin
(all from Gibco-BRL) in a 95% humidified air and 5% CO2
atmosphere at 37°C.
Ovarian cancer mouse xenograft model
All procedures for handling and euthanizing the
animals used in the present study were performed in strict
compliance with the guidelines of the Korean animal protection law
and were approved by the Institutional Animal Care and Use
Committee (IACUC) of Ewha Womans University School of Medicine.
SK-OV-3 cells (2×106) suspended in culture media were
intraperitoneally injected into 10 female nude mice (BALB/c, 4–6
weeks of age). Four weeks after inoculation, the xenograft mice
were sacrificed, and at least four implants adhering to the
mesothelial surface of each mouse were harvested.
RNA preparation and quantitative
reverse-transcription polymerase chain reaction (qRT-PCR)
Total RNA was extracted from the metastatic implants
of the ovarian cancer mouse xenografts and SK-OV-3 cells using the
RNeasy Mini kit (Qiagen) according to the manufacturer’s protocol.
One microgram of total RNA was converted to cDNA using SuperScript
II reverse transcriptase and oligo-(dT)12–18 primers
(both from Invitrogen) according to the manufacturer’s
instructions. qRT-PCR was performed in a 20-μl reaction mixture
containing 1 μl cDNA, 10 μl SYBR Premix Ex Taq, 0.4 μl ROX
Reference Dye (50X) (both from Takara Bio), and 200 nM primers for
each gene. The primer sequences were: IFITM1 (forward),
5′-CGCCAAGTGCCTGAACATCT-3′ and IFITM1 (reverse),
5′-TACCAGTAACAGGATGAATCCAATG-3′; GAPDH (forward),
5′-AATCCCATCACCATCTTCCA-3′ and GAPDH (reverse),
5′-TGGACTCCACGACGTACTCA-3′. The reactions were run on a 7500 Fast
Real-Time PCR System (Applied Biosystems) at 95°C for 30 sec,
followed by 40 cycles of 95°C for 3 sec and 60°C for 30 sec, and a
single dissociation cycle of 95°C for 15 sec, 60°C for 60 sec, and
95°C for 15 sec. All PCR reactions were performed in triplicate,
and the specificity of the reaction was detected by melting-curve
analysis at the dissociation stage. Comparative quantification of
each target gene was performed based on cycle threshold
(Ct) normalized to GAPDH using the
ΔΔCt method.
Messenger RNA microarray chip processing
and analysis of gene expression data
Total RNA was extracted from the harvested
metastatic implants of the ovarian cancer mouse xenografts and
SK-OV-3 cells using the RNeasy Mini kit, and 1 μg of total RNA was
amplified and labeled according to the Affymetrix GeneChip Whole
Transcript Sense Target Labeling protocol. The resulting labeled
cDNA was hybridized to Affymetrix Human Gene 1.0 ST arrays
(Affymetrix). The scanned raw expression values were background
corrected, normalized and summarized using the Robust Multi-array
Average approach in the Bioconductor ‘affy’ package (Affymetrix).
The resulting log2-transformed data were used for
further analyses.
To identify differentially expressed genes (DEGs),
we applied moderated t-statistics based on an empirical Bayesian
approach (11). Significantly
upregulated and downregulated DEGs were defined as genes with at
least a 2-fold difference in expression level between the xenograft
cells and the wild-type SK-OV-3 cells after correction for multiple
testing [Benjamini-Hochberg-false discovery rate (BH-FDR)-adjusted
p-value <0.05] (18). Finally,
we excluded genes with a low expression level (maximum
log2 expression level in a total of 8 samples <7.0)
from the list of DEGs. The DAVID bioinformatics resource was used
to detect overrepresented GO clusters from the identified DEGs
(13).
Bisulfite sequencing PCR (BSP)
Genomic DNA was extracted from the harvested
metastatic implants of the ovarian cancer mouse xenografts and
SK-OV-3 cells using the QIAmp DNA Mini kit (Qiagen) according to
the manufacturer’s protocol. Bisulfite treatment of genomic DNA was
performed using the EpiTect Bisulfite kit (Qiagen) according to the
manufacturer’s instructions. For bisulfite sequencing of the target
promoter region of IFITM1, BSP was carried out using
conventional PCR in a 50-μl reaction mixture containing 10 ng
bisulfite-modified genomic DNA, 1.5 mM MgCl2, 200 μM
dNTP, 1 U Platinum Taq polymerase (Invitrogen), 1X Platinum
Taq buffer, and 200 nM specific BSP forward and reverse
primers for each gene. The BSP primers were designed using the
MethPrimer software (http://www.urogene.org/methprimer). For IFITM1,
the BSP product was 522 bp (position in the human GRCh37/hg19
assembly: ch11 313,635-314,156) and contained 9 CpG sites. The BSP
primer sequences were: (forward), 5′-GGATTGTAGTTTGAGGAAAGAGTAAG-3′
and (reverse), 5′-AAAAAAAATATTAAATAAAAATTTA AAAAA-3′. The reaction
was run at 95°C for 5 min, followed by 30 cycles of 95°C for 30
sec, 50–55°C for 30 sec, and 72°C for 30 sec and a final elongation
step at 72°C for 5 min.
The BSP products were purified using the QIAquick
Gel Extraction kit (Qiagen) according to the manufacturer’s
protocols and ligated into the yT&A cloning vector (Yeastern
Biotech). The ligation products were used to transform competent
DH5α Escherichia coli cells (RBC Bioscience) using standard
procedures. Blue/white screening was used to select bacterial
clones, and BSP product-positive clones were confirmed by colony
PCR using the BSP primers to verify the insert size. Plasmid DNA
was then extracted from at least 15 insert-positive clones using
the QIAprep Spin Miniprep kit (Qiagen) and sequenced using the M13
primer to analyze the methylation status at specific CpG sites.
Quantitative methylation-specific PCR
(qMSP)
Quantitative MSP was carried out with
bisulfite-modified genomic DNA as the template and specific primer
sequences designed to detect the methylated and unmethylated forms
of IFITM1. The following methylated/unmethylated-specific
primers were used: (forward), 5′-TAGGAAGTTATTAGTTTTGATTTG AGT-3′;
(methylated reverse), 5′-TAAAACCTCCTTTCCC CTATCG-3′ and
(unmethylated reverse), 5′-TAAAACCTCC TTTCCCCTATCA-3′. For qMSP, a
20-μl reaction mixture containing 2 μl (10–100 ng/μl)
bisulfite-treated DNA, 10 μl SYBR Premix Ex Taq (Takara Bio), 0.4
μl ROX Reference Dye (50X; Takara Bio), and 200 nM each primer were
reacted using a 7500 Fast Real-Time PCR system (Applied
Biosystems). The amplification reaction conditions were: 95°C for
30 sec, followed by 40 cycles of 95°C for 3 sec, and 62°C for 30
sec. The PCR product was then reacted at 95°C for 15 sec, 60°C for
1 min, and 95°C for 15 sec to examine the specificity. Methylation
and non-methylation of the specific CpG sites were calculated as
follows (Ct represents the threshold cycle): Percent methylation =
100/[1 + 2(ΔCtmeth − ΔCtnonmeth)].
Treatment with 5-aza-2′-deoxycytidine
(5-aza-dC)
To demethylate the methylated CpG sites, SK-OV-3
cells were treated with an increasing concentration (0, 5 and 10
μM) of 5-aza-dC (Sigma-Aldrich) for 3 days. The medium was replaced
daily.
Transient transfection
To establish a transient expression system, SK-OV-3
cells were transfected with pCMV6-XL5-IFITM1 (Origene) or pEGFP-N3
(Clontech) plasmid DNAs using Lipofectamine™ 2000 (Invitrogen).
Briefly, the cells were plated at a density of 6×105
cells/well in 6-well plates and allowed to grow overnight. Two
micrograms of each plasmid DNA and 5 μl Lipofectamine 2000 were
diluted separately in Opti-MEM medium to a total volume of 250 μl.
The diluted plasmid DNAs and Lipofectamine 2000 were mixed and
incubated at room temperature for 20 min to generate the
transfection mixtures. The cells were washed with serum-free
McCoy’s 5A medium, and then the transfection mixtures were added to
each well of the 6-well plates containing complete growth medium
and incubated at 37°C for 24 h in a 5% CO2
incubator.
Transwell migration and in vitro invasion
assay
After 24 h of transfection, the transfected cells
were starved by serum deprivation. The cell migration assay was
performed in 24-well Transwell plates containing inserts with a
polycarbonate membrane with an 8.0-μm pore size (Corning). After 24
h of serum deprivation, the cells were detached from the plates and
resuspended in serum-free medium at a density of 2×106
cells/ml. One hundred microliters of the SK-OV-3 cell suspension
was added to the upper compartment of the Transwell chamber. For
each experiment, both chemotactic migration to medium containing
15% FBS and random migration in serum-free medium were assessed in
parallel Transwell plates for 6 h at 37°C in a 5% CO2
incubator.
The in vitro invasion assay was performed
using a BD BioCoat Matrigel Invasion Chamber (Becton-Dickinson).
After 24 h of serum deprivation, SK-OV-3 cells were detached from
the plates and resuspended in serum-free medium at a density of
1×106 cells/ml. One hundred microliters of the SK-OV-3
cell suspension was added to the upper compartment of the invasion
chamber, and 500 μl McCoy’s 5A medium containing 10% FBS was added
to the lower compartment of the chamber. The migration through the
Matrigel chamber was allowed to proceed at 37°C for 24 h in a 5%
CO2 incubator. After the incubation period, the cells
that had not migrated from the upper side of the filter were
carefully scraped away with cotton swabs. The cells on the lower
side of the filter were fixed for 2 min using Diff-Quick kit
solution (Fisher Scientific), stained with 1% crystal violet for 2
min and washed twice with distilled water at room temperature. The
images of the stained cells on the lower side of the membrane were
acquired at ×200 magnification in six different fields. For
quantitative analysis, the stained cells were subsequently
extracted with 10% acetic acid, and colorimetric measurement was
performed at 590 nm.
Results
Identification of DEGs
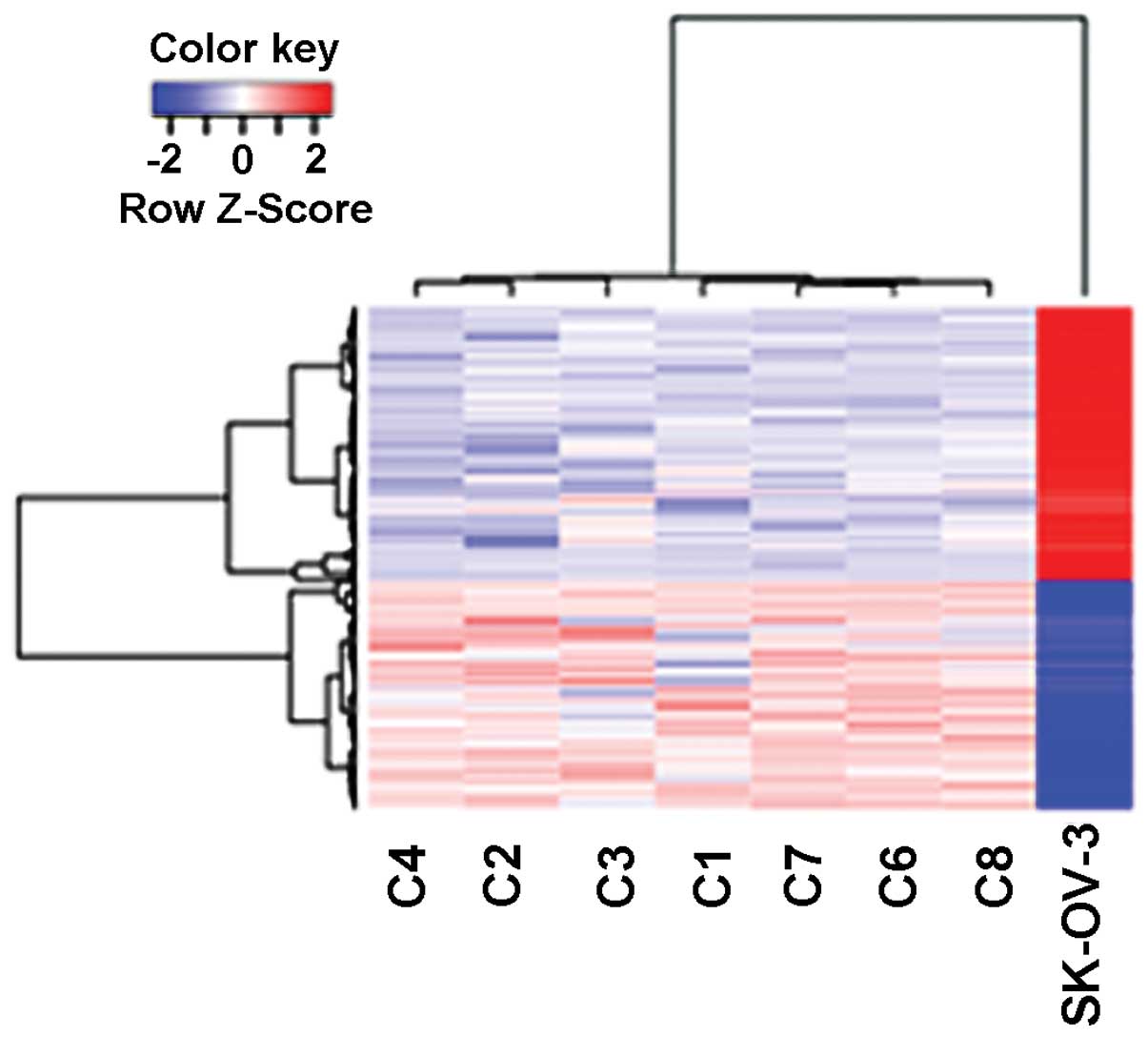
A total of 973 genes were found to be differentially
expressed in the xenografts relative to the SK-OV-3 cells (Fig. 1). The 444 DEGs that were upregulated
were enriched with genes involved in cell adhesion, blood
coagulation and wound healing, response to steroid hormone
stimulus, blood vessel development and cell mobility (Table I). The 529 DEGs that were
downregulated were enriched with genes related to the inflammatory
response, regulation of programmed cell death and response to
endoplasmic reticulum stress (Table
I).
 | Table ITop 10 enriched clusters of gene
ontology (GO) in the differentially expressed genes. |
Table I
Top 10 enriched clusters of gene
ontology (GO) in the differentially expressed genes.
| Annotation
cluster | Enrichment
score | Representative two
GO terms in the cluster (GOTERM_BP_FAT) | Count | P-value | BH P-value |
|---|
| Upregulated |
| Cluster 1 | 6.8 | Cell adhesion | 43 | 1.54E-08 | 2.86E-05 |
| | Biological
adhesion | 43 | 1.60E-08 | 1.48E-05 |
| Cluster 2 | 4.1 | Blood
coagulation | 12 | 2.48E-05 | 0.0046 |
| | Wound healing | 16 | 4.11E-05 | 0.0069 |
| Cluster 3 | 4.1 | Extracellular
structure organization | 19 | 4.59E-08 | 2.83E-05 |
| | Homophilic cell
adhesion | 16 | 3.85E-07 | 1.78E-04 |
| Cluster 4 | 3.2 | Response to
drug | 18 | 1.23E-05 | 0.0025 |
| | Response to steroid
hormone stimulus | 16 | 4.36E-05 | 0.0062 |
| Cluster 5 | 2.2 | Nucleosome
assembly | 10 | 1.43E-04 | 0.016 |
| | Chromatin
assembly | 10 | 1.87E-04 | 0.020 |
| Cluster 6 | 1.8 | Cell-substrate
adhesion | 8 | 0.0079 | 0.34 |
| | Cell-matrix
adhesion | 7 | 0.018 | 0.49 |
| Cluster 7 | 1.6 | Ectoderm
development | 11 | 0.018 | 0.50 |
| | Epidermis
development | 10 | 0.028 | 0.60 |
| Cluster 8 | 1.6 | Blood vessel
development | 14 | 0.0050 | 0.26 |
| | Vasculature
development | 14 | 0.0060 | 0.29 |
| Cluster 9 | 1.5 | Localization of
cell | 14 | 0.028 | 0.60 |
| | Cell motility | 14 | 0.028 | 0.60 |
| Cluster 10 | 1.4 | Collagen metabolic
process | 4 | 0.027 | 0.59 |
| | Multicellular
organismal macromolecule metabolic process | 4 | 0.034756 | 0.646305 |
| Downregulated |
| Cluster 1 | 4.9 | Response to
wounding | 36 | 3.36E-06 | 0.0041 |
| | Inflammatory
response | 25 | 2.08E-05 | 0.010 |
| Cluster 2 | 4.4 | Organic acid
biosynthetic process | 18 | 2.07E-06 | 0.0050 |
| | Carboxylic acid
biosynthetic process | 18 | 2.07E-06 | 0.0050 |
| Cluster 3 | 4.1 | Serine family amino
acid metabolic process | 8 | 6.00E-06 | 0.0049 |
| | Cellular amino acid
biosynthetic process | 10 | 1.20E-05 | 0.0073 |
| Cluster 4 | 2.6 | Regulation of
cytokine biosynthetic process | 10 | 2.45E-04 | 0.045 |
| | Regulation of
cytokine production | 14 | 0.002082 | 0.15 |
| Cluster 5 | 2.4 | Regulation of
programmed cell death | 43 | 1.32E-04 | 0.035 |
| | Regulation of cell
death | 43 | 1.44E-04 | 0.035 |
| Cluster 6 | 2.4 | tRNA
aminoacylation | 8 | 2.99E-04 | 0.051 |
| | Amino acid
activation | 8 | 2.99E-04 | 0.051 |
| Cluster 7 | 2.2 | Response to
endoplasmic reticulum stress | 7 | 3.55E-04 | 0.056 |
| | Endoplasmic
reticulum unfolded protein response | 5 | 0.0027 | 0.17 |
| Cluster 8 | 2.1 | Serine family amino
acid metabolic process | 8 | 6.00E-06 | 0.0049 |
| | Cysteine metabolic
process | 4 | 4.32E-04 | 0.064 |
| Cluster 9 | 1.6 | Vitamin metabolic
process | 9 | 0.0013 | 0.14 |
| | Cellular hormone
metabolic process | 7 | 0.0065 | 0.31 |
| Cluster 10 | 1.6 | Neuron projection
development | 17 | 0.0028 | 0.17 |
| | Neuron
development | 19 | 0.0087 | 0.35 |
Validation of altered IFITM1
expression
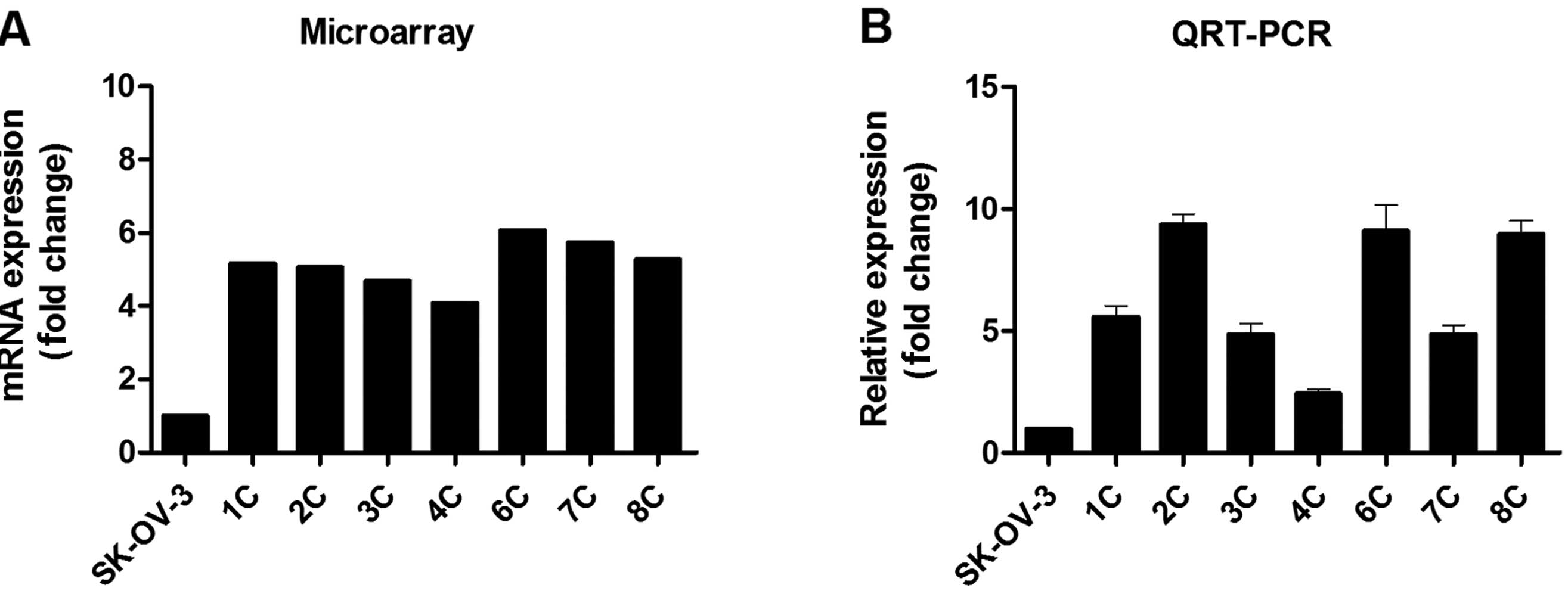
From among the 444 upregulated DEGs, we selected
IFITM1 as a representative gene to validate the expression
microarray results, and we confirmed its expression with qRT-PCR.
In agreement with the expression microarray results, IFITM1
mRNA expression was profoundly (2.4- to .4-fold) upregulated in the
metastatic implants from the ovarian cancer xenografts (n=7)
compared with that in the wild-type SK-OV-3 cells (Fig. 2).
DNA methylation regulates the expression
of IFITM1
The promoter-region CpG sites in the ovarian
metastatic implants were found to be hypomethylated when compared
with those in the SK-OV-3 cells, particularly the CpG sites at
−272, +54, +84 and +96 (Fig. 3A).
Further analyses of DNA methylation using MSP revealed
significantly reduced methylation at the +54 CpG site in the
ovarian metastatic implants (Fig.
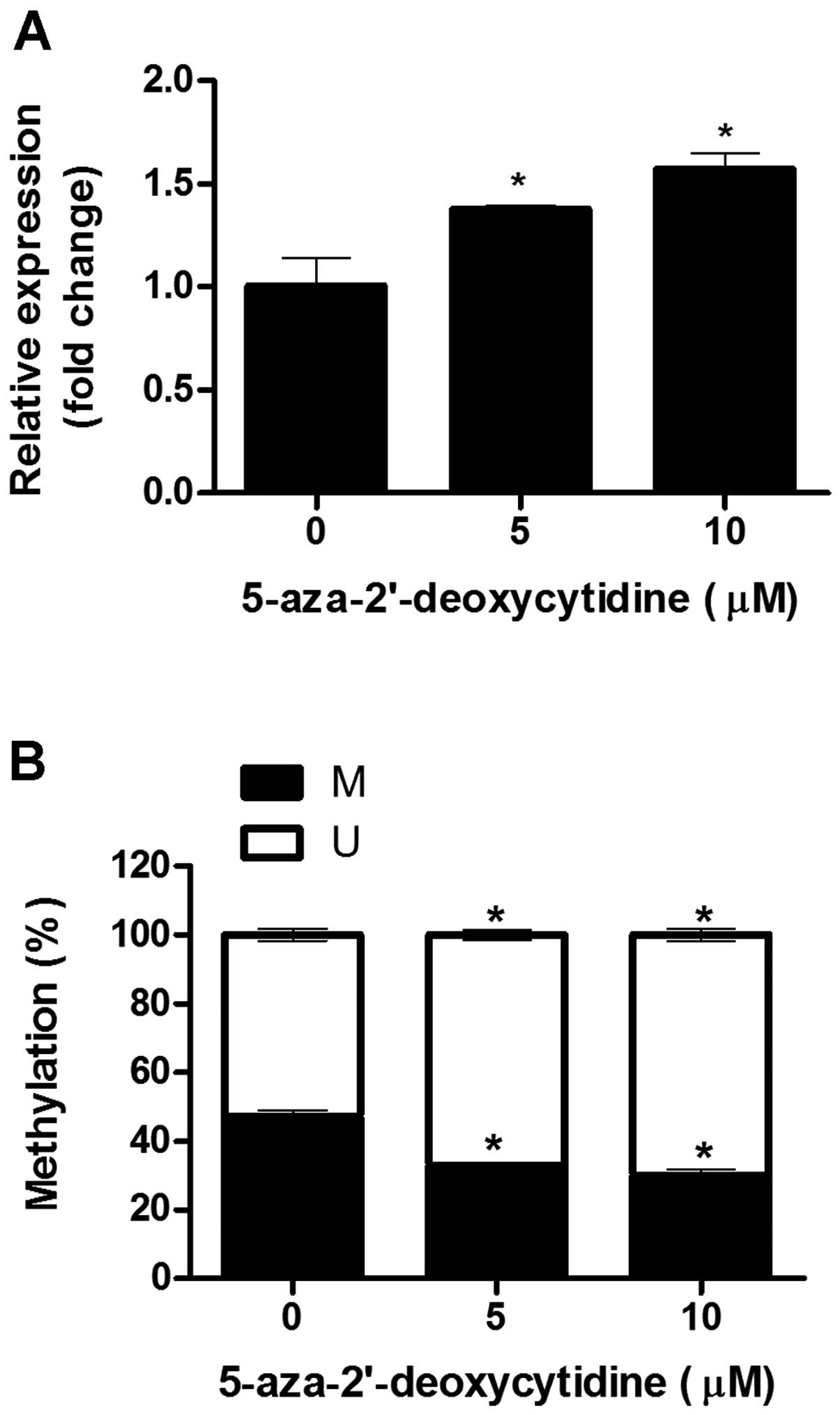
3B). Following treatment with 0, 5 and 10 μM 5-aza-dC,
decreased methylation activity at the +54 CpG site was confirmed
using MSP; and in parallel, the expression of IFITM1 mRNA
was significantly increased in a dose-dependent manner (Fig. 4).
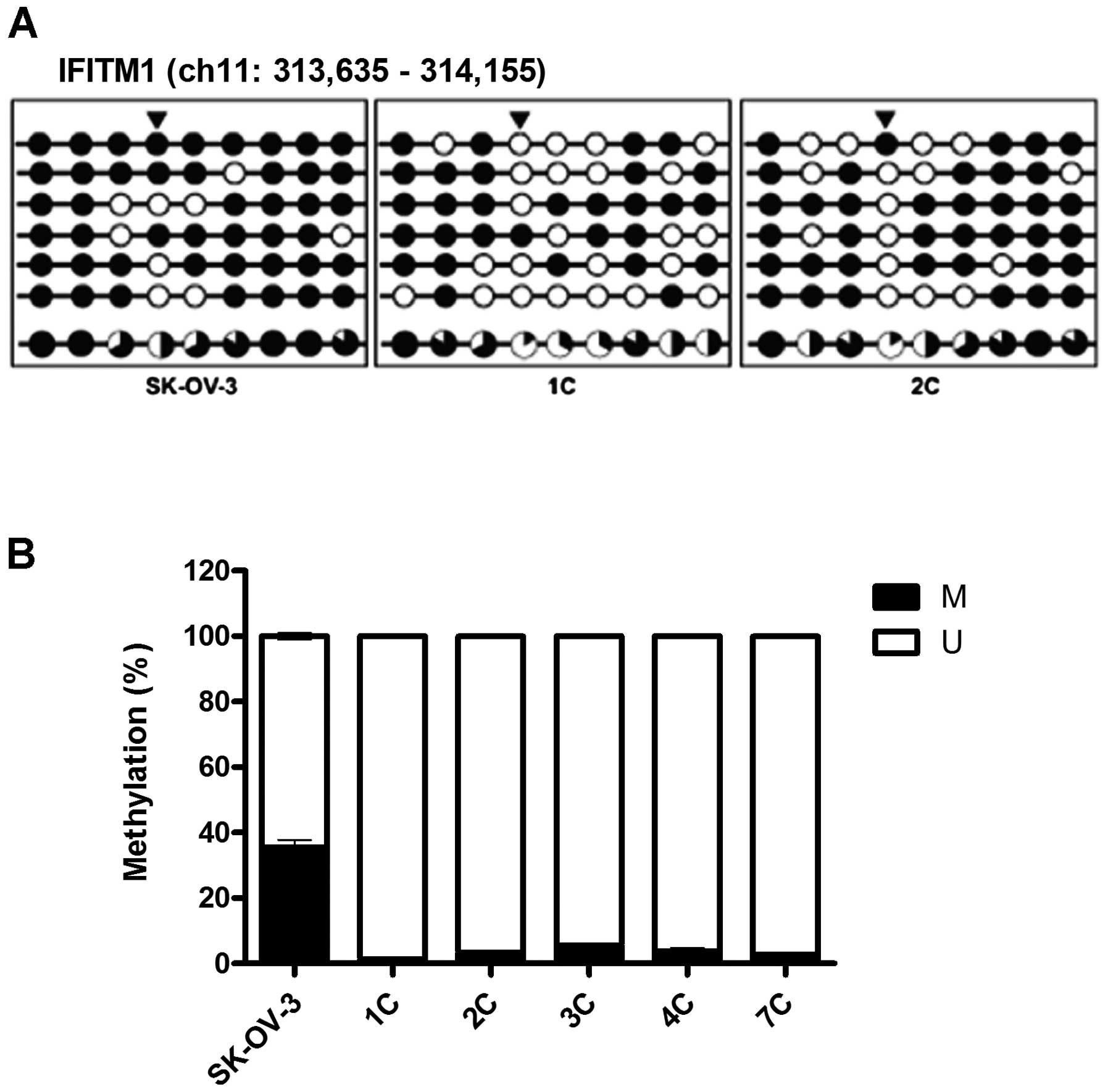 | Figure 3DNA methylation is altered at CpG
sites in the IFITM1 promoter in metastatic implants from the
mouse xenografts. (A) The DNA methylation status was analyzed using
Bisulfite sequencing analysis. The IFITM1 promoter region is
located at positions 313,635 - 314,156 in the human GRCh37/hg19
assembly and contains nine CpG residues within chromosome 11. The
nine CpGs are located at positions −310, −272, −236, +54, +84, +96,
+108, +116 and +123 from the transcription start site. Each circle
represents CpG dinucleotides. The methylation status of each CpG
site is illustrated by black (methylated) and white (unmethylated)
circles, and the total percentage of methylation at each site is
indicated by a pie graph on the bottom line. The black segment of
the pie graph indicates the methylated CpG percentage, whereas the
white segment represents the unmethylated CpG percentage. (B) The
DNA methylation status at the +54 CpG site was analyzed using qMSP.
Triangles above the circles in A indicate the specific CpG site
used for qMSP. M, the percentage of methylated CpGs; U, the
percentage of unmethylated CpGs. IFITM1, interferon-induced
transmembrane protein 1; qMSP, quantitative methylation-specific
PCR. |
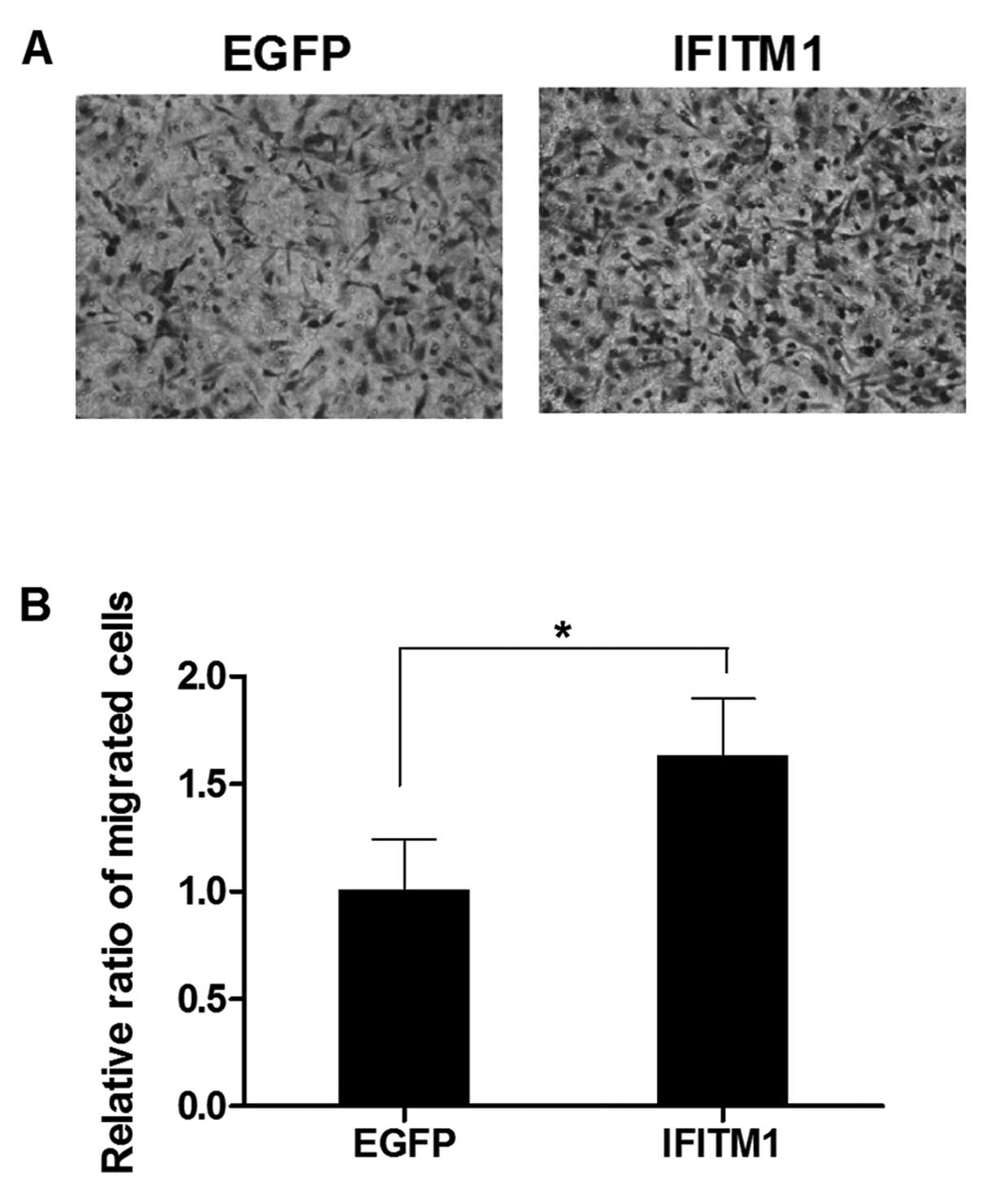
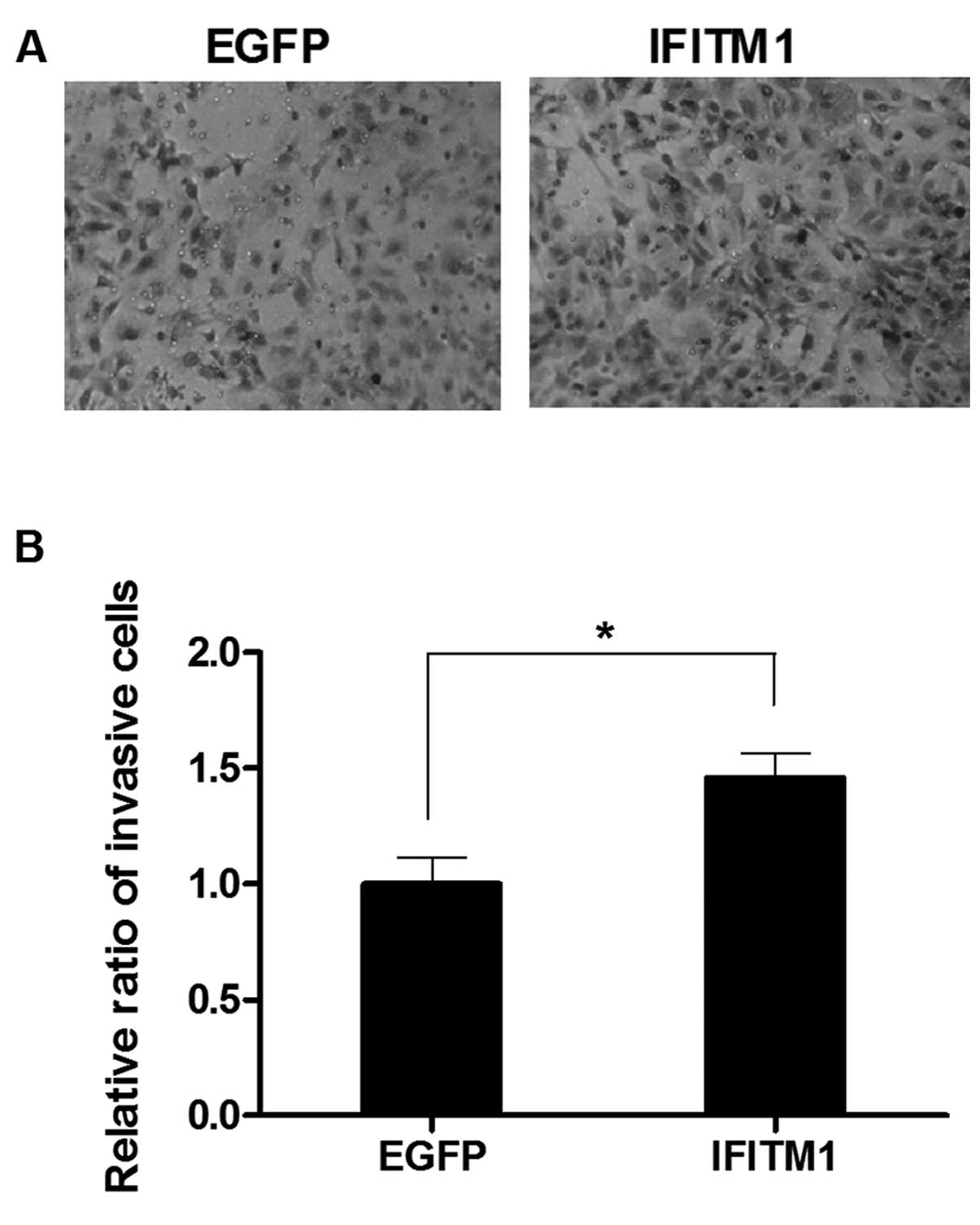
IFITM1 overexpression induces the
migration and invasion of the ovarian cancer cells
To examine the effect of altered IFITM1
expression on metastasis, we performed in vitro migration
and invasion assays using Transwell chambers. SK-OV-3 cells were
transfected with full-length IFITM1 or enhanced green
fluorescence protein (EGFP) cDNA. IFITM1 mRNA
expression in the transfected cells was confirmed by qRT-PCR (data
not shown). The migration capacity of the IFITM1-transfected
cells was increased compared with that of the
EGFP-transfected cells (Fig.
5). Invasiveness was also markedly increased in the
IFITM1-transfected cells compared with that in the
EGFP-transfected cells (Fig.
6).
Discussion
Metastasis is a complex biological process at the
molecular level involving adhesion, migration, invasion, growth,
proliferation and apoptosis. Metastasizing ovarian cancer cells
exhibit unique biological behavior that differs from the classical
patterns of metastasis arising at other organ sites (3). Ovarian carcinoma can spread directly
to adjacent organs via local invasion, and exfoliated tumor cells
can be transported into the intraperitoneal fluid and subsequently
implanted onto mesothelial surfaces. Implantation in the peritoneal
cavity is associated with the accumulation of ascites resulting
from obstruction of peritoneal lymphatic drainage and the secretion
of vascular permeability factors by tumor cells (19).
To better understand the molecular mechanisms
involved in ovarian cancer metastasis in vivo, we analyzed
transcriptional expression levels in metastatic implants from human
ovarian carcinoma xenografts in mice. The expression of 937 genes
was significantly altered in the xenografts by at least 2-fold
compared with that in wild-type SK-OV-3 cells (Fig. 1). The upregulated genes were
enriched with functions involved in cell adhesion, blood
coagulation and wound healing, response to steroid hormone
stimulus, blood vessel development and cell mobility (Table I). On the other hand, the
downregulated genes were enriched with functions involved in the
inflammatory response, the regulation of programmed cell death and
response to endoplasmic reticulum stress (Table I).
During the progression of metastasis, ovarian tumor
cells exfoliate from their origin and attach to a new location,
requiring the remodeling of cell-cell junctions and the alteration
of cellular adhesive properties. Our gene ontology (GO) analysis
showed that the expression of 43 genes, including
P-cadherin, cadherin 16, protocadherin 18 and
protocadherin β cluster, involved in cell adhesion (GO
terms: cell, biological, hemophilic cell, calcium-dependent
cell-cell and cell-cell adhesion) were significantly upregulated in
the metastatic cells (data not shown). In addition to the cadherin
and protocadherin families, the expression of certain
integrin-family genes (ITGB4, ITGB6 and ITGA7)
was upregulated in the metastatic cells (data not shown). These
genes encode adhesion receptors that function in signaling from the
extra-cellular matrix to the cell. Integrin proteins form dimers
composed of α and β chains; integrin heterodimers can bind to
fibronectin, collagen VI, laminin and transforming growth factor 1
(TGF1), promoting cell growth and matrix production by providing
physical adhesion between the cytoskeletal structure and the
extracellular matrix (20,21).
In human ovarian cancer cell lines, ITGB4
expression is highly elevated in cells expressing aggressive
phenotypes. Previously, immunostaining of ITGB4 in 196 human
ovarian serous carcinoma samples reveled that high levels of ITGB4
expression were related to tumor aggressiveness (22). Recently, Cheon et al
identified 10 collagen-remodeling genes, including COL6A2,
that are regulated by TGF-β signaling and are associated with
metastasis and poor survival among patients with serous ovarian
cancer (23). In line with that
study, the expression profile of collagen superfamily genes,
including COL6A2, was extensively modified in our ovarian
metastatic implants (data not shown). The chemokine receptor
CXCR4 was also upregulated in the xenografts (data not
shown). Scotton et al previously showed that among 14
chemokine receptors, CXCR4 was the only one expressed in
ovarian tumor cells (24). The
CXCR4 ligand CXCL12 is detectable in ascites in patients with
ovarian cancer and is secreted by peritoneal mesothelial cells. The
CXCR4-CXCL12 interaction could direct cancer cell migration in the
peritoneum, leading to the spread of ovarian cancer (24). We also observed the upregulation of
S100 family members (S100A1, S100A2, S100A3
and S100A4) in the metastatic implants (data not shown).
S100 family members are overexpressed in many types of cancers and
promote metastasis by interacting with many different proteins,
including matrix metalloproteinase and by acting as
chemoattractants (25). S100A4 has
been suggested as a biomarker for the high risk of metastasis and
mortality among subgroups of patients with solid tumors, including
bladder (26) and breast cancer
(27), esophageal squamous cell
carcinomas (28), pancreatic cancer
(29) and colorectal carcinomas
(30).
The expression patterns of certain genes in our
xenografts reflect, to some extent, the pathophysiological
condition of human metastatic ovarian cancer. IFITM1 is a member of
the interferon-induced transmembrane protein family. The
upregulation of IFITM1 has been reported in diverse tumor tissues
and in cell lines including colorectal (31), head and neck (17) and gastric cancer (16) and glioma (14). IFITM1 overexpression is
associated with clinicopathological features in colorectal cancer,
making it a potential biomarker for clinical diagnosis. The
involvement of IFITM1 in cancer progression via the promotion of
cell migration and invasion has been demonstrated in gastric and
head and neck types of cancers (17). Recently, the epigenetic regulation
of IFITM1 by aberrant promoter methylation was explored in
gastric cancer (16); however, DNA
methylation-dependent epigenetic regulation of IFITM1 has
not been investigated in ovarian cancer.
Our results revealed that IFITM1 expression
was profoundly upregulated in metastatic cells, and the methylation
of specific CpG sites within the IFITM1 promoter was highly
reduced in the metastatic cells compared with that in the wild-type
SK-OV-3 cells (Figs. 2 and 3). Treating wild-type SK-OV-3 cells with
the demethylating agent 5-aza-dC caused dose-dependent enhancement
of IFITM1 expression, implying transcriptional regulation by
promoter methylation. IFITM1 overexpression also increased
cell migration and invasiveness, suggesting that aberrant
upregulation of IFITM1 is strongly associated with the
acquisition of metastatic phenotypes in ovarian carcinomas.
In conclusion, we used a mouse xenograft model of
human ovarian carcinoma to demonstrate that IFITM1 could be
a novel metastasis-promoting gene that enhances the metastatic
phenotype in ovarian cancer via epigenetic transcriptional
regulation. Although further clinical research is warranted, our
findings suggest that the status of DNA methylation within the
IFITM1 promoter region may be a biomarker indicating
metastatic progression in ovarian cancer.
Acknowledgements
This study was supported by a grant of the Korean
Health Technology R&D Project, the Ministry of Health and
Welfare, Republic of Korea (HI12C0050).
References
|
1
|
Ozols RF: Update on the management of
ovarian cancer. Cancer J. 8(Suppl 1): S22–S30. 2002.PubMed/NCBI
|
|
2
|
Tummala MK and McGuire WP: Recurrent
ovarian cancer. Clin Clin Adv Hematol Oncol. 3:723–736. 2005.
|
|
3
|
Šale S and Orsulic S: Models of ovarian
cancer metastasis: murine models. Drug Discov Today Dis Models.
3:149–154. 2006.
|
|
4
|
Sallinen H, Anttila M, Narvainen J, et al:
A highly reproducible xenograft model for human ovarian carcinoma
and application of MRI and ultrasound in longitudinal follow-up.
Gynecol Oncol. 103:315–320. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hamilton TC, Young RC, Louie KG, et al:
Characterization of a xenograft model of human ovarian carcinoma
which produces ascites and intraabdominal carcinomatosis in mice.
Cancer Res. 44:5286–5290. 1984.PubMed/NCBI
|
|
6
|
Molpus KL, Koelliker D, Atkins L, et al:
Characterization of a xenograft model of human ovarian carcinoma
which produces intraperitoneal carcinomatosis and metastases in
mice. Int J Cancer. 68:588–595. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Takahashi S, Doss C, Levy S and Levy R:
TAPA-1, the target of an antiproliferative antibody, is associated
on the cell surface with the Leu-13 antigen. J Immunol.
145:2207–2213. 1990.PubMed/NCBI
|
|
8
|
Bradbury LE, Kansas GS, Levy S, Evans RL
and Tedder TF: The CD19/CD21 signal transducing complex of human B
lymphocytes includes the target of antiproliferative antibody-1 and
Leu-13 molecules. J Immunol. 149:2841–2850. 1992.PubMed/NCBI
|
|
9
|
Matsumoto AK 1, Martin DR, Carter RH,
Klickstein LB, Ahearn JM and Fearon DT: Functional dissection of
the CD21/CD19/TAPA-1/Leu-13 complex of B lymphocytes. J Exp Med.
178:1407–1417. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Brass AL, Huang IC, Benita Y, et al: The
IFITM proteins mediate cellular resistance to influenza A H1N1
virus, West Nile virus, and dengue virus. Cell. 139:1243–1254.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Smyth GK: Linear models and empirical
Bayes methods for assessing differential expression in microarray
experiments. Stat Appl Genet Mol Biol. 3:32004.PubMed/NCBI
|
|
12
|
Watts GS, Futscher BW, Holtan N, DeGeest
K, Domann FE and Rose SL: DNA methylation changes in ovarian cancer
are cumulative with disease progression and identify tumor stage.
BMC Med Genomics. 1:472008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009.PubMed/NCBI
|
|
14
|
Yu F, Ng SS, Chow BK, et al: Knockdown of
interferon-induced transmembrane protein 1 (IFITM1) inhibits
proliferation, migration, and invasion of glioma cells. J
Neurooncol. 103:187–195. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yang Y, Lee JH, Kim KY, et al: The
interferon-inducible 9–27 gene modulates the susceptibility to
natural killer cells and the invasiveness of gastric cancer cells.
Cancer Lett. 221:191–200. 2005.
|
|
16
|
Lee J, Goh SH, Song N, et al:
Overexpression of IFITM1 has clinicopathologic effects on gastric
cancer and is regulated by an epigenetic mechanism. Am J Pathol.
181:43–52. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hatano H, Kudo Y, Ogawa I, et al:
IFN-induced transmembrane protein 1 promotes invasion at early
stage of head and neck cancer progression. Clin Cancer Res.
14:6097–6105. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: a practical and powerful approach to
multiple testing. J Roy Stat Soc Ser B (Stat Method). 57:289–300.
1995.
|
|
19
|
Naora H and Montell DJ: Ovarian cancer
metastasis: integrating insights from disparate model organisms.
Nat Rev Cancer. 5:355–366. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Howe A, Aplin AE, Alahari SK and Juliano
RL: Integrin signaling and cell growth control. Curr Opin Cell
Biol. 10:220–231. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Aplin A, Howe A, Alahari S and Juliano RL:
Signal transduction and signal modulation by cell adhesion
receptors: the role of integrins, cadherins, immunoglobulin-cell
adhesion molecules, and selectins. Pharmacol Rev. 50:197–263.
1998.PubMed/NCBI
|
|
22
|
Choi YP, Kim BG, Gao MQ, Kang S and Cho
NH: Targeting ILK and β4 integrin abrogates the invasive potential
of ovarian cancer. Biochem Biophys Res Commun. 427:642–648.
2012.
|
|
23
|
Cheon DJ, Tong Y, Sim MS, et al: A
collagen-remodeling gene signature regulated by TGFβ signaling is
associated with metastasis and poor survival in serous ovarian
cancer. Clin Cancer Res. 20:711–723. 2014.PubMed/NCBI
|
|
24
|
Scotton CJ, Wilson JL, Milliken D, Stamp G
and Balkwill FR: Epithelial cancer cell migration: a role for
chemokine receptors? Cancer Res. 61:4961–4965. 2001.PubMed/NCBI
|
|
25
|
Salama I 1, Malone PS, Mihaimeed F and
Jones JL: A review of the S100 proteins in cancer. Eur J Surg
Oncol. 34:357–364. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Davies BR, O’Donnell M, Durkan GC, et al:
Expression of S100A4 protein is associated with metastasis and
reduced survival in human bladder cancer. J Pathol. 196:292–299.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Rudland PS, Platt-Higgins A, Renshaw C, et
al: Prognostic significance of the metastasis-inducing protein
S100A4 (p9Ka) in human breast cancer. Cancer Res. 60:1595–1603.
2000.PubMed/NCBI
|
|
28
|
Ninomiya I, Ohta T, Fushida S, et al:
Increased expression of S100A4 and its prognostic significance in
esophageal squamous cell carcinoma. Int J Oncol. 18:715–720.
2001.PubMed/NCBI
|
|
29
|
Ai KX, Lu LY, Huang XY, Chen W and Zhang
HZ: Prognostic significance of S100A4 and vascular endothelial
growth factor expression in pancreatic cancer. World J
Gastroenterol. 14:1931–1935. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gongoll S, Peters G, Mengel M, et al:
Prognostic significance of calcium-binding protein S100A4 in
colorectal cancer. Gastroenterology. 123:1478–1484. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
He JD 1, Luo HL, Li J, Feng WT and Chen
LB: Influences of the interferon induced transmembrane protein 1 on
the proliferation, invasion, and metastasis of the colorectal
cancer SW480 cell lines. Chin Med J. 125:517–522. 2012.PubMed/NCBI
|